Advances in Microbiology
Vol. 2 No. 1 (2012) , Article ID: 18055 , 7 pages DOI:10.4236/aim.2012.21001
Destruction of Escherichia coli and Broad-Host-Range Plasmid DNA in Treated Wastewater by Dissolved Ozone Disinfection under Laboratory and Field Conditions
Department of Crop, Soil, and Environmental Sciences, University of Arkansas, Fayetteville, USA
Email: msavin@uark.edu
Received December 17, 2011; revised January 5, 2012; accepted January 16, 2012
Keywords: Disinfection; Wastewater Treatment; Ozone; qPCR; Plasmid; Broad-Host-Range Plasmid; DNA; Escherichia coli
ABSTRACT
Broad-host-range plasmids are frequently associated with antibiotic resistance genes and can quickly spread antibiotic resistant phenotypes among diverse bacterial populations. Wastewater treatment plants have been identified as reservoirs for broad-host-range plasmids carrying resistance genes. The threat of broad-host-range plasmids released into the environment from wastewater treatment plants has identified the need for disinfection protocols to target broad-hostrange plasmid destruction. Here we evaluate the efficacy of dissolved ozone at 2 and 8 mg∙L–1 as a primary means for the destruction of broad-host-range plasmid and chromosomal DNA in simulated effluent. Pilot-scale tests using an experimental unit were carried out in municipal wastewater treatment plant effluent and compared with ultraviolet (UV)-irradiation and chlorination methodologies. Genes specific to Escherichia coli (uidA) and IncP broad-host-range plasmids (trfA) were monitored using real-time quantitative polymerase chain reaction (qPCR), and total DNA was monitored using absorbance spectroscopy. In wastewater treatment plant experiments, E. coli qPCR results were compared to a recognized culture-based method (Colilert®) for E. coli. In laboratory experiments, dissolved ozone at 8 mg∙L–1 significantly destroyed 93% total, 98% E. coli, and 99% of broad-host-range plasmid DNA. Ozonation, UV-irradiation, and chlorination significantly reduced DNA concentrations and culturable E. coli in wastewater treatment plant effluent. Chlorination and UV disinfection resulted in 3-log decreases in culture-based E. coli concentrations in wastewater treatment plant effluent while changes were not significant when measured with qPCR. Only ozonation significantly decreased the IncP broad-host-range plasmid trfA gene, although concentrations of 2.2 × 105 copies trfA∙L–1 remained in effluent. Disinfection processes utilizing high dissolved ozone concentrations for the destruction of emerging contaminants such as broad-host-range plasmid and total DNA may have utility as methods to ensure downstream environmental health and safe water reuse become more important.
1. Introduction
Municipal and regional wastewater treatment facilities remain a cornerstone of urban and economic development. While the processes employed at modern wastewater treatment facilities in the United States and in other industrialized countries are considerably evolved, the final effluent leaving these plants is not completely free of biological and chemical materials that are potentially harmful to humans and the environment [1-4]. Of developing concern are bioreactive compounds that are not targeted by regulations governing discharge of wastewater. This emerging group of contaminants includes compounds such as antibiotic resistance genes, disinfection by-products, pharmaceuticals, and hormones [5-7].
Biological contaminants pose a special set of threats among the compounds leaving wastewater treatment plants [8]. An obvious threat to public health is disease from pathogens that may persist in the environment after release. A less obvious threat is genetic determinants that can provide antimicrobial resistance phenotypes to pathogenic populations, such as antibiotic resistance genes contained on bacterial plasmids and other mobile genetic elements [1,9,10]. In particular, broad-host-range plasmids have several attributes contributing to concerns about their control: they can carry genes coding for resistance to multiple antibiotics and metals [11]; they can be stably maintained within and transferred among diverse bacterial taxa [12-14]; and they can persist outside of the cell for possible later uptake and expression by bacterial hosts [15]. As human population density increases, drinking water resources are increasingly likely to be downstream from wastewater treatment plant discharges. Thus, to protect drinking water resources and other downstream uses, limiting the release of mobile genetic elements and antibiotic resistance genes along with pathogens from wastewater treatment plants may need to become a cornerstone of municipal wastewater treatment and corresponding regulations for discharge.
Typically the final step in wastewater treatment, disinfection of wastewater in the US is focused entirely on reducing numbers of fecal coliforms present in polished effluent to levels below the USEPA mandated criterion. Chlorination and UV-irradiation are the most commonplace because of the relative cost and equipment problems when using ozone as a disinfectant [16,17]. However, ozone is gaining renewed interest in wastewater treatment due to decreasing water supplies, ensuing stricter discharge limits, and increasing concern regarding pharmaceutical and personal care products along with other emerging contaminants [18].
While inactivation kinetics of indicator bacteria and several pathogens using ozone have received attention in scientific literature historically, little is known about the effectiveness of ozone in the destruction of bacterial and plasmid DNA [19-23]. Several studies have shown the efficacy of ozone disinfection through non-reversible reactions with amine groups, aromatic rings and double bonds on compound molecules like pharmaceuticals, microbial membranes and cellular contents, but these studies have given less attention to the overall destruction of chromosomal and plasmid DNA by either ozone or hydroxyl radicals in wastewater treatment plant disinfection [24,25].
To determine if ozone utilized in municipal disinfection would improve destruction of indicator bacterial and broad-host-range plasmid DNA, controlled laboratory tests compared two ozone concentrations (approximately 2.0 and 8.0 mg∙L–1) in simulated effluent to a no-ozone treatment. The destruction of DNA by ozone was characterized using qPCR. To deliver ozone in a wastewater treatment plant, a proprietary, experimental pilot-scale dissolved ozone delivery system (HyDOZ®, BlueInGreen LLC, Fayetteville, AR, USA) was deployed in a municipal wastewater treatment plant sidestream. The effectiveness of the HyDOZ® ozonation system was measured using both culturable E. coli and a culture-independent method to measure DNA destruction. Results were compared to those obtained with UV-irradiation and chlorination.
2. Materials and Methods
A schematic of the dissolved ozone production system for laboratory use is shown in Figure 1. Briefly, oxygen gas (USP Grade, Airgas, Inc., Radnor, PA, USA) was converted to ozone gas using a CD2000P ozone generator
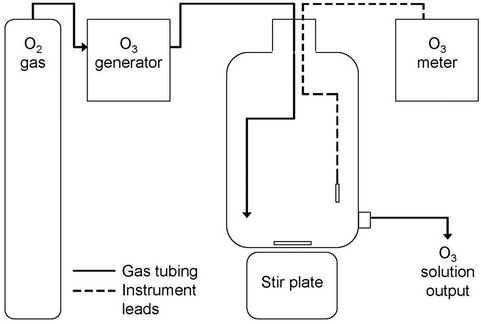
Figure 1. Diagram of the apparatus used to generate dissolved ozone solution from oxygen gas in the laboratory.
(Clearwater Tech LLC, Lewiston, ID, USA) and bubbled into a 20-L polypropylene carboy (Nalgene-Thermo Fisher Scientific, Rochester, NY, USA) containing 15 L of reagent-grade water. To ensure homogeneous dissolved ozone concentrations throughout the carboy, a large polytetrafluoroethylene-coated magnetic stir bar and stir plate (Corning Life Sciences, Lowell, MA, USA) were utilized to mix the contents. Dissolved ozone concentrations in the carboy were monitored continuously using a Q-45H dissolved ozone meter (Ozone Solutions Inc., Hull, IA, USA) and output concentrations from the system were periodically verified throughout testing using a DR/890 portable colorimeter and the indigo colorimetric method (Hach method 8311, APHA equivalent 4500-O3 B) and AccuVac® ampules (Hach Company, Loveland, CO, USA) (APHA, 1989).
Dissolved ozone treatments were carried out in sterile, sealed 1-L polycarbonate reactor vessels (VWR International, Radnor, PA, USA). To produce the two ozone treatment concentrations, differing amounts of water containing dissolved ozone were dispensed into prepared sample solutions containing a buffer, test organisms and background organic carbon to a final volume of 1 L. Sterile, stock organic carbon solution for assembly of the sample matrix was prepared from a garden soil using the soil extract broth method [26] and the total organic carbon concentration determined by combustion using a Shimadzu TOC-V (Shimadzu Corporation, Kyoto, Japan). Stock organic carbon solution was diluted into sterile phosphate buffered saline at pH 7.4 to yield a final concentration of 10.00 mg organic carbon L–1 in each sample, representing a maximum background dissolved organic carbon concentration expected in secondary effluent. Escherichia coli DH5α™ (stabilized cloning vector, Invitrogen Corporation, Carlsbad, CA, USA) harboring the broad-host-range plasmid R751 (IncP-1β subgroup; [27]) was cultured overnight in LB broth (Difco, Beckton Dickinson and Co., Sparks, MD, USA) containing trime-
thoprim (Sigma-Aldrich Corporation, St. Louis, MO, USA) at a concentration of 20 µg∙mL–1 for use as a seed organism. Just prior to addition of dissolved ozone, each reactor was seeded with 0.5 mL of liquid culture E. coli DH5α/R751. Triplicate 0.5-mL aliquots of the overnight seed culture were also transferred to 2-mL Lysing Matrix-E tubes (Qbiogene Inc., Carlsbad, CA) for total DNA extraction and use as a control.
Reactors were immediately sealed after the addition of dissolved ozone, mixed by inverting 10 times, and allowed to incubate at room temperature for 15 minutes. Treated samples were then promptly subjected to vacuum membrane filtration through sterile 47-mm diameter, 0.22-µm pore-size nitrocellulose membranes (Whatman GmbH, Dassel, Germany). Upon completion of filtration, membranes were stored at −80˚C until DNA extraction. Individual membranes were aseptically shredded and subjected to DNA extraction using the FastDNA® SPIN Kit for Soil and the FastPrep® Instrument (Qbiogene Inc., Carlsbad, CA). Total DNA extracts were cleaned by ethanol precipitation and quantified by UV-Vis spectroscopy on a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA) as described previously [2].
A pilot-scale version of the experimental HyDOZ® dissolved ozone delivery system (BlueInGreen LLC, Fayetteville, AR, USA) designed to treat approximately 3.8 million liters per day (MLD) of wastewater was installed in a temporary sidestream just before the disinfection stage at Springdale municipal wastewater treatment plant in Springdale, AR, USA. Between October 2010 and March 2011, treated wastewater was collected for filtration at the Springdale wastewater treatment plant (chlorination and ozonation) and from a wastewater treatment plant in Fayetteville, Arkansas (UV-irradiation). Water samples (1 L) were collected in sterile polycarbonate 1-L vessels (VWR International, Radnor, PA, USA) and subjected to serial filtration through 5.0- and 0.22-µm pore-size nitrocellulose membranes (Whatman GmbH, Dassel, Germany) and DNA extraction as described previously [2]. An average ozone treatment condition of 3.7 mg∙min∙L–1 (range 2 - 6 mg∙min∙L–1, equivalent to 0.37 mg∙L–1 sustained for 10 min) was reported (G. S. Osborn, personal communication).
The real-time quantitative polymerase chain reaction (qPCR) method to quantify a 187-bp region of the E. coli-specific uidA gene (encodes β-glucuronidase enzyme) for E. coli enumeration was modified for use in this study [28]. We also optimized a qPCR assay for quantification of a 241-bp fragment of the trfA gene specific to IncP plasmids based on previously published primer pairs used in the study of broad-host-range plasmids [2,9,29]. Assays for uidA and trfA had lower detection limits of 64 and 23 gene copies, respectively. Real-time qPCR runs were performed on an Applied Biosystems StepOnePlus™ real-time PCR system (Applied Biosystems, Life Technologies Corp., Carlsbad, CA, USA). Real-time qPCR reactions (20 µL each) consisted of SYBR® GreenER™ master mix, 200 nmol each of primer, 0.04% bovine serum albumin, 1000 nM ROX (Invitrogen, Life Technologies Corp., Carlsbad, CA, USA) passive reference dye and 1 µL of DNA-extracts or dilutions. Amplification run temperature profiles, melt curve analyses, and quality control factors were performed as recommended by the manufacturers.
Cultured-based E. coli most probable number determination (USEPA-approved) was carried out by diluting subsamples in sterile phosphate buffered saline and using Colilert® reagents with Quantitray®/2000 (IDEXX Laboratories, Inc., Westbrook, ME, USA) test materials according to manufacturer directions.
Statistical comparisons of appropriate log-transformed values were made using Oneway Analysis in JMP 9.0 software (SAS Institute Inc., Cary, NC, USA) and Tukey’s HSD (α = 0.05).
3. Results
In laboratory experiments, we found that only the higher 8.0 mg∙L–1 dose of dissolved ozone significantly reduced total DNA, E. coli chromosomal DNA, and IncP broadhost-range plasmid DNA in 1-L simulated wastewater samples containing dissolved organic carbon to provide a background O3 demand (Table 1). Reductions of 1.6 × 103 ng∙L–1 total DNA, 2.1 × 108 copies uidA∙L–1, and 1.2 × 109 copies trfA∙L–1 corresponded to respective decreases of approximately 93%, 98%, and 99% in the 8.0 mg ozone L–1 treatment. No significant differences were observed for any of the parameters among the 2.0 mg∙L–1, 0.0 mg∙L–1, and direct extraction control.
In wastewater treatment plant experiments, DNA concentrations were significantly reduced by 71% - 80% across all three disinfection processes (Table 2). Culturable E. coli were reduced 95% by ozonation, 99.8% by chlorination, and 99.9% by UV-irradiation. Interestingly, culture-independent qPCR assays did not indicate the same level of reduction in E. coli as the culture-dependent analysis (Table 2). Greater than 18,000 copies L–1 of the E. coli uidA amplicon remained after ozone disinfection compared to culturable E. coli concentrations of 2000 cells L–1. While three-log reductions occurred in culturable E. coli numbers in chlorination and UV treatments, only non-significant one-log reductions were detected in uidA gene fragments. In addition, we measured a significant reduction of 95% of trfA broad-host-range amplicons after ozone disinfection, while neither chlorination nor UV-iradiation significantly reduced trfA.
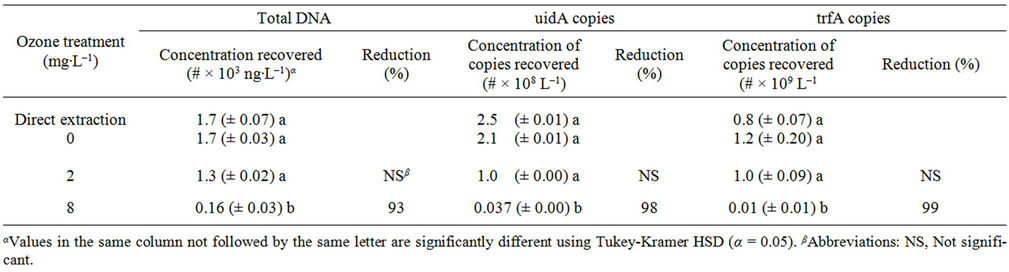
Table 1. Mean bacterial DNA remaining (± standard error of the mean) and percent reduction with respect to ozone concentrations present at 0 mg∙L–1 after ozone disinfection (n = 3).

Table 2. Enumeration (mean ± standard error of the mean) and percent reduction of Escherichia coli (Colilert® and uidA gene fragment), total DNA and IncP broad-host-range plasmids (trfA gene fragment) in wastewater treatment plant effluent.
4. Discussion
Chlorination and UV-irradiation, the primary disinfection processes utilized by the two wastewater treatment plants tested in this study, effectively reduced culturable E. coli 99.8% and 99.9%, respectively, meeting US EPA-mandated criterion related to the discharge of treated effluent. Ozone disinfection of culturable E. coli also resulted in significant reductions in E. coli. Furthermore, reduction using ozone was not significantly different than UV-irradiation on a percentage basis, indicating the possible utility of ozone disinfection for meeting disinfection regulations. Results from laboratory experiments had indicated that dissolved ozone treatments as high as 8.0 mg∙L–1 may be necessary to significantly reduce DNA in wastewater. Reductions in total DNA in the wastewater treatment plant were lower than percent reductions in laboratory experiments using 8.0 mg ozone L–1, which is consistent with the lower concentration of ozone reported in the pilot-scale HyDoz. However, percent reductions of total DNA in the wastewater treatment plant were significant by each respective disinfection process investigated.
Our results for total DNA remaining after disinfection in laboratory and wastewater treatment plant experiments are similar to results of previous studies examining the release of DNA into receiving streams. Akiyama et al. [2] found a mean of 3150 ng∙L–1 total DNA from multiple sites in water of two streams receiving effluent from wastewater treatment plants, one of which was Springdale wastewater treatment plant, the site of the pilot-scale system evaluated in this study. Using an APHA approved most probable number procedure, Tripathi et al. [30] reported removals of approximately 95% and greater than 99% of viable E. coli from wastewater samples after ozone treatment at 5 and 10 mg∙min∙L–1 (10 minute exposure times), respectively. While we measured lower DNA reductions of 93% in our laboratory study, our results of 98% reduction in the uidA gene as a measure to enumerate E. coli is in agreement with the reduction percentage measured in Tripathi et al. [30].
Despite these agreements in results across studies, the wastewater treatment plant results suggest that culturable E. coli cell counts will provide greater reductions compared to qPCR results targeting the uidA gene. The assumption in utilizing the uidA gene for E. coli enumeration is that one copy is equivalent to one cell, a ratio that has been demonstrated previously [28]. The disparity we observed between culture-based and culture-independent assays for E. coli may have been a result of disinfection effects on E. coli culturability (Table 2). Disinfection processes may inhibit short-term culturability of organisms while allowing viable cells to persist through disinfection and subsequent release into receiving streams. When evaluating fluorescent in situ hybridization results and a viability testing technique in contrast to a common culture-based most probable number method, Servais et al. [31] found a disparity similar to our observations. They noted that ratios of viable-to-culturable E. coli can vary drastically depending on the nutrient status of wastewater treatment plant effluents. Additionally, Arana et al. [32] noted that after chlorination, E. coli cells showed a tendency to aggregate, which could thereby affect the accuracy of conventional culture-based methods. A similar tendency for culture-based methods to enumerate Enterococci yielded lower values compared to qPCR methods [33]. More investigations evaluating the relationship between culture-dependent and culture-independent assays for fecal indicator bacteria such as E. coli will be necessary to achieve true calibration between cultivationdependent and cultivation-independent methods.
The lower ozone concentration in the wastewater treatment plant (reported 3.7 mg∙min∙L–1) compared to the results in the laboratory using 8.0 mg ozone L–1 may explain the lack of significant reduction in the uidA gene in the wastewater treatment plant. The primary target sites of microorganisms during ozone disinfection may be the outer membranes and cell surfaces [34,35]. While other disinfection agents may first diffuse through (chlorine-based oxidants) or penetrate (UV-irradiation) cell walls and cytoplasmic membranes before inactivating cellular components and inhibiting growth, ozone likely behaves differently [36,37]. E. coli membrane permeability may be altered after exposure to residual ozone levels as low as 0.1 - 0.2 mg∙L–1 [35]. Likewise, Komanapalli and Lau [34] also found initial damage to E. coli membranes using ozone, but 30 min exposure times to ozone were necessary to damage intracellular components, including DNA. Our observed reductions in DNA may have taken place after a portion of the available ozone and hydroxyl radicals were consumed during cellular membrane destruction. These consumptive reactions of available ozone and hydroxyl radicals may have reduced the amount necessary to further degrade DNA from E. coli cells.
We do not expect that the lack of significant reduction in the uidA gene after disinfection in the wastewater treatment plant is because the targeted uidA (or trfA) gene is more refractory than total DNA. The 98% reduction for the individual gene compared to the 93% reduction in total DNA in the laboratory indicated that percent reductions should be greater for individual genes. In fact, a significant reduction in the trfA gene was detected as a result of ozone disinfection in the wastewater treatment plant.
In contrast to ozonation, trfA genes were not reduced using chlorination and UV-irradiation in the wastewater treatment plant. The potential of co-extraction of compounds that interfere with the qPCR reactions contributes to variability in environmental samples. Not all replications produced reliable qPCR results, which reduced our final sample size (Table 2), and this contributes to whether significant differences are detected after each disinfection process. However, even with a significant 95% reduction by ozonation, 2.2 × 105 copies L–1 of the trfA gene specific to IncP broad-host-range plasmids remained in effluent. Akiyama et al. [2] also showed the introduction of broad-host-range plasmids into receiving streams by wastewater treatment plants using chlorination or UV-irradiation, but the results were not quantitative for broad-host-range plasmids.
The lack of a regulatory standard for broad-host-range plasmids makes determination of an adequate level of disinfection difficult to ascertain. In theory, as few as one mobile genetic element containing antibiotic resistance determinants is all that is needed to give rise to a resistant population. Babic et al. [38] observed that a single cell harboring a mobile genetic element was capable of transferring genes to >50% of surrounding cells. Furthermore, 81% of recipient cells became donors within 30 minutes of the first conjugation event, indicating the speed at which horizontal evolution can spread genes amongst a population given a small initial abundance. Because IncP plasmids are particularly promiscuous [39], often associated with multiple resistance genes [40], and are often present in wastewater treatment plants [9], this lack of complete removal may represent a potential concern for downstream users of streams receiving discharged effluent. However, epidemiological studies are required to assess the final health risk of broad-host-range plasmid discharge into surface water. To what extent broad-hostrange plasmids persist, are disseminated, and stably maintained in the environment is still not well understood.
5. Conclusion
Our results confirm that ozonation, UV-irradiation, and chlorination all significantly reduce DNA concentrations and culturable E. coli. Our results indicate the efficacy for high ozone concentrations for DNA destruction as measured by total DNA or specific genes targeting chromosomal or plasmid DNA. Mobile genetic elements such as broad-host-range plasmids can be considered emerging contaminants. More investigations will be required to determine the level of reduction of broad-hostrange plasmids needed to ensure downstream environmental health and safe water reuse. Additionally, disparities exist between culture-based and culture-independent assays for the detection of bacteria, and molecular methods need to be calibrated to culture-based methods for quantification of fecal indicator bacteria.
6. Acknowledgements
This research was funded by the National Institutes of Health STTR research grant #2R42ES014137-02-UA, the University of Arkansas and the Division of Agriculture. Real-time PCR data were obtained on an instrument provided by the P3 Center, funded through the Arkansas ASSET Initiative II (EPS-1003970) by the National Science Foundation. We would like to thank J. Enos (Springdale Water) for information regarding the Springdale wastewater treatment plant, Dr. G. S. Osborn, G. Richardson and colleagues with BlueInGreen LLC for their help during wastewater treatment plant testing, and Dr. K. Korth and Dr. J. T. Scott for use of instrumentation. We thank T. Akiyama for critical discussion and P. Tomlinson, F. Taggart and S. Potter for technical assistance.
REFERENCES
- T. Akiyama and M. C. Savin, “Populations of antibiotic-resistant coliform bacteria change rapidly in a wastewater effluent dominated stream,” Science of the Total Environment, Vol. 408, No. 24, 2010, pp. 6192- 6201. doi:10.1016/j.scitotenv.2010.08.055
- T. Akiyama, K. L. Asfahl and M. C. Savin, “Broad-hostrange plasmids in treated wastewater effluent and receiving streams,” Journal of Environmental Quality, Vol. 39, No. 6, 2010, pp. 2211-2215. doi:10.2134/jeq2010.0228
- M. F. da Silva, I. Tiago, A. Veríssimo, R. A. R. Boaventura, O. C. Nunes and C. M. Manaia, “Antibiotic resistance of enterococci and related bacteria in an urban wastewater treatment plant,” FEMS Microbiology Ecology, Vol. 55, No. 2, 2005, pp. 322-329. doi:10.1111/j.1574-6941.2005.00032.x
- B. E. Haggard, J. M. Galloway, W. R. Green and M. T. Meyer, “Pharmaceuticals and other organic chemicals in selected North-Central and Northwestern Arkansas streams,” Journal of Environmental Quality, Vol. 35, No. 4, 2006, pp. 1078-1087. doi:10.2134/jeq2005.0248
- S. Richardson, “Disinfection by-products and other emerging contaminants in drinking water,” Trends in Analytical Chemistry, Vol. 22, No. 10, 2003, pp. 666-684. doi:10.1016/S0165-9936(03)01003-3
- S. Richardson and T. A. Ternes, “Water Analysis: Emerging Contaminants and Current Issues,” Analytical Chemistry, Vol. 77, No. 12, 2005, pp. 3807-3838. doi:10.1021/ac058022x
- A. Pruden, R. T. Pei, H. Storteboom and K. H. Carlson, “Antibiotic Resistance Genes as Emerging Contaminants: Studies in Northern Colorado,” Environmental Science & Technology, Vol. 40, No. 23, 2006, pp. 7445-7450. doi:10.1021/es060413l
- World Health Organization (WHO), “Guidelines for the safe use of wastewater, excreta and greywater. Volume 3: wastewater and excreta use in aquaculture,” WHO, Geneva, 2006.
- M. Dröge, A. Pühler and W. Selbitschka, “Phenotypic and molecular characterization of conjugative antibiotic resistance plasmids isolated from bacterial communities of activated sludge,” Molecular and General Genetics, Vol. 263, No. 3, 2000, pp. 471-482. doi:10.1007/s004380051191
- E. Kristiansson, J. Fick, A. Janzon, R. Grabic, C. Rutgersson, B. Weijdegård, H. Söderström and D. G. J. Larsson, “Pyrosequencing of Antibiotic-Contaminated River Sediments Reveals High Levels of Resistance and Gene Transfer Elements,” Public Library of Science, Vol. 6, No. 2, 2011, Article ID: e17038. doi:10.1371/journal.pone.0017038.
- A. Schlüter, R. Szczepanowski, A. Pühler and E. M. Top, “Genomics of IncP-1 Antibiotic Resistance Plasmids Isolated from Wastewater Treatment Plants Provides Evidence for a Widely Accessible Drug Resistance Gene Pool,” FEMS Microbiology Reviews, Vol. 31, No. 4, 2007, pp. 449-477. doi:10.1111/j.1574-6976.2007.00074.x
- K. Smalla, H. Heuer, A. Götz, D. Niemeyer, E. Krögerrecklenfort and E. Tietze, “Exogenous Isolation of Antibiotic Resistance Plasmids from Piggery Manure Slurries Reveals a High Prevalence and Diversity of IncQ-Like Plasmids,” Applied and Environmental Microbiology, Vol. 66, No. 11, 2000, pp. 4854-4862. doi:10.1128/AEM.66.11.4854-4862.2000
- A. Siddique and D. H. Figurski, “The Active Partition Gene incC of IncP Plasmids Is Required for Stable Maintenance in a Broad Range of Hosts,” Journal of Bacteriology, Vol. 184, No. 6, 2002, pp. 1788-1793. doi:10.1128/JB.184.6.1788-1793.2002
- R. I. Aminov and R. I. Mackie, “Evolution and ecology of Antibiotic Resistance Genes,” FEMS Microbiology Letters, Vol. 271, No. 2, 2007, pp. 147-161. doi:10.1111/j.1574-6968.2007.00757.x
- J. Sikorski, N. Teschner and W. Wackernagel, “Highly Different Levels of Natural Transformation Are Associated with Genomic Subgroups within a Local Population of Pseudomonas stutzeri from Soil,” Applied and Environmental Microbiology, Vol. 68, No. 2, 2002, pp. 865- 873. doi:10.1128/AEM.68.2.865-873.2002
- A. Kontana, C. A. Papadimitriou, P. Samaras, A. Zdragas and M. Yiangou, “Effectiveness of ozonation and chlorination on Municipal Wastewater Treatment Evaluated by a battery of bioassays and biomarkers,” Water Science and Technology, Vol. 60, No. 6, 2009, pp. 1497-1505. doi:10.2166/wst.2009.478
- J. Kuo and L. Abustan, “Disinfection and antimicrobial processes,” Water Environment Research, Vol. 81, No. 10, 2009, pp. 1361-1375. doi:10.2175/106143009X12445568399695
- M. A. Oneby, C. O. Bromley, J. H. Borchardt and D. S. Harrison, “Ozone Treatment of Secondary Effluent at U.S. Municipal Wastewater Treatment Plants,” Ozone Science & Engineering, Vol. 32, No. 1, 2010, pp. 43-55. doi:10.1080/01919510903482780
- G. B. Wickramanayake, A. J. Rubin and O. J. Sproul, “Inactivation of Giardia lamblia cysts with ozone,” Applied and Environmental Microbiology, Vol. 48, No. 3, 1984, pp. 671-672.
- G. R. Finch, E. K. Black, L. Gyürék and M. Belosevic, “Ozone inactivation of Cryptosporidium parvum in Demand Free Phosphate Buffer Determined by in vitro excystation and animal infectivity,” Applied and Environmental Microbiology, Vol. 59, No. 12, 1993, pp. 4203-4210.
- N. K. Hunt and B. J. Mariñas, “Kinetics of Escherichia coli inactivation with ozone,” Water Research, Vol. 31, No. 6, 1997, pp. 1355-1362. doi:10.1016/S0043-1354(96)00394-6
- J. L. Rennecker, B. J. Mariñas, J. H. Owens and E. W. Rice, “Inactivation of Cryptosporidium parvum oocysts with ozone,” Water Research, Vol. 33, No. 11, 1999, pp. 2481-2488. doi:10.1016/S0043-1354(99)00116-5
- A. Driedger, E. Staub, U. Pinkernell, B. Mariñas, W. Köster and U. von Gunten, “Inactivation of Bacillus subtilis spores and formation of bromate during ozonation,” Water Research, Vol. 15, No. 12, 2001, pp. 2950-2960. doi:10.1016/S0043-1354(00)00577-7
- U. von Gunten, “Ozonation of Drinking Water: Part I. Oxidation Kinetics and Product Formation,” Water Research, Vol. 37, No. 7, 2003, pp. 1443-1467. doi:10.1016/S0043-1354(02)00457-8
- M. M. Huber, A. Göbel, A. Joss, N. Hermann, D. Löffler, S. Mcardell, A. Ried, H. Siegrist, T. A. Ternes and U. von Gunten, “Oxidation of Pharmaceuticals during Ozonation of Municipal Wastewater Effluents: A Pilot Study,” Environmental Science and Technology, Vol. 39, No. 11, 2005, pp. 4290-4299. doi:10.1021/es048396s
- N. James, “Soil extract in soil microbiology,” Canadian Journal of Microbiology, Vol. 4, No. 4, 1958, pp. 363- 370. doi:10.1139/m58-038
- R. S. Jobanputra and N. Datta, “Trimethoprim R Factors in Enterobacteria from Clinical Specimens,” Journal of Medical Microbiology, Vol. 7, No. 2, 1974, pp. 169-177. doi:10.1099/00222615-7-2-169
- L. Heijnen and G. Medema, “Quantitative detection of E. coli, E. coli O157 and other Shiga Toxin Producing E. coli in water Samples Using a Culture Method Combined with Real-Time PCR,” Journal of Water & Health, Vol. 4, No. 4, 2006, pp. 487-498.
- A. Götz, R. Pukall, E. Smit, E. Tietze, R. Prager, H. Tschäpe, J. D. van Elsas and K. Smalla, “Detection and Characterization of Broad-Host-Range plasmids in environmental Bacteria by PCR,” Applied and Environmental Microbiology, Vol. 62, No. 7, 1996, pp. 2621-2628.
- S. Tripathi, V. Pathak, D. M. Tripathi and B. D. Tripathi, “Application of Ozone Based Treatments of Secondary Effluents,” Bioresource Technology, Vol. 102, No. 3, 2011, pp. 2481-2486. doi:10.1016/j.biortech.2010.11.028
- P. Servais, J. Prats, J. Passerat and T. Garcia-Armisen, “Abundance of culturable versus viable Escherichia coli in freshwater,” Canadian Journal of Microbiology, Vol. 55, No. 7, 2009, pp. 905-909. doi:10.1139/W09-043
- I. Arana, P. Santorum, A. Muela and I. Barcina, “Chlorination and ozonation of Waste-Water: Comparative Analysis of efficacy through the effect on Escherichia coli membranes,” Journal of Applied Microbiology, Vol. 86, No. 5, 1999, pp. 883-888. doi:10.1046/j.1365-2672.1999.00772.x
- R. L. Whitman, Z. Ge, M. B. Nevers, A. B. Boehm, E. C. Chern, R. A. Haugland, A. M. Lukasik, M. Molina, K. Przybyla-Kelly, D. A. Shively, E. M. White, R. G. Zepp and M. N. Byappanahalli, “Relationship and Variation of qPCR and Culturable Enterococci Estimates in Ambient Surface Waters Are Predictable,” Environmental Science & Technology, Vol. 44, No. 13, 2010, pp. 5049-5054. doi:10.1021/es9028974
- I. R. Komanapalli and B. H. S. Lau, “Ozone-induced damage of Escherichia coli K-12,” Applied Environmental and Biotechnology, Vol. 46, No. 5-6, 1996, pp. 610-614. doi:10.1007/s002530050869
- F. Zuma, J. Lin and S. B. Jonnalagadda, “Ozone-initiated disinfection kinetics of Escherichia coli in water,” Journal of Environmental Science and Health, Vol. 44, No. 1, 2009, pp. 48-56. doi:10.1080/10934520802515335
- R. L. Wolfe, “Ultraviolet disinfection of potable water— Current Technology and Research Needs,” Envrionmental Science and Technology, Vol. 24, No. 6, 1990, pp. 768- 773. doi:10.1021/es00076a001
- J. Y. Maillard, “Bacterial target sites for biocide action,” Journal of Applied Microbiology, Vol. 92, No. 1, 2002, pp. 16S-27S. doi:10.1046/j.1365-2672.92.5s1.3.x
- A. Babic, M. B. Berkmen, C. A. Lee and A. D. Grossman, “Efficient Gene Transfer in Bacterial Cell Chains,” mBio, Vol. 2, No. 2, 2011. doi:10.1128/mbio.00027-11
- M. Sota, H. Yano, J. M. Hughes, G. W. Daughdrill, Z. Abdo, L. J. Forney and E. M. Top, “Shifts in the Host Range of a Promiscuous Plasmid through Parallel Evolution of its Replication Initiation Protein,” The ISME Journal, Vol. 4, No. 12, 2010, pp. 1568-1580. doi:10.1038/ismej.2010.72
- R. Szczepanowski, I. Krahn, N. Bohn, A. Pühler and A. Schlüter, “Novel Macrolide Resistance Module Carried by the IncP-1β Resistance Plasmid pRSB111, Isolated from a Wastewater Treatment Plant,” Antimicrobial Agents and Chemotherapy, Vol. 51, No. 2, 2005, pp. 673-678. doi:10.1128/AAC.00802-06

