Open Journal of Medical Microbiology
Vol.2 No.1(2012), Article ID:18192,7 pages DOI:10.4236/ojmm.2012.21001
Blastomyces dermatitidis: Chitinase Homology Model, in Silico Docking, and Inhibition Assay
Department of Biological Science, Idaho State University, Idaho, USA
Email: hillama2@isu.edu
Received January 4, 2012; revised January 20, 2012; accepted February 4, 2012
Keywords: Blastomyces dermatitidis; chitinase; homology modeling; acetazolamide; inhibition
ABSTRACT
Blastomyces dermatitidis is a thermally dimorphic fungus that causes the disease blastomycosis. Currently there are a limited number of effective treatments, many of which have harsh side effects. Chitin, a component of the fungal cell wall is often broken down and recycled for cell wall remodeling and growth. Chitinase is the digestive enzyme capable of chitin hydrolysis. By inhibiting the chitinase we predicted that cells wouldn’t be able to divide and multiply normally, thereby leading to possible anti-fungal treatments. For this study we modeled the structure of B. dermatitidis chitinase, using homology modeling. By predicting a three-dimensional structure we were able to do additional analyses of the active site of the chitinase and predict the binding of a possible small molecule, acetazolamide, in silico. This binding allowed us to predict that this molecule might be capable of inhibiting the chitinase of B. dermatitidis. This inhibition was tested in vivo. No difference in the growth curves of the test and control organisms was observed, however there was a difference within the cell walls of the yeast cells. The cell walls appeared thicker with additional differences in cell wall orderly growth. These changes are consistent with changes that may occur as B. dermatitidis chitinases are inhibited.
1. Introduction
Blastomyces dermatitidis is a fungus that grows in moist, slightly acid soil and is endemic to eastern North America and causes blastomycosis [1-3]. B. dermatitidis is dimorphic and when growing naturally in the environment it grows in the mycelial phase. During this phase it produces spores that can be inhaled by humans or other animals, particularly dogs [4-7]. Within the moist, warm environment of the lung the fungus begins to grow in its yeast form. In some cases, the disease will resolve on its own; however, this depends on the dose and the immune status of the host. The fungus may disseminate and patients may have skin, bone, gastrointestinal tract, and central nervous system involvement [2,4,6-9]. B. dermatitidis is remarkable at resisting the antimicrobial activity of the alveolar macrophages [10] allowing it to progress and spread throughout the body. Without treatment this infection may have a fatal outcome [5,11].
The antifungal drugs Amphotericin B, ketoconazole, itraconazole, fluconazole, or posaconazole may be used to treat this disease [7,11-14]. These drugs inhibit the cell membrane synthesis of ergosterol, but are only partially effective. While humans do not have ergosterol they have a similar cell membrane component, cholesterol, which can lead to severe side effects in some cases [13,15]. Due to the side effects of these drugs and the fact that some fungal organisms are beginning to show resistance [16, 17], there is a push to develop additional drugs that would aid in resolving blastomycosis and other fungal disease. One potential target for drug design is the fungal chitinase.
Chitinases are enzymes involved in the breakdown and turnover of fungal cell walls. The major components of the fungal cell wall are chitin, β-1,3-glucan, and man- noproteins which are highly glycosylated [18]. Chitin provides rigidity and stability to the wall [19]. Chitinase breaks down the glycosidic bonds of chitin. For fungal growth there must be cell wall remodeling, as well as chitin broken down to be placed in the new cell wall. Fungal cell remodeling must take place when yeast cells are budding from the mother cell and if there is damage to the cell wall [20]. Chitinase, of the glycosyl hydrolase family 18, breaks down chitin in the wall, allowing for localized weakening during the budding process, and for additional remodeling [20-23]. Therefore, chitinase is an important element in fungal survival.
Today a large part of science is being done in silico. This is especially true for predicting and testing small molecules that may effect or bind to enzymes. By using a three-dimensional structure of both the ligand and receptor, binding pockets and energies can be generated using a variety of different in silico programs. Structures for most small molecules are readily available. The threedimensional structure of proteins are experimentally determined by crystallography and deposited in the Protein Data Bank (pdb.org). However, currently, the structure a B. dermatitidis chitinase is unknown. Another method of determining a protein structure is by homology modeling. This uses a structure that is known, and homologous, as a template for constructing the three-dimensional structure of the unknown protein [24].
The current study modeled the B. dermatitidis chitinase using homology modeling. This model was then used to predict the binding of acetazolamide. Acetazolamide is a small molecule that is being examined for chitinase inhibition of the fungal organism Aspergillus fumigatus [19]. Because of it being referenced in recent literature, our study was to test acetazolamide’s effectiveness on the fungus B. dermatitidis. Before testing this agent in vivo, we decided to model the docking of acetazolamide on the B. dermatitidis chitinase homology model. This prediction was then tested in vivo on B. dermatitidis.
2. Materials and Methods
2.1. Homology Modeling
Homology modeling was preformed using MODELLER [25]. Homology modeling uses a known three-dimensional protein structure to generate a three-dimensional model of the protein sequence while satisfying spatial restraints. The input protein sequence used was the gene chitinase from B. dermatitidis ATCC found at locus BDDG_05679.1. The three-dimensional protein structure template was found using an Hmmsearch of all experimentally determined three-dimensional files, with the B. dermatitidis chitinase used for the input. Hmmsearch used a hidden Markov model to identify the protein family, and then identified the most homologous sequence from all three-dimensional protein files [26]. The significant hit was 2XVN, a chitinase (CHIA1) from A. fumigatus. The alignment that was created using Hmmer was then used for the modeling.
2.2. Comparative Analysis
To visualize and compare models the graphical interface of Visual Molecular Dynamics (VMD) was used. The MultiSeq function of VMD was used to deduce structural alignments [27]. To visualize the docking interactions Jmol, a 3D chemical structure viewer, was used [28].
2.3. Docking Acetazolamide and Chitin
Chitin and acetazolamide were docked on the predicted three-dimensional structure of B. dermatitidis chitinase using AUTO DOCK TOOLS (ADT). This program predicts how small molecules bind to a three-dimensional structure. For each docking, ADT predicts bond energies for these docking and interactions that likely take place [29]. The chitin oligosaccharide and acetazolamide structural files were obtained using PRODRG web interface [30].
2.4. Inhibition Assay
Acetazolamide was dissolved in NaOH and adjusted to a pH of 8.2 using McIlvaines buffer, forming a 22.3 mM solution of acetazolamide. For the control McIlvaines buffer was also used and adjusted to a pH of 8.2 with NaOH. B. dermatitidis strain B5896 (human isolate from Mountain Iron, Minnesota) were grown as yeast cells on chemically defined medium at 37˚C [31]. Acetazolamide solution or control solution, 5 ml, was added to 25 ml chemically defined media and 0.5 ml B. dermatitidis (OD of 1.328). Absorbance was measured at 600 nm. Cells were also collected and stained with lactol-phenol cotton blue. These were observed under light microscopy 100X oil immersion phase contrast.
3. Results
3.1. Modeling B. dermatitidis Chitinase
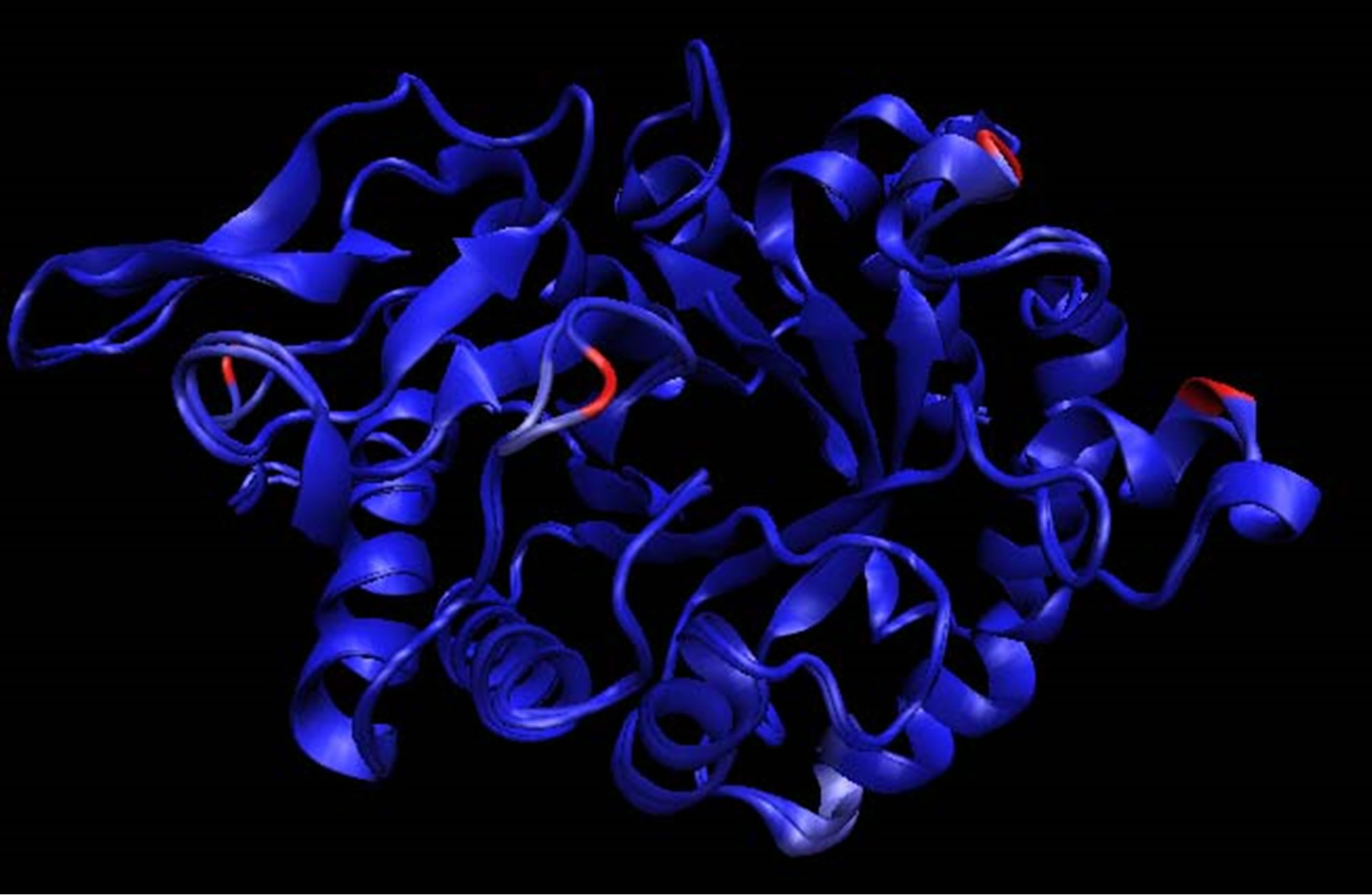
The homology model of B. dermatitidis ATCC glycosyl hydrolase family 18 chitinase is shown in Figure 1. It is aligned with the structure of CHIA1 of A. fumigatus, the homologous protein that was used as a template. The e-value for this alignment was 2.1e−119, reported by Hmmsearch. The alignment of the two genes that were used is shown in Figure 1(c). The B. dermatitidis chitinase had a 59.6% amino acid identity and 74.3% amino acid similarity to A. fumigatus CHIA1. The homology model generated had a ga341 score of 1, as reported by Modeller.
3.2. Docking of Chitin on the Model
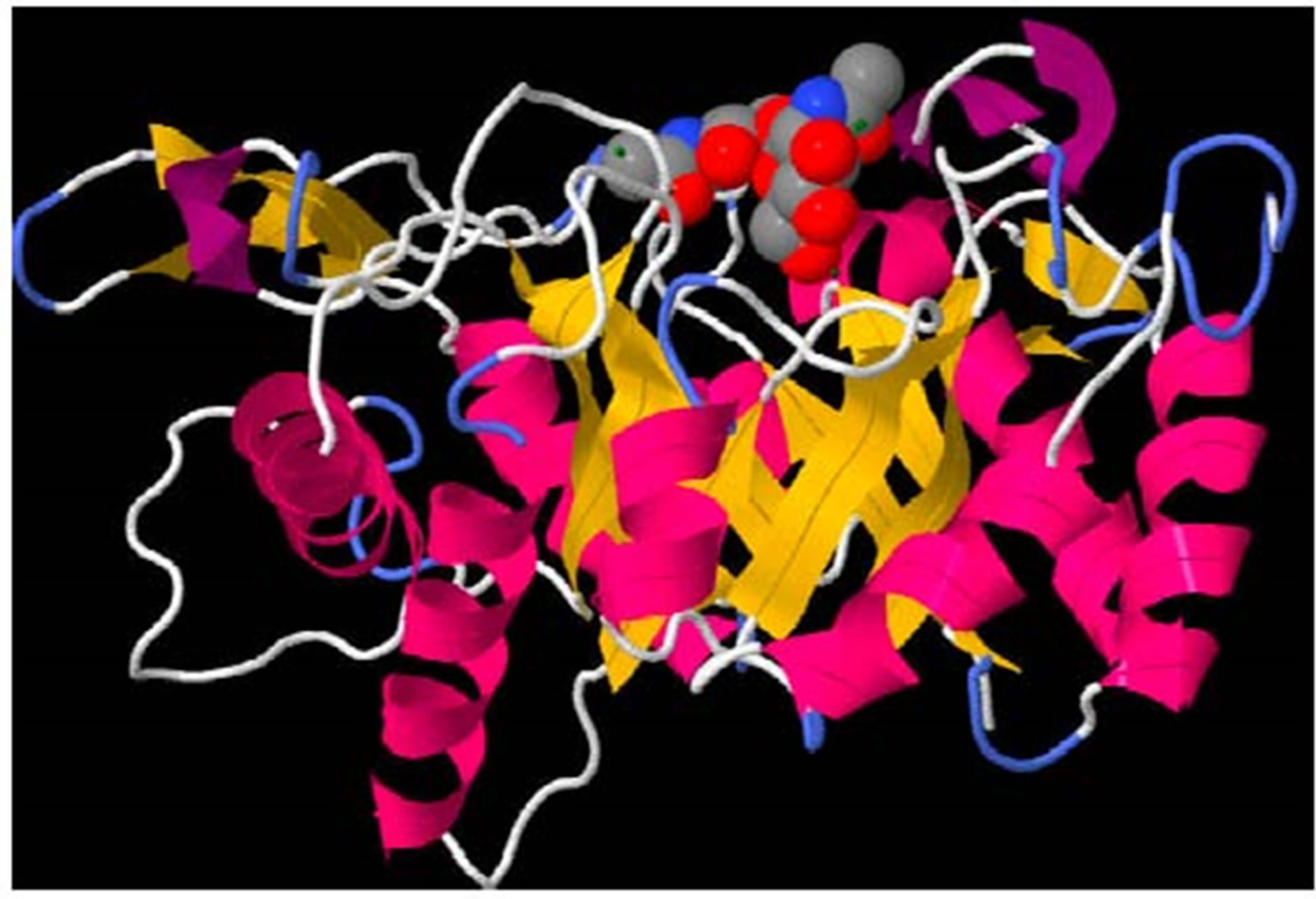
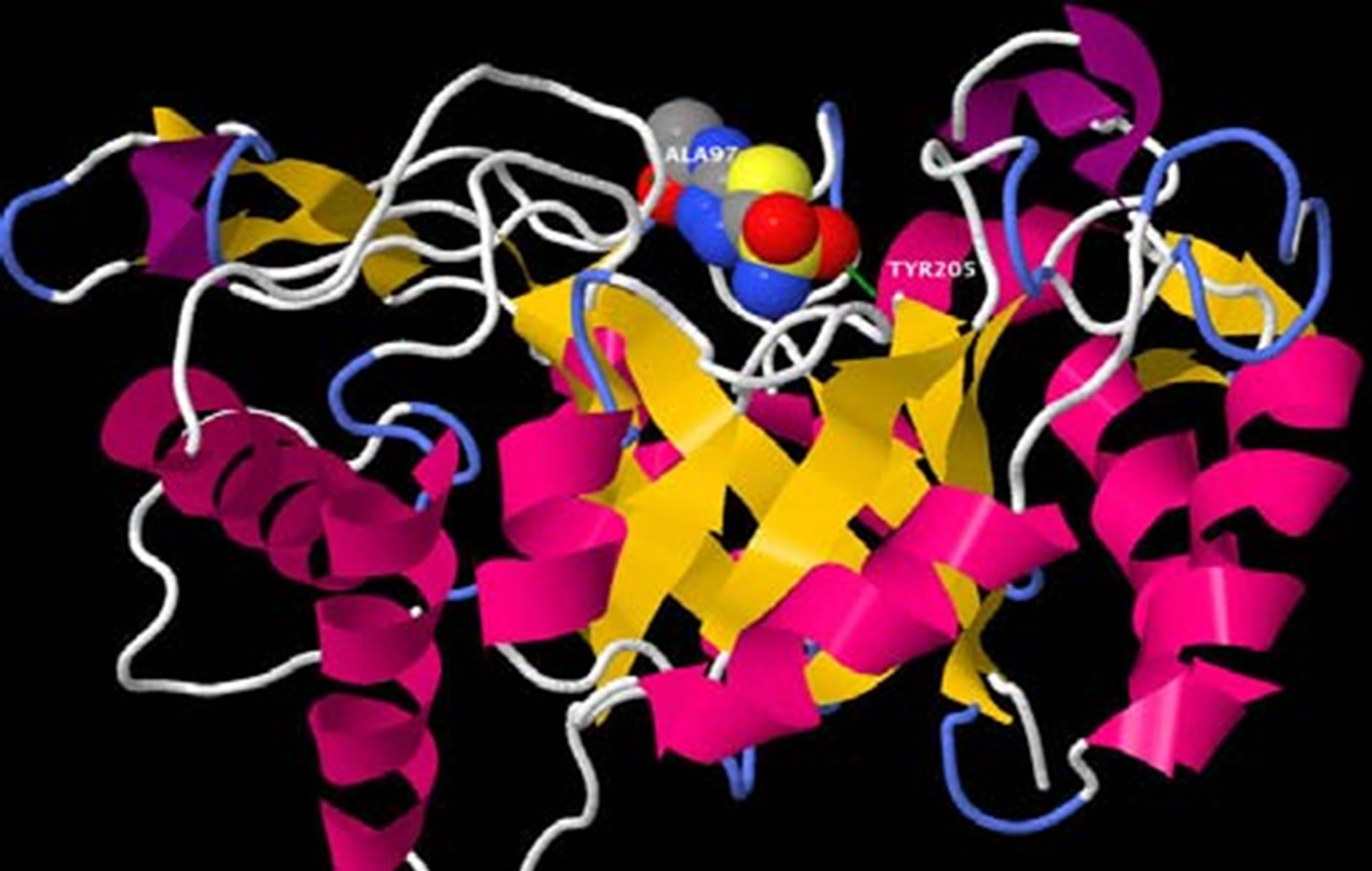
An oligosaccharide of chitin was docked with the B. dermatitidis chitinase model to demonstrate the active site of the enzyme. This docking is shown in Figure 2. The docking energies calculated for the 10 best binding sites of chitin are shown in Table 1. Once the chitin oligosaccharide was docked we observed the amino acids that interacted with the compound. These amino acids were Ala97, Thr98, Tyr205, Asn206, and Trp288.
3.3. Docking of Acetazolamide on the Model
Acetazolamide was docked on the B. dermatitidis model to examine possible binding and location. This docking is shown in Figure 3. The acetazolamide appeared to in-
 (a)
(a)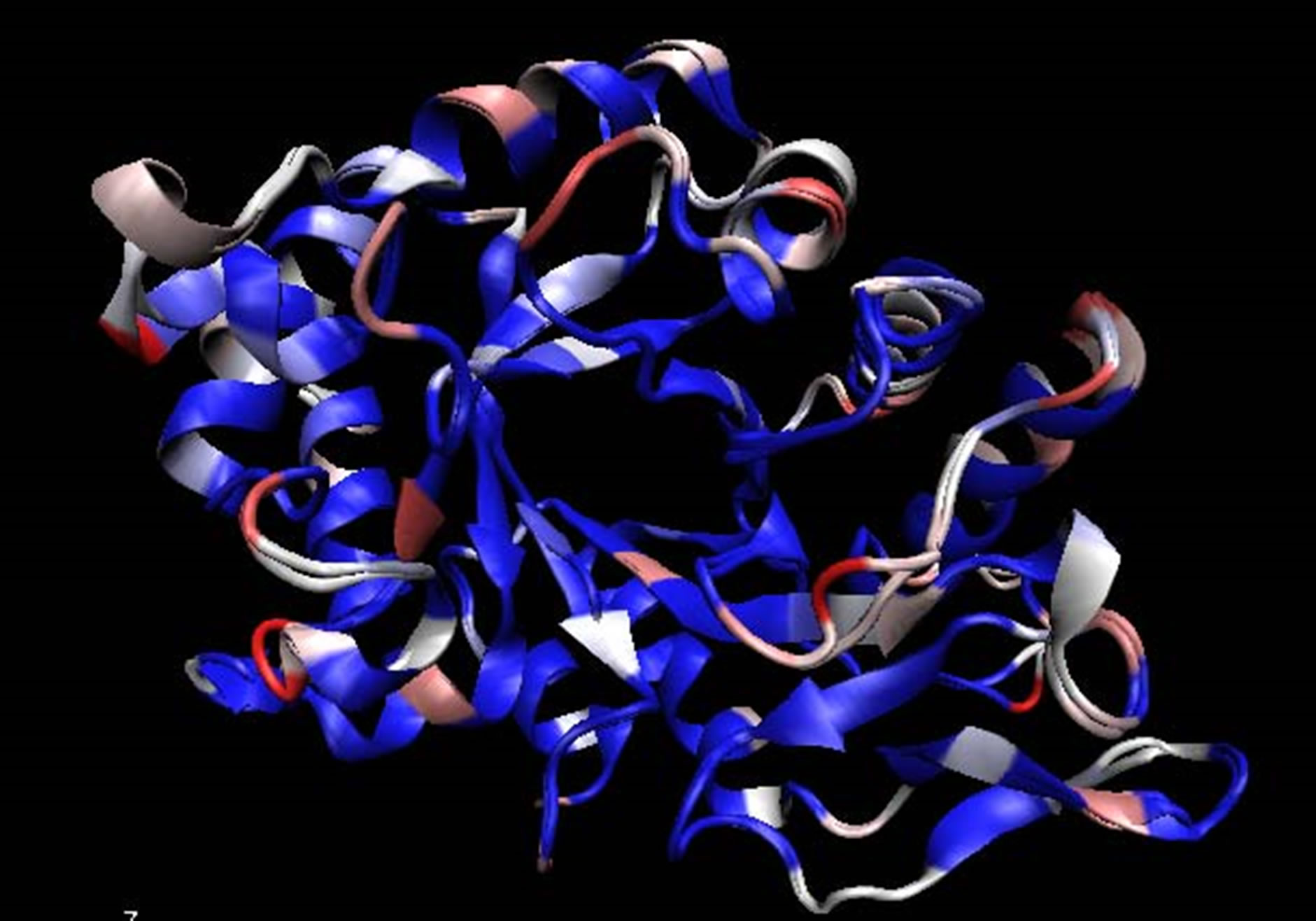 (b)
(b) (c)
(c)
Figure 1. (a) Aligned structure of A. fumigatus CHIA1 with B. dermatitidis chitinase. Identical amino acids in blue, amino acids that are similar in white, and amino acids that are different are shown in pink to red based on their dissimilarity, in accordance with the Blosum 60; (b) Aligned structures of A. fumigatus CHIA1 with B. dermatitidis chitinase. Identical folds are shown in blue while areas that fold differently are shown in red. The blue areas represent molecules that are structure conserved. If there is no correspondence in structural proximity at points, they appear red. These images were made with VMD; (c) Amino Acid alignment generated by Hmmsearch of B. dermatitidis chitinase with CHIA1 of A. fumigatus. The top sequence is the B. dermatitidis chitinase and the bottom sequence is the A. fumigatus CHIA1 chitinase. The line of amino acids shown between the two is the hidden Markov model of the glycosyl hydrolase protein family 18, to which these chitinases belong. The amino acids circled correspond with the active site of both enzymes. Area 1 represents an alanine, A, that is in the active site of both enzymes; this alanine corresponds to Ala97 from B. dermatitidis and Ala124 from A. fumigatus. Following the Ala from area 1 there is a threonine, T, in the B. dermatitidis sequence and a tyrosine, Y, in the A. fumigatus sequence; these correspond to Thr98 and Tyr125. Area 2 highlights a tyrosine followed by an asparagine for both sequences; these are Tyr232 and Asn233 for A. fumigatus while for B. dermatitidis they are Tyr205 and Asn206. Area 3 highlights a tryptophan, W, in both sequences; this is Trp288 for B. dermatitidis and Trp312 for A. fumigatus.
teract with Ala97 and Tyr205. Some interaction was also predicted with Thr98, and Trp288, pictures of these less favorable conformations are not shown. The docking energies calculated for the 10 best binding sites of acetazolamide are shown in Table 1.
3.4. Inhibition Assay of Acetazolamide
The growth curve of B. dermatitidis yeast cells in the presence of acetazolamide showed no statistically significant difference from that of the control, data not shown. However, there was a difference in the morphology of
 (a)
(a)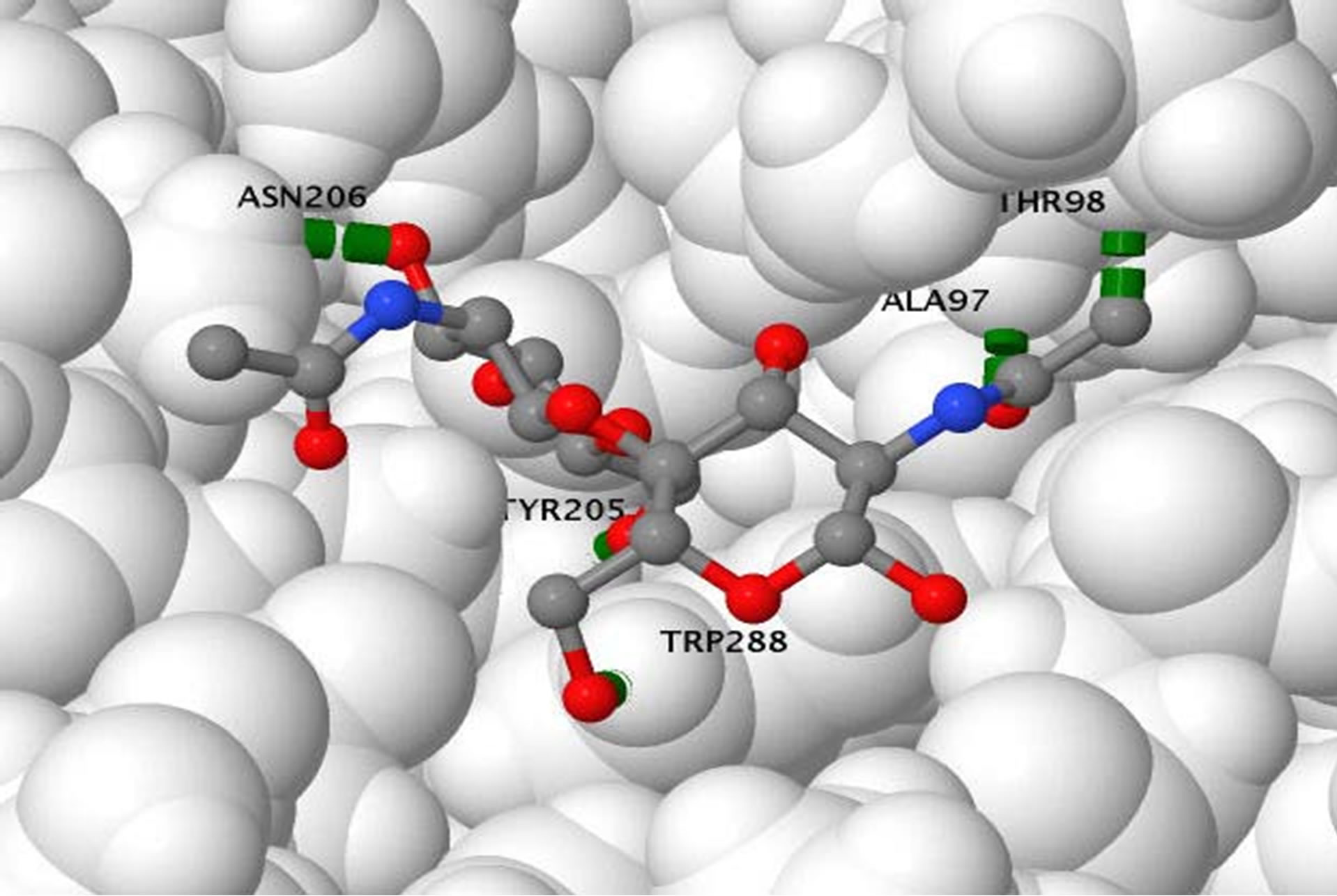 (b)
(b)
Figure 2. (a) Oligosaccharide of chitin docked in opening of the beta-barrel of the B. dermatitidis chitinase model. The oligosaccharide of chitin is shown in spacefill and colored according to atoms; with oxygen in red, cabon in gray, and nitrogen in blue. The green lines shown are illustrating a few of the hydrogen bonds present. The protein is colored according to the type of secondary structure. Alpha helixes are shown in pink ribbon, beta-sheets are shown in yellow ribbon, and loop regions are shown in white and blue; (b) Oligosaccharide of chitin docked in opening of the beta-barrel of the B. dermatitidis chitinase model. This picture is rotated showing the image looking down on the molecule with respect to the previous picture. The protein is displayed in spacefill, showing the opening the chitin oligosaccharide fits in. The chitin oligosaccharide is shown in stick and ball structure to allow visual of the five hydrogen bonds formed. Hydrogen bonds are shown in green and the amino acid they form with is labeled in black.
 (a)
(a) (b)
(b)
Figure 3. (a) Acetazolamide docked in opening of the beta-barrel of the B. dermatitidis chitinase model. The oligosaccharide of chitin is shown in spacefill and colored according to atoms; with oxygen in red, cabon in gray, and nitrogen in blue. The green lines shown are illustrating a few of the hydrogen bonds present. The protein is colored according to the type of secondary structure. Alpha helixes are shown in pink ribbon, beta-sheets are shown in yellow ribbon, and loop regions are shown in white and blue; (b) Acetazolamide docked in opening of the beta-barrel of the B. dermatitidis chitinase model. This picture is rotated showing the image looking down on the molecule with respect to the previous picture (showing the same prespective as in Figure 2(b). The protein is displayed in spacefill, showing where acetazolamide binds. Acetazolamide is shown in stick and ball structure to allow visual of two hydrogen bonds formed. Hydrogen bonds are shown in green and the amino acid they form with is labeled in black.
the yeast cells. The cells grown with acetazolamide are shown in Figure 4 and the growth of the control cells are shown in Figure 5. There appeared to be a difference in the cell wall. The cells grown with acetazolamide appear to have a thicker cell wall, larger cells with irregular shape, and did not form the characteristic budding cells that are seen in the control culture. As observed microscopically, approximately 75% of the yeast cells treated with acetazolamide displayed the altered characteristics listed above compared to the control culture.
4. Conclusions
To identify the template that was used for modeling an Hmmsearch was used. This allowed us to search for the best alignment amongst three-dimensional structures, but

Table 1. Docking energies on B. dermatitidis. (kJ/mol)
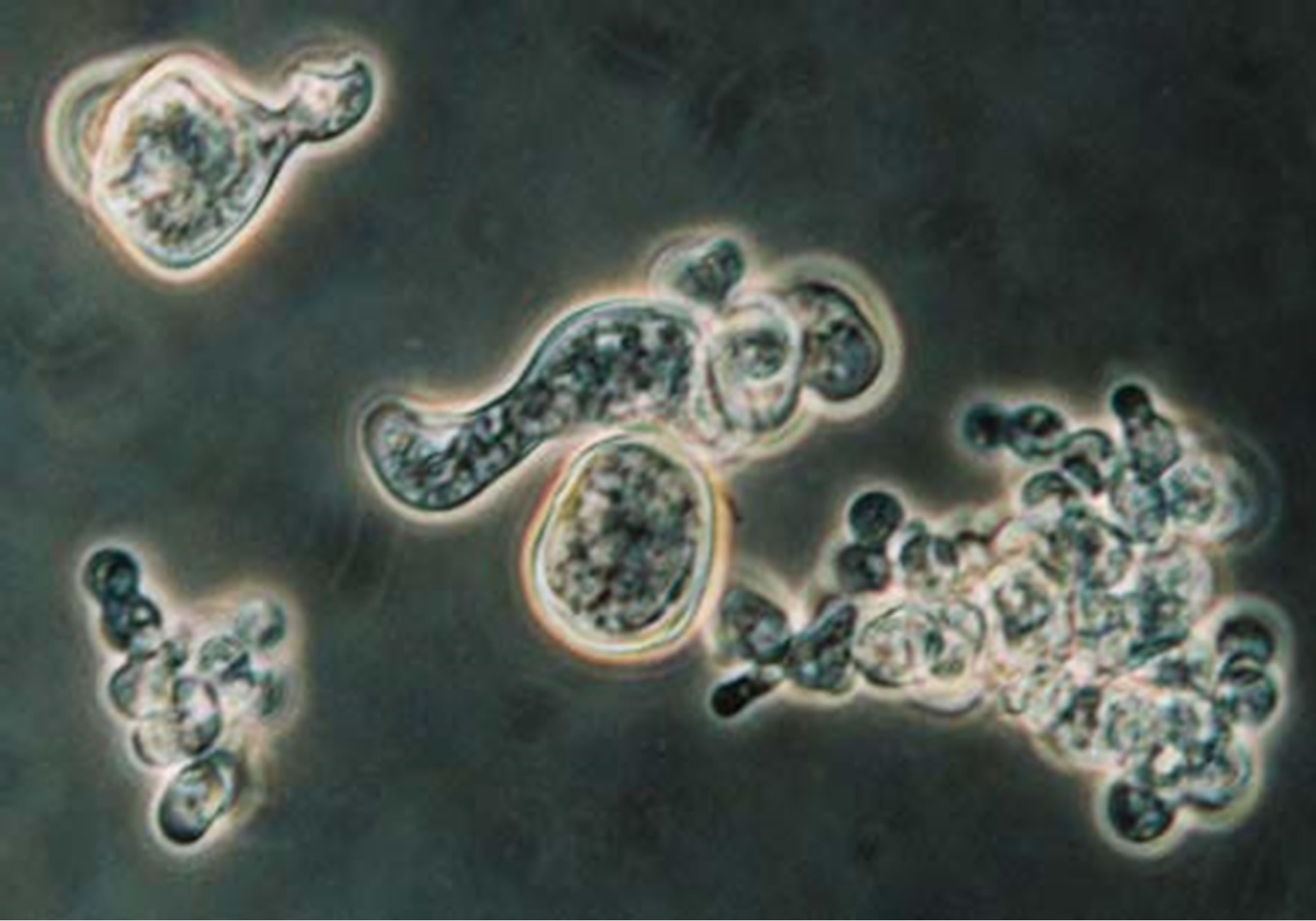
Figure 4. B. dermatitidis cells treated with acetazolamide, collected and stained with lacto phenol blue under 100X oil immersion. Highlighting an elongated budding cell in the middle.
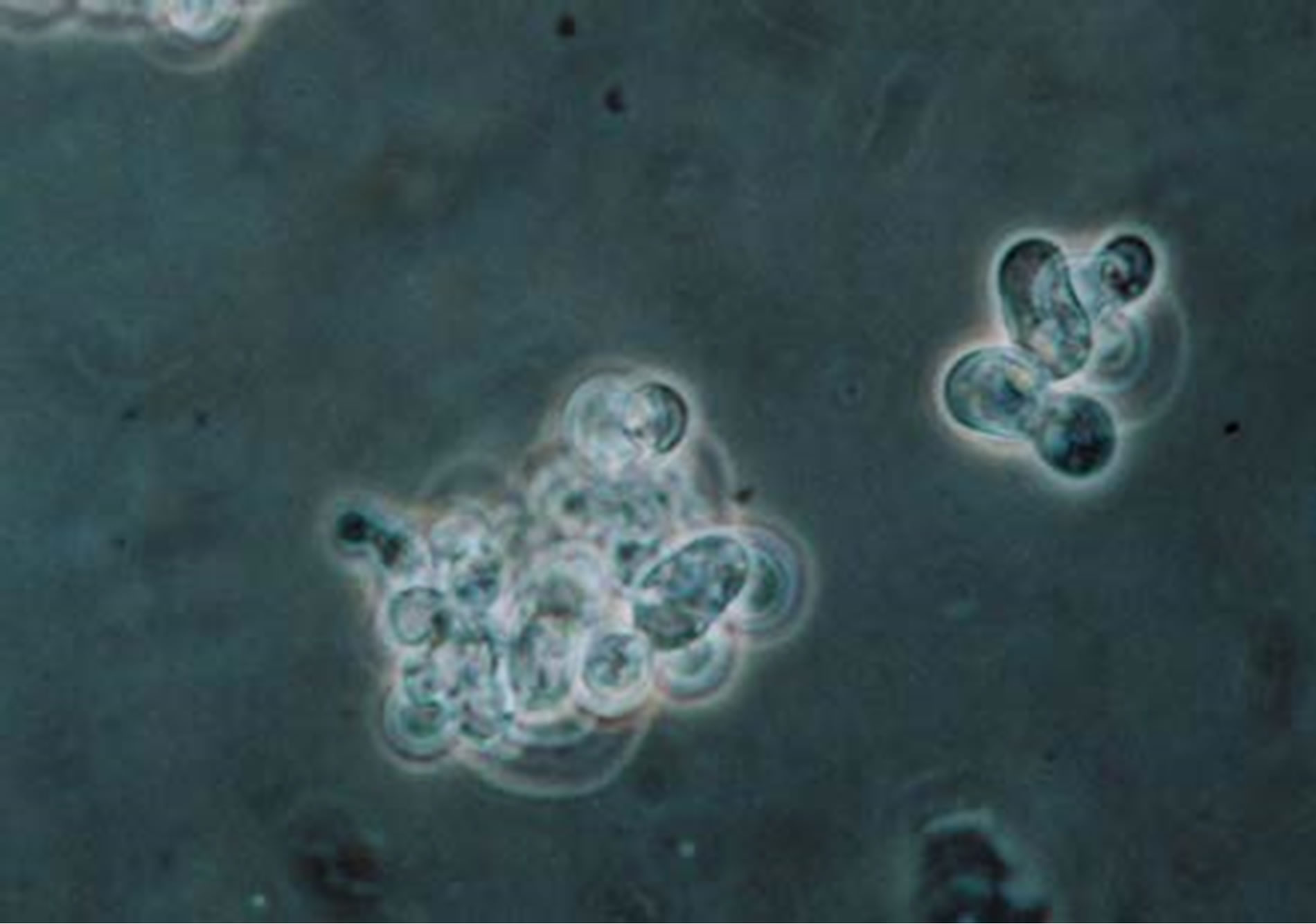
Figure 5. Additional B. dermatitidis cells treated with control buffer, collected and stained with lacto phenol blue under 100X oil immersion. Highlighting a normal budding cell in the right, small cluster.
did so by searching with an underlying hidden Markov model and by protein families [26]. The template found was that of ChIA1 of A. fumigatus. This chitinase is also of the glycosyl hydrolase family 18. The template and the target sequence should have a high degree of similarity and identity when performing the modeling; otherwise the structure may not be predicted accurately [24]. With CHIA1 as a template we achieved 74.3% amino acid similarity and 59.6% amino acid identity. Amino acids are considered similar when the side chains have the same properties. For instance, threonine and tyrosine both have uncharged polar side chains, thus they are considered similar; whereas histidine and valine are not similar because the first is polar and charged while the later is nonpolar. Due to the fact that both the A. fumigatius CHIA1 and the B. dermatitidis chitinase are of the same protein family [19,21,32], the three-dimensional structures should have a global fold that is nearly identical, however, the catalytic activity is governed by specific details and amino acids of the structure [33]. We observed this high degree of identity in Figure 1(b), this figure is colored to show areas that fold differently; as there are only a few red folding areas, thus the model and template do have a global folding pattern. Areas are colored blue by Qres of the Multiseq within VMD, here blue areas are where there is conservation of structural molecules and red areas indicated no correspondence in structural identity [27]. We observed amino acid conservation on the inside of the b-barrel in Figure 1(a), represented by the blue coloring of folding areas compared to the A. fumigatus ChIA1. This should enable the active site of the enzyme to behave in a similar manner and maintain a global folding pattern. The modeling program produced a ga341 score. This score is indicative if the model created is reasonable or if it should be discarded. If the ga341 score is less than 0.6 the model should be discarded, while a score of 1 distinguished a reasonable model [25]. Based on these points the model of B. dermatitidis chitinase provided us with an accurate representation of the three-dimensional structure of the enzyme for further testing.
Within the current literature, there have been reports of acetazolamide and acetazolamide-like molecules being inhibitors of chitinase within other fungal species [19]. Prior to experimenting with its inhibition of B. dermatitidis we performed in silico docking on the chitinase to ensure similar results are theoretically possible. This was done using a suite of MGL tools known as Auto Dock [29]. Using our model of B. dermatitidis chitinase we first tested the binding of a small chitin oligosaccharide for comparison. This oligosaccharide bound in a small groove barely inside the barrel composing the middle of the enzyme, as shown in Figure 2(a). This agreed with literature, which reports the barrel as being the catalytic domain of the enzyme [19,33-35]. The active site of A. fumigatus CHIA1 is reported to contain Ala 124, Tyr125, Tyr232, Asn233, and Trp312 [19]. B. dermatitidis chitinase contained Ala97, Thr98, Tyr205, Asn206, and Trp288. These amino acids correspond with the alignment shown in Figure 1(c). The main difference observed in these active sites is this enzyme is a Thr replaced a Tyr from the A. fumigatus sequence. This difference should not affect the active site greatly due to the similarity of these amino acids; both are uncharged polar side chains containing an OH group. The binding energies associated with the binding conformations are reported in Table 1. Binding energies refer to affinity the protein has toward the small molecule, higher binding energies reflect a lower affinity for the small molecule.
Following this docking, acetazolamide was tested, displayed in Figure 3. The results showed that binding within the same groove as the chitin oligosaccharide. If acetazolamide binds to the same area chitin would bind, acetazolamide may act as a competitive inhibitor. Thus, when acetazolamide is bound in the active site of the enzyme chitin would be blocked from binding, inhibiting the enzyme. We observed some of the same amino acid side chains interacting with the actetazolamide. These amino acids were Tyr205, Thr98, and Trp288. When comparing the binding energies of the chitin oligosaccharide to acetazolamide we observed the acetazolamide docking energies were higher, meaning the docking was not as favorable as the docking of the chitin oligosaccharide. However, we concluded that it is theoretically possible to inhibit the B. dermatitidis chitinase with acetazolamide.
When testing the inhibition of the acetazolamide on B. dermatitidis, we observed no inhibition or statistical difference in the growth curve of the test group verse the control group (data not shown). However, there was a difference observed when examining the B. dermatitidis cells microscopically. The majority of the yeast cells treated with acetazolamide had cell wall alterations. It appeared that the yeast cells were not budding in their normal manner. The way a budding cell should appear is shown in Figure 5, with a broad base bud coming off the mother cell. With the treated cells there also appeared to be a disturbance in wall synthesis or orderly growth and the presence of elongated cells (Figure 4). When comparing the treated cells to the control cells, the treated cell wall was thicker and there may be more than one set of cytoplasm and nuclei within one cell wall. Further more, testing needs to be done to confirm this observation.
These yeast cell growth changes are consistent with our hypothesis that acetazolamide may inhibit B. dermatitidis chitinase. Chitinase is used to weaken the cell wall and remodel for cellular budding and it seems relevant that if this were inhibited the cells would have a difference in shape and growth. It is known that chitinase is required for cell separation and growth in S. cerevisiae [20]; however, B. dermatitidis showed no difference in the rate of growth. One hypothesis about this observation is that acetazolamide is only partially inhibiting the chitinase, still allowing for limited cell separation. To specifically identify this activity a chitinase enzymatic assay would need to be conducted. Nonetheless, our results showed that by adding acetazolamide, cell wall alterations were observed. If this effect was enhanced, it could lead to the death of B. dermatitidis yeast cells. Perhaps when used in a combination therapy with additional antifungal drugs, acetazolamide may produce better antifungal effects.
5. Acknowledgements
This research was supported by the University Research Committee Grant No. S11-6U and the department of Biological Sciences at Idaho State University, Pocatello, Idaho.
REFERENCES
- D. J. Baumgardner and D. P. Paretsky, “The in vitro isolation of Blastomyces dermatitidis from a woodpile in north central Wisconsin,” Medical Mycology, Vol. 37, No. 3, 1999, pp. 163-168. doi:10.1080/j.1365-280X.1999.00214.x
- A. F. DiSalvo, “Topley and Wilson’s Microbiology and Microbial Infections,” 9th Edition, Arnold Publishers, London, 1998.
- B. S. Klein, J. M. Vergeront, R. J. Weeks, U. N. Kumar, G. Mathai, B. Varkey, L. Kaufman, R. W. Bradsher, J. F. Stoebig and J. P. Davis, “Isolation of Blastomyces dermatitidis in soil associated with a large out-break of blastomycosis in Wisconsin,” New England Journal of Medicine, Vol. 314, No. 9, 1986, pp. 529-534. doi:10.1056/NEJM198602273140901
- D. J. Baumgardner and D. P. Paretsky, “Identification of Blastomyces dermatitidis in the stool of a dog with acute pulmonary blastomycosis,” Journal of Medical and Veterinary Mycology, Vol. 35, No. 6, 1997, pp. 419-421. doi:10.1080/02681219780001521
- R. W. Bradsher, “Clinical features of blastomycosis,” Seminars in Respiratory Infections, Vol. 12, No. 3, 1997, pp. 229-234.
- J. F. Shurley and G. M. Scalarone, “Isoelectric focusing and ELISA evaluation of a Blastomyces dermatitidis human isolate,” Mycopathologia, Vol. 164, No. 2, 2007, pp. 73-76. doi:10.1007/s11046-007-9033-8
- M. A. Pfaller and D. J. Diekema, “Epidemiology of invasive mycoses in North America,” Critical Reviews in Microbiology, Vol. 36, No. 1, 2010, pp. 1-53. doi:10.3109/10408410903241444
- M. C. Dobre, W. R. K. Smoker and P. Kirby, “A case of solitary Blastomyces dermatitidis meningitis,” Clinical Neurology and Neurosurgery, Vol. 113, No. 8, 2011, pp. 665-667. doi:10.1016/j.clineuro.2011.03.012
- B .S. Klein, R. A. Squires, J. K. Lloyd, D. R. Ruge and A. M. Legendre, “Canine antibody response to Blastomyces dermatitidis WI-1 antigen,” American Journal of Veterinary Research, Vol. 61, No. 5, 2000, pp. 554-558. doi:10.2460/ajvr.2000.61.554
- N. M. Rocco, J. C. Carmen and B. S. Klein, “Blas-tomyces dermatitidis yeast cells inhibit nitric oxide production by alveolar macrophage inducible nitric oxide synthase,” Infection and Immunity, Vol. 79, No. 6, 2011, pp. 2385-2395. doi:10.1128/IAI.01249-10
- J. E. Cutler, G. S. Deepe Jr and B. S. Klein, “Advances in combating fungal diseases: vaccines on the threshold,” Nature Reviews Microbiology, Vol. 5, No. 1, 2007, pp. 13- 28. doi:10.1038/nrmicro1537
- J. A. McKinnell and P. G. Pappas, “Blastomycosis: New insights into diagnosis, prevention, and treatment,” Clinics in Chest Medicine, Vol. 30, No. 2, 2009, pp. 227-239. doi:10.1016/j.ccm.2009.02.003
- National Center for Biotechnology Information, “Blastomycosis,” US National Library of Medicine, 2010, http://www.ncbi.nlm.nih.gov/pubmedhealth/PMH0001163/
- K. E. Brick and W. A. Agger, “Successful treatment of brainstem blastomycosis with fluconazole,” Clinical Medicine & Research, Vol. 9, 2011, pp. 1-10.
- H. V. Bossche, “Biochemical targets for antifungal azole derivatives: hypothesis on the mode of action,” Current Topics in Medical Mycology, Vol. 1, 1985, pp. 313-351. doi:10.1007/978-1-4613-9547-8_12
- J. W. M. van der Linden, R. R. Jansen, D. Bresters, C. E. Visser, S. E. Geerlings, E. J. Kuijper, W. J. G. Melchers and P. E. Verweij, “Azole-resistant central nervous system aspergillosis,” Clinical Infectious Diseases, Vol. 48, No. 8, 2009, pp. 1111-1113. doi:10.1086/597465
- E. Snelders, H. A. van der Lee, J. Kuipers, A. J. Riji, J. Varga, R. A. Samson, E. Mellado, A. R. Donders, W. J. Melchers and P. E. Verweij, “Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism,” PLoS Medicine, Vol. 5, No. 11, 2008, p. 219. doi:10.1371/journal.pmed.0050219
- G. W. Hudler, “Magical Mushrooms, Mischievous Molds,” Princeton University Press, Princeton, 1998.
- A. W. Schuettelkopf, L. Gros, D. E Blair, J. A. Frearson, D. M. F. van Aalten and I. Gilbert, “Acetazolamide-based fungal chitinase inhibitors,” Bioorganic & Medicinal Chemistry, Vol. 18, No. 23, 2010, pp. 8334-8340. doi:10.1016/j.bmc.2010.09.062
- M. J. Kuranda and P. W. Robbins, “Chitinase is required for cell separation during growth of Saccharomyces cerevisiae,” Journal of Biological Chemistry, Vol. 266, No. 29, 1991, pp. 19758-19767.
- B. L. Cantarel, P. M. Coutinho, C. Rancurel, T. Bernard, V. Lombard and B. Henrissat, “The carbohydrate-active enzymes database (crzy): an expert resource for glycogenomics,” nucleic acids research, Vol. 37, No. 1, 2009, pp. 233-238. doi:10.1093/nar/gkn663
- T. Roemer, L. G. Vallier and M. Snyder, “Selection of polarized growth sites in yeast,” Trends in Cell Biology, Vol. 6, No. 11, 1996, pp. 434-441. doi:10.1016/S0962-8924(96)10039-8
- L. Ni and M. Snyder, “A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae,” molecular biology of the cell, Vol. 12, 2001, pp. 2147-2170.
- J. Xiong, “Essential Bioinformatics,” Cambridge University Press, Cambridge, 2006
- A. Šali and T. L. Blundell, “Comparative protein modeling by satisfaction of spatial restraints,” Journal of Molecular Biology, Vol. 234, No. 3, 1993, pp. 779-815. doi:10.1006/jmbi.1993.1626
- Howard Hughs Medical Institute, “Hmmer: Biosequence analysis using profile hidden Markov models,” 2011. http://hmmer.janelia.org/
- Bechman Institute for Advance Science and Technology, “Theoretical and Computational Biophysics Group: VMD,” 2011. http://www.ks.uiuc.edu/Research/vmd/
- Jmol, “Jmol: an open-source Java viewer for chemical structures in 3D,” 2011. http://www.jmol.org/
- G. M. Morris, D. S. Goodsell, R. S. Halliday, R. Huey, W. E. Hart, R. K. Belew and A. J. Olson, “Automated Docking Using a Lamarckian Genetic Algorithm and Empirical Binding Free Energy Function,” Journal of Computational Chemistry, Vol. 19, No. 14, 1998, pp. 1639-1662. dio:10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B
- A. W. Schüttelkopf and D. M. F. van Aalten, “PRODRG: a tool for high-throughput crystallography of proteinligand complexes,” Acta Crystallographica, Vol. 60, Pt. 8, 2004, pp. 1355-1363.
- S. M. Johnson, and G. M. Scalarone, “Preparation and ELISA evaluation of Blastomyces dermatitidis yeast phase lysate antigens,” Diagnostic Microbiology and Infectious Disease, Vol. 11, No. 2, 1988, pp. 81-86. dio:10.1016/0732-8893(88)90076-4
- Broad Institute, “Blastomyces dermatitidis database,” 2010. http://www.broadinstitute.org/annotation/genome/blastomyces_dermatitidis/MultiHome.html
- G. Davies and B. Henrissat, “Structure and mechanisms of glycosyl hydrolases,” Structure, Vol. 3, No. 9, 1995, pp. 853- 859. dio:10.1016/S0969-2126(01)00220-9
- A. Perrakis, I. Tews, Z. Dauter, A. B. Oppenheim, I. Chet, K. S. Wilson and C. E. Vorgias, “Crystal structure of a bacterial chitinase at 2.3 A resolution,” Structure, Vol. 2, No. 12, 1994, pp. 1169-1180. dio:10.1016/S0969-2126(94)00119-7
- C. Songsiriritthiqul, S. Pantooms, A. H. Aquda, R. C. Robin-son and W. Suginta, “Crystal structures of Vibrio harveyi chitinase a complexed with chitooligo-saccharides: implications for the catalytic mechanism,” Journal of Structural Biology, Vol. 162, No. 3, 2008, pp. 491-499. dio:10.1016/j.jsb.2008.03.008

