Open Journal of Medical Microbiology
Vol.3 No.4(2013), Article ID:41009,6 pages DOI:10.4236/ojmm.2013.34031
Detection of Extended Spectrum β-Lactamase Producing Klebsiella pneumoniae and Escherichia coli in Two Hospitals in the Federal Capital Territory, Abuja, Nigeria
Department of Biological Sciences, University of Abuja, Nigeria
University of Abuja Teaching Hospital, Abuja, Nigeria
Email: tosinakanbi2@yahoo.co.uk
Copyright © 2013 Bolaji Oluwatosin Akanbi et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accordance of the Creative Commons Attribution License all Copyrights © 2013 are reserved for SCIRP and the owner of the intellectual property, Bolaji Oluwatosin Akanbi et al. All Copyright © 2013 are guarded by law and by SCIRP as a guardian.
Received June 11, 2013; revised July 12, 2013; accepted August 19, 2013
Keywords: Extended Spectrum Beta-Lactamase; Klebsiella pneumoniae; Escherichia coli; Nigeria; Multidrug Resistance
ABSTRACT
In this study, the prevalence of Extended Spectrum Beta-lactamase (ESBL) producing Klebsiella pneumoniae and Escherichia coli isolates from the University of Abuja Teaching Hospital and the National Hospital was determined. A total of two hundred and fifteen (215) clinical isolates were examined, of which 60% were E. coli and 40% K. pneumoniae respectively. The isolates were collected from various samples namely: Stool, Urine, Pus, High Vagina Swab, Sputum and Wound swab. Out of these isolates, 54 of K. pneumoniae were screened to be ESBL negative and 32 as ESBL positive isolates, while 88 and 40 E. coli were also screened as ESBL negative and ESBL positive isolates respectively. These represent 37.9% of all K. pneumoniae isolates and 31.25% of E. coli isolates respectively. The prevalence of ESBL among the species was not however statistically different (p > 0.05). Multiple resistance in these isolates was common and there is the need for routine screening of ESBL in our hospitals to guide rational and effective use of antibiotics.
1. Introduction
Antimicrobial resistance has arisen across the globe in both nosocomial and community settings as a consequence of widespread antibiotics consumption [1]. The Beta-lactam antibiotics are among the most widely used antimicrobial agents worldwide. Destruction of these antibiotics by the bacterial enzyme, beta-lactamase is the most frequently encountered mechanism of resistance among Gram-negative microorganisms [2]. Multidrug resistant gram negative bacilli belonging to the family Enterobacteriaceae have been increasingly responsible for infections in many countries [3]. The emergence of multiresistance in the Enterobacteriaceae family needs attention, because these are important causative agents of hospital infections, typically associated with pneumonias, blood stream infections, urinary tract infections, bacteremia and other intra-abdominal infections [4,5]. By definition, ESBLs are β-lactamases capable of conferring bacterial resistance to the penicillins, first-, second-, and third-generation cephalosporins, and aztreonam (but not the cephamycins or carbapenems) by hydrolysis of these antibiotics, which are inhibited by β-lactamase inhibitors such as clavulanic acid [6].
The presence of ESBLs has tremendous clinical significance due to the fact that ESBLs are frequently plasmid encoded and also in most cases the plasmids responsible for ESBL production frequently carry genes encoding resistance to other drug classes therefore limiting antibiotic options in the treatment of ESBL-producing organisms [6]. The selection pressure that drives the emergence of ESBLs has usually been attributed to the intense use of oxyimino-beta lactams, mainly the third generation cephalosporins in addition to extensive use of broad spectrum antibiotics, prolonged hospitalization, indwelling devices and severe underlying diseases [7-9].
Several studies have demonstrated that ESBL-producing bacteria are isolated with increasing frequency in many parts of the world [3,10-12] and a number of reports are also available in other parts of this country [13,14]. The aim of this study was to detect ESBL-producing K. pneumoniae and E. coli from clinical isolates from both inpatients and outpatients to evaluate the risk factors that may be inherent in this location. This is necessary because the prevalence of resistant strains of these organisms varies from one geographical location to another. Two hospitals receiving the highest number of patients from the federal capital territory and its environs namely, the University of Abuja Teaching Hospital and National Hospital were chosen for this study. To our knowledge, no published study exists on ESBL in Klebsiella pnuemoniae and E. coli in this locality, probably reflecting the lack of appreciation of the problem.
2. Materials and Methods
2.1. Isolation and Identification of Klebsiella pneumoniae and Escherichia coli
The isolates were collected from the University of Abuja Teaching Hospital Gwagwalada and National Hospital, Abuja. The organisms were isolated from stool, pus, urine, sputum as well as wound and high vaginal swabs (HVS) samples of both out-patients and In-patients. Isolates presumed to be the etiologic agent responsible for the disease condition were used for the study. Isolates considered to be contaminants were not included in the screening for ESBL. Isolates from pus and wound swabs were however included in the screening even though the organisms are not usually associated with infection at these sites.
Briefly, swabs and clinical specimens were inoculated on eosin methylene blue agar (EMB, Oxoid) and MacConkey agar (Oxoid). After inoculation, the plates were incubated at 37˚C for 24 hr. The mucoids and smooth colonies suggesting K. pneumoniae strains were Gram stained. Routinely, Indian ink was used to detect the presence of capsules and isolates were also inoculated onto the screening media for biochemical identification: TSI (triple sugar iron), SIM (sulphate⁄indole⁄motility) and citrate agar (Oxoid), and incubated at 37˚C for 24 h. Colonies showing green metallic sheen on EMB and non mucoid round pinkish colonies on MacConkey were tested for production of indole, methyl red, Voges Proskaeur and citrate utilization (IMVIC).
2.2. Antimicrobial Susceptibility Tests
The K. pneumoniae and E. coli strains isolated were submitted to antimicrobial susceptibility testing according to the recommendations of the Clinical and Laboratory Standards Institute [15]. The turbidity of the suspensions used for sensitivity testing was adjusted to 0.5 McFarland standard and inoculated onto Mueller-Hinton agar medium followed by incubation at 35˚C ± 1˚C for 18 - 24 hrs. The following antimicrobial discs were used: ceftriaxone (CRO) (30 µg), ceftazidime (CAZ) (30 µg), cefepime (FEP) (30 µg), gentamicin (GEN) (10 µg), amikacin (AK) (30 µg), ciprofloxacin (CIP) (5 µg), chloramphenicol (C) (30 µg) and trimethoprim⁄sulfamethoxazole (SXT) (1.25⁄23.75 µg).
Isolates that exhibited a zone of inhibition of growth for ceftazidime and ceftriaxone ≤22 mm and ≤25 mm, respectively, were submitted to the combined disc test in order to check for ESBL-producing strains. The combined disc methodology used to detect ESBLproducing K. pneumoniae and E. coli was performed as recommended by CLSI [15]. The antimicrobials used were: cefotaxime (30 µg) and cefotaxime (30 µg) plus clavulanic acid (10 µg), and ceftazidime (30 µg) and ceftazidime (30 µg) plus clavulanic acid (10 µg). Results were interpreted according to the criteria established by the CLSI [16]. A 5 mm increase in a zone of inhibition of growth for cefotaxime plus clavulanic acid as compared with the zone around the cefotaxime disc, and a 5 mm increase in the zone diameter for ceftazidime plus clavulanic acid as compared with the zone formed by the ceftazidime disc, were confirmatory for the result of ESBL-producing strains.
Interpretation of results for other antibacterial agents was as per the guidelines of The European Committee on Antimicrobial Susceptibility Testing (EUCAST) for enterobacteriaceae [16].
2.3. Statistical Analysis
Chi square was used to analyze data on gender distribution of the isolates, site distribution of the isolates and frequency of ESBL production by K. pneumoniae and E. coli using the software Smith’s Statistical Package (SSP) version 2.80 copyright© 1995-2005 Gary Smith.
3. Results
A total of 215 isolates were collected from the University of Abuja Teaching Hospital and the National Hospital Abuja to determine ESBL production. Out of 215 isolates 128 were E. coli (60%) and 87 were K. pneumoniae (40%). The age range of these patients was between 2 - 78 years.
Figure 1 shows that more females were infected by these pathogens than men with values of 40.63% (52) for E. coli infections in men which was less than the 59.37% (76) for women. Similarly in K. pneumoniae infections the frequency was higher in females n = 52, (55.170%) compared to males n = 39, (44.83%). The observed differences in male and female proportions of the two pathogens were not significant with a p value of 0.74 χ2, 1 df, (p > 0.5).
The distribution of K. pneumoniae and E. coli in the different specimens examined is shown in Figure 2. The number of isolates obtained from different sites was not uniform. The highest numbers of both E. coli as well as K. pneumoniae were obtained from urine at 60 and 36 respectively. For E. coli diarrheic fecal samples, pus, HVS wound swabs and sputum in decreasing order accounted for the remainder of the isolates at 37, 17, 10 and 0 respectively. K. pneumoniae was isolated in decreasing order from sputum (30), pus (9), HVS (8), wound swabs (3) and stool (0). The distribution of E. coli and K. pneumoniae isolates was significantly different at these sites p value of 0.00000 χ2, 5 df, (p < 0.05).
Table 1 shows the antimicrobial susceptibilities of K. pneumoniae isolates to different classes of antibiotics namely third generation cephalosporins (ceftriaxone and ceftazidime), fourth generation cephalosporins (cefepime), aminoglycosides (gentamicin and amikacin),
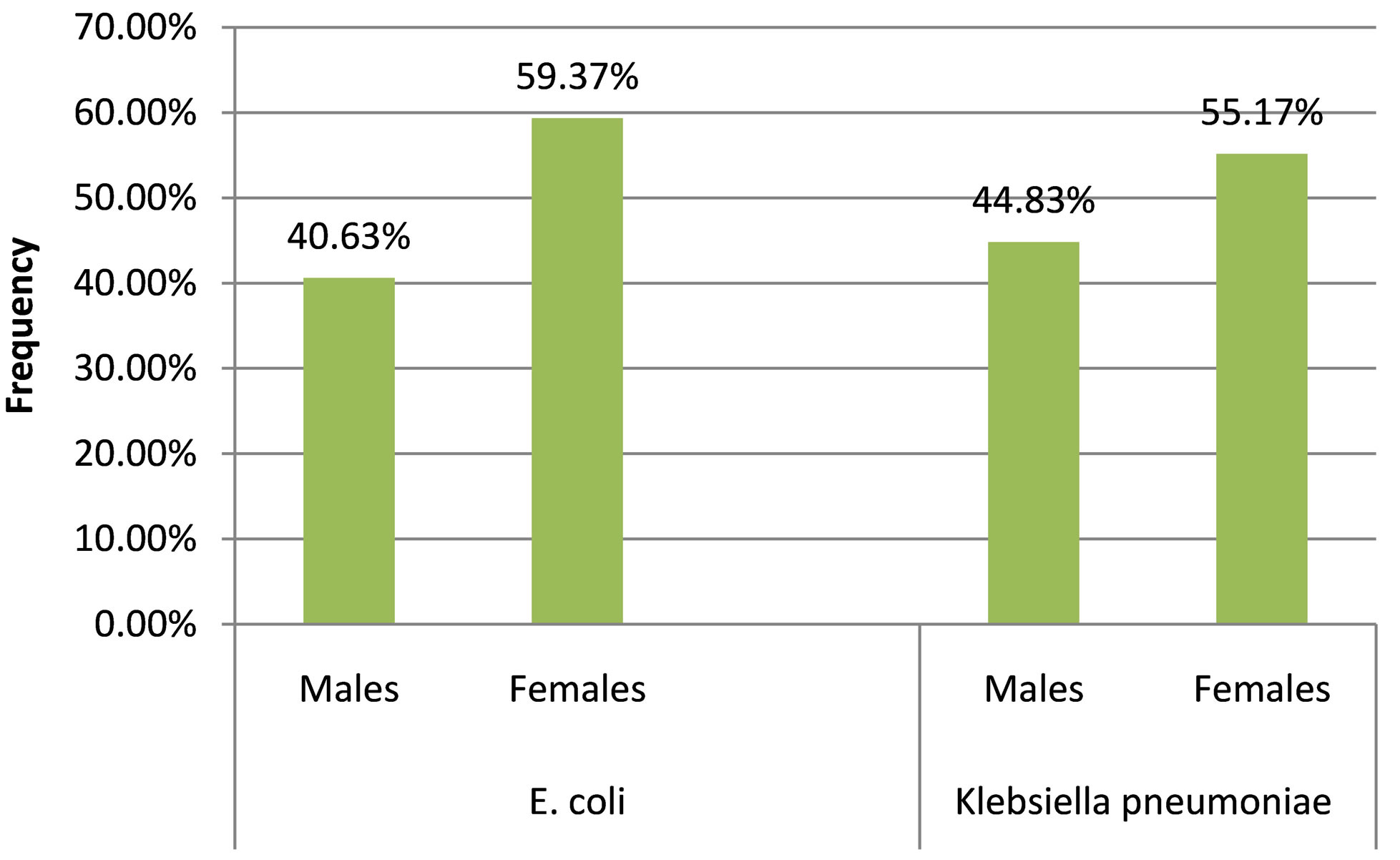
Figure 1. Gender Distribution of the isolates.
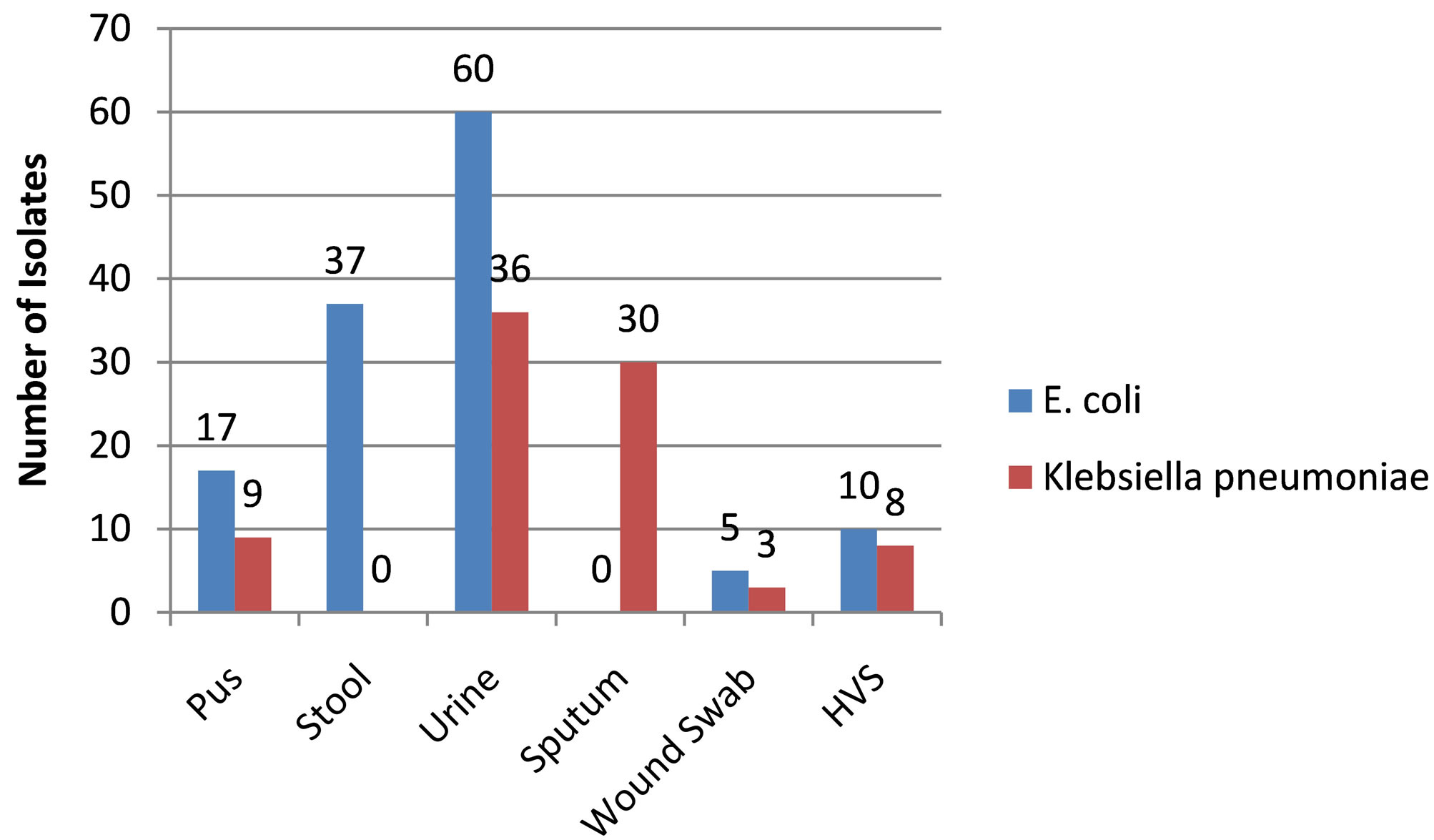
Figure 2. Site Distribution of the isolates.
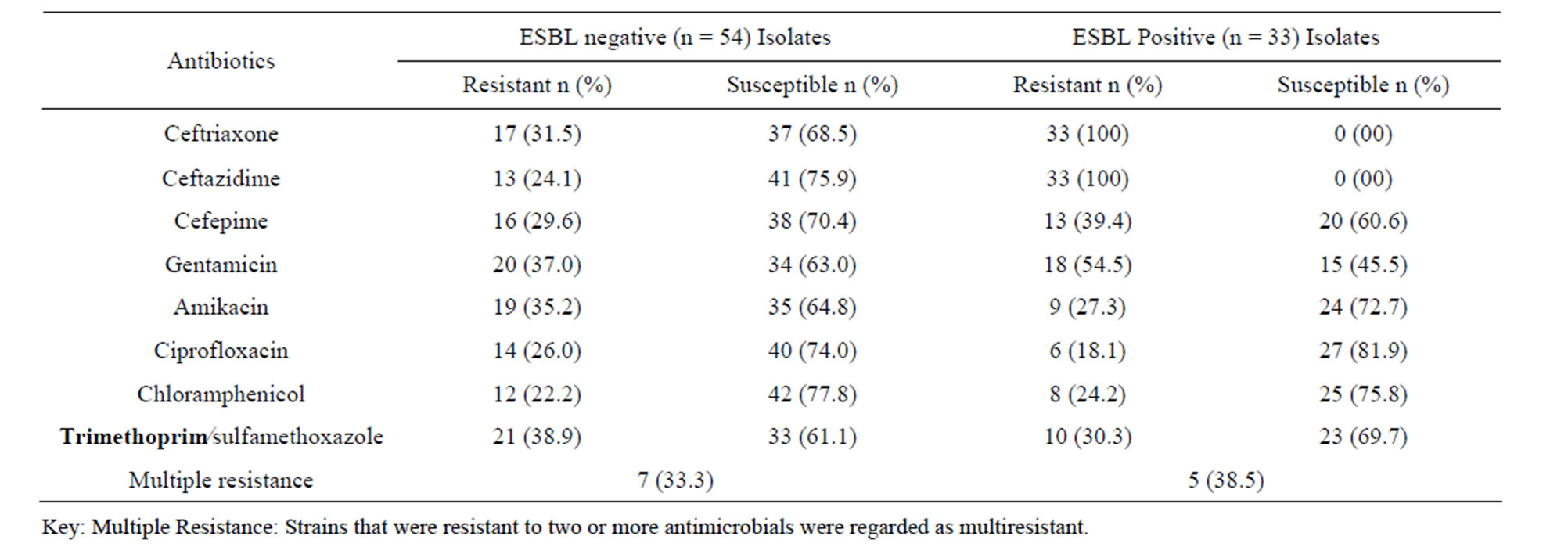
Table 1. Antimicrobial susceptibilities of Klebsiella pneumoniae isolates.
fluoroquinolone (ciprofloxacin), sulfonamide/trimethoprim combination (trimethoprim/sulfamethoxazole) and chlo ramphenicol.
A large number of ESBL-producing Klebsiella pneumoniae strains isolated also showed resistance to other classes of antimicrobials tested. Out of the tested isolates, 54.5% were resistant to gentamicin whereas resistance was lower to other antibacterial agents with values of 27.3%, 18.1%, 24.2% and 30.3% resistance to amikacin, ciprofloxacin, chloramphenicol and trimethprim/sulfamethoxazole respectively. Resistance of a strain to 2 or more antimicrobials was considered multiple resistance and in this case 38.5% were multiple resistant. Comparatively non-ESBL producing strains were less resistant with resistant values of 37.0%, 35.2%, 26.0%, 22.2% and 38.9% to gentamicin, amikacin, ciprofloxacin, chloramphenicol and trimethprim/sulfamethoxazole respectively. Seven of the non-ESBL (33.3%) were multiple resistant which is slightly lower than that of ESBL producing strains.
Table 2 shows the antimicrobial susceptibilities of E. coli isolates to different classes of antibiotics. Of the ESBL-producing E. coli strains isolated, 32.5%, 20.0%, 25.0%, 27.5% and 35.0% were resistant to gentamicin, amikacin, ciprofloxacin, chloramphenicol, and trimethprim/sulfamethoxazole respectively. In addition 42.9% were multiple resistant to the test antibiotics as previously defined. Resistance to these antimicrobials by non-ESBL producing strains was variable being higher with amikacin (29.5%) but lower with respect to gentamicin (23.9%), ciprofloxacin (22.7%), chloramphenicol (23.9%) and, trimethoprim/sulfamethoxazole (31.8%).
Comparatively more isolates of K. pneumoniae (37.9% of all K. pneumoniae isolates) were ESBL producing when compared to ESBL producing E. coli (31.25% of all E. coli isolates). These values were not however statistically different (p > 0.05). χ2, 1 df, p = 0.31
4. Discussion
K. pneumoniae and E. coli infections are some of the most commonly encountered ones in clinical medicine, causing a wide range of clinical conditions from relatively mild to serious, sometimes life-threatening conditions that can lead to death. In the present study, both organisms were isolated from different age groups and equally from both male and female genders. No differences were apparent which is similar to observations in a study that found no relationship between ESBL-producing E. coli or K. pneumonia infection with age or sex [17]. However in another study that assessed risk factor for mortality, most patients infected with ESBL producers were elderly and with a slight male predominance (59%) in the group studied [18].
The distribution of isolates and sites of infection were significantly different in this study and supports in a similar report were out of a total of 33 patients with ESBL-producing E. coli or K. pneumonia infection. 25 (75.8%) of them had infections due to K. pneumonia and 8 (24.2%) had infections due to E. coli [17].
Moreover, the distribution of the two pathogens studied reflects differences in their pathogenicity and associated disease conditions. K. pneumoniae is known to cause suppurative infections, bacteremia, and a substantial percentage of nosocomal infections. However, urinary tract infections and infections of the respiratory tract predominate [19]. E. coli strains are known to cause a wide variety of diseases. The isolates from this study were not, however, further characterized into patho groups.
About 38% of K. pneumoniae were ESBL-producing compared to 31% of E. coli isolates; this may indicate that ESBL production is more common among K. pneumoniae isolates as reported in other studies such as Lautenbach et al. (2001) with K. pneumoniae accounting for 75.8% of ESBL production compared to 24.2% for E.coli [17]. Also Serefhanoglu et al. (2009) reported 60.6% of ESBL producing isolates to be K. pneumoniae whereas 39.4% were E. coli in a total of 94 bloodstream infections examined [20]
A large number of ESBL-producing K. pneumoniae isolates also showed resistance to other classes of antimicrobials tested particularly gentamicin (54.5%) and trimethprim/sulfamethoxazole (30.3%). This is consistent with observations that ESBL-producing bacteria are associated with the transfer of conjugative plasmids, which also carry genes of resistance to aminoglycosides and sulfonamides, giving the bacteria multiresistance attributes [21].
Importantly, conjugative plasmids can be easily transferred across species as demonstrated in transfer from clinical isolates of Klebsiella pneumoniae to Escherichia coli involving transfer of resistance to ceftazidime, cefotaxime, ceftriaxone, gentamicin, amikacin, ciprofloxacin, aztreonam, cefoxitin and ticarcillin/CA and intermediate resistance to piperacillin/tazobactam, cefoperazone/sulbactam and cefepime [22].
Similarly, a substantial number of ESBL-producing E. coli strains isolated were also resistant to other antibiotics although the rates appeared lower than those of Klebsiella pneumonia and the same mechanisms of resistance have been noted earlier. Comparatively, resistance to these antimicrobials by non-ESBL producing strains was lower probably reflecting lower rates of transfer of multiple transfers of resistance genes in non ESBL producers.
Our results show a very high incidence of ESBL producing clinical isolates. The implications of which are the necessity for circumspection in prescription of anti-
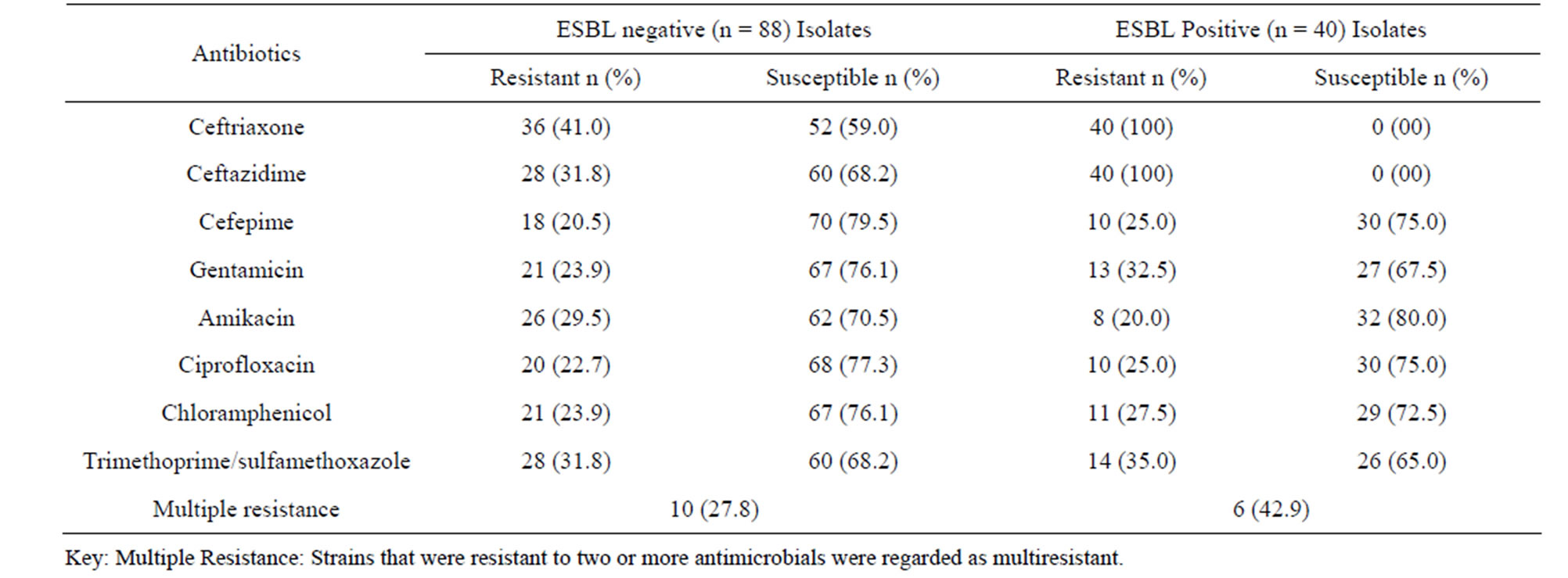
Table 2. Antimicrobial susceptibilities of E. coli isolates
biotics, appropriate hospital waste disposal and efficient hospital sewage treatment to prevent and limit environmental spread. Furthermore, regular surveillance is necessary including more detailed molecular studies as well as research in the prevalence studies on carbepenemase producing Klebsiella pneumoniae in this region to get a fuller picture of the extent of the problem.
REFERENCES
- E. A. Ashley, Y.Lubell, N. J. White and P. Turner, “Antimicrobial Susceptibility of Bacterial Isolates from Community Acquired Infections in Sub-Saharan Africa and Asian Low and Middle Income Countries,” Tropical Medicine and International Health, Vol. 16, No. 9, 2011, pp. 1167-1179. http://dx.doi.org/10.1111/j.1365-3156.2011.02822.x
- D. Faith, M. Villanueva, E. Thelma, D. Tupasi, G. Homer, M. D.Abiad, Q. Benilda, M. Baello, C. Rowena and D. Cardano, “Extended Spectrum β-Lactamase Production among Escherichia coli and Klebsiella spp.,” Philippines Journal Microbiological Infectious Diseases, Vol. 32, No. 3, 2003, pp. 103-108.
- R. Canton, A. Novais, A. Valverde, E. Machado, L. Peixe, F. Baquero and T. M. Coque, “Prevalence and Spread of Extended-Spectrum Betalactamase-Producing Enterobacteriaceae in Europe,” Clinical Microbiology and Infection, Vol. 14, Suppl. 1, 2008, pp. 144-153. http://dx.doi.org/10.1111/j.1469-0691.2007.01850.x
- S. Kotapati, J. L. Kuti and C. H. Nightingale, “Clinical Implications of Extended Spectrum b-Lactamase (ESBL) Producing Klebsiella Species and Escherichia coli on Cefepime Effectiveness,” Journal of Infection, Vol. 51, No. 3, 2005, pp. 211-217.
- H. Y. Sun, S. Y. Chen, S. C. Chang, S. C. Pan, C. P. Su, and Y. C. Chen, “Community-Onset Escherichia coli and Klebsiella pneumoniae bacteremia: Influence of Health Care Exposure on Antimicrobial Susceptibility,” Diagnostic Microbiology and Infectious Disease, Vol. 55, No. 2, 2006, pp. 135-141. http://dx.doi.org/10.1016/j.diagmicrobio.2006.01.004
- D. L. Paterson and R. A. Bonomo, “Extended-Spectrum β-Lactamases: A Clinical Update,” Clinical Microbiology Reviews, Vol. 18, No. 4, 2005, pp. 657-686. http://dx.doi.org/10.1128/CMR.18.4.657-686.2005
- M. Gniadkowski, “Evolution and Epidemiology of Extended Spectrum β-Lactamases (ESBLs) and ESBL-Producing Microorganisms,” Clinical Microbiology and Infection, Vol. 7, No. 11, 2001, pp. 597-608. http://dx.doi.org/10.1046/j.1198-743x.2001.00330.x
- S. Nathisuwan, D. S. Burgess and J. S. Lewis, “ExtendedSpectrum Beta-Lactamases: Epidemiology, Detection, and Treatment,” Pharmacotherapy, Vol. 21, No. 8, 2001, pp. 920-928. http://dx.doi.org/10.1592/phco.21.11.920.34529
- M. F. Lin, M. L. Huang and S.H. Lai, “Risk Factors in the Acquisition of Extended-Spectrum Beta-Lactamase Klebsiella pneumoniae: A Case-Control Study in a District Teaching Hospital in Taiwan,” The Journal of Hospital Infection, Vol. 53, No. 1, 2003, pp. 39-45. http://dx.doi.org/10.1053/jhin.2002.1331
- J. Tham, I. Odenholt, M. Walder, A. Brolund, J. Ahl and E. Melander, “Extended-Spectrum Beta-Lactamase-Producing Escherichia coli in Patients with Travellers’ Diarrhoea,” Scandavanian Journal of Infectious Diseases, Vol. 42, No. 4, 2010, pp. 275-280. http://dx.doi.org/10.3109/00365540903493715
- F. Ndugulile, R. Jureen, S. H. W. Urassa and N. Langeland, “Extended Spectrum β-Lactamases among GramNegative Bacteria of Nosocomial Origin from an Intensive Care Unit of a Tertiary Health Facility in Tanzania,” BMC Infectious Diseases, Vol. 5, 2005, p. 86. http://dx.doi.org/10.1186/1471-2334-5-86
- K. Bush, “Extended-Spectrum Beta-Lactamases in North America, 1987-2006,”Clinical Microbiology and Infection, Vol. 14, Suppl. 1, 2008, pp. 134-143. http://dx.doi.org/10.1111/j.1469-0691.2007.01848.x
- O. O. Soge, A. M. Queenan, K. K. Ojo, B. A. Adeniyi and M. C. Roberts, “CTX-M-15 Extended-Spectrum (beta)-Lactamase from Nigerian Klebsiella pneumoniae,” The Journal of Antimicrobial Chemotherapy, Vol. 57, No.1, 2006, pp. 24-30. http://dx.doi.org/10.1093/jac/dki429
- R. A. Afunwa, D. C. Odimegwu, R. I. Iroha and C. O. Esimone, “Antimicrobial Resistance Status and Prevalence Rates of Extended Spectrum Beta-Lactamase Producers Isolated from a Mixed Human Population,” Bosnian Journal of Basic Medical Sciences, Vol. 11, No. 2, 2011, pp. 91-96.
- Clinical and Laboratory Standards Institute (CLSI), “Performance Standards for Antimicrobial Susceptibility Testing,” 17th Information Supplement, 17th Edition, Approved Standard M100-S17, Clinical and Laboratory Standards Institute, Wayne, 2007.
- The European Committee on Antimicrobial Susceptibility Testing—EUCAST. http://www.eucast.org
- E. Lautenbach, J. B. Patel, W. B. Bilker P. H. Edelstein and N. O. Fishman, “Extended-Spectrum β-LactamaseProducing Escherichia coli and Klebsiella pneumoniae: Risk Factors for Infection and Impact of Resistance on Outcomes,” Clinical Infectious Diseases, Vol. 32, No. 8 2001, pp. 1162-1171. http://dx.doi.org/10.1086/319757
- R. Tsay, L. K. Siu, C. Fung and F. Chang, “Characteristics of Bacteremia Between Community-Acquired and Nosocomial Klebsiella pneumoniae Infection Risk Factor for Mortality and the Impact of Capsular Serotypes as a Herald for Community-Acquired Infection,” Archives of Internal Medicine, Vol. 162, No. 9, 2002, pp. 1021-1027. http://dx.doi.org/10.1001/archinte.162.9.1021
- C. Struve and K. A. Krogfelt, “Pathogenic Potential of Environmental Klebsiella pneumoniae Isolates,” Environmental Microbiology, Vol. 6, No. 6, 2004, pp. 584- 590. http://dx.doi.org/10.1111/j.1462-2920.2004.00590.x
- K. Serefhanoglu, H. Turan, F. E. Timurkaynak and H. Arslan, “Bloodstream Infections Caused by ESBL-Producing E. coli and K. pneumoniae: Risk Factors for Multidrug-Resistance,” Brazilian Journal of Infectious Diseases, Vol. 13, No. 6, 2009 pp. 403-407. http://dx.doi.org/10.1590/S1413-86702009000600003
- D. L. Paterson, “Resistance in Gram-Negative Bacteria: Enterobacteriaceae,” American Journal of Medicine, Vol. 119, No. 6, 2006, pp. S20-28.
- Z. Q. Wei, Y. G. Chen, Y. S. Yu, W. X. Lu and L. J. Li. “Nosocomial Spread of Multi-Resistant Klebsiella pneumoniae Containing a Plasmid Encoding Multiple β-Lactamases,” Journal of Medical Microbiology, Vol. 54 No. 9, 2005 pp. 885-888. http://dx.doi.org/10.1099/jmm.0.46151-0

