CellBio
Vol.2 No.1(2013), Article ID:29387,10 pages DOI:10.4236/cellbio.2013.21001
Expression and Localization of NANOS1 in Spermatogenic Cells during Spermatogenesis in Rat
Section of Functional Morphology, Faculty of Pharmaceutical Sciences, Nagasaki International University, Nagasaki, Japan
Email: syokota@niu.ac.jp
Received December 13, 2012; revised January 15, 2013; accepted January 25, 2013
Keywords: NANOS1; Nuage; Chromatoid Body; Satellite Body; Non-Nuage Compartments; Immunocytochemistry
ABSTRACT
Expression and localization of NANOS1 in the spermatogenic cells of rat testis were studied by immunofluorescence and immunoelectron microscopy. Using immunofluorescence techniques, NANOS1 was localized in the cytoplasm and discrete granules of spermatocytes and spermatids. The staining intensity of NANOS1 signal varied depending upon the stage of the cycle of seminiferous epithelium. Double immunofluorescence staining with antibodies against NANOS1 and DDX4 showed that several DDX4-positive compartments of nuage were also stained for NONOS1. Immunoelectron microscopy revealed that the major subcellular localization sites for NANOS1 were the chromatoid body (CB) and satellite body (SB), and the minor sites were the rest of the nuage compartments, including the irregularly-shaped perinuclear granules (ISPG), and intermitochondrial cement (IMC). Non-nuage structures such as mitochondria-associated granules (MAG), granulated body (GB), and reticulated body (RB) were also labeled by theNANOS1 antibody. In addition, NANOS1 was found in the outer dense fibers of flagella of spermatids at steps 12-19, and in the head cap of late spermatids after step 15. These results suggest that NANOS1 is one of the nuage proteins and functions mainly in the CBs as well as in the cytoplasm. NANOS1 may also have additional functions in non-nuage structures such as MAGs, GBs and RBs.
1. Introduction
The nanos gene has been extensively studied in Drosophila, and has been shown to play multiple roles in the development and maintenance of the germ line [1-3], embryonic patterning [4-7], migration of the primordial germ cells to the embryonic gonad [1,8-10], and inhibition of apoptosis of germ line cells in embryo [11]. There are three homologous genes, NANOS1, NANOS2 and NANOS3, in mouse [10,12]. NANOS1 is not expressed in the primordial germ cells (PGCs) but its expression can be detected in the spermatogenic cells of mature testis. During embryogenesis, the level of maternally expressed NANOS1 is found to be rapidly reduced after fertilization. The expression of NANOS1 can be detected in the central nervous system, and continues to occur in the adult brain [12]. NANOS2 is predominantly expressed in the male germ cells, and the elimination of this gene causes the complete loss of spermatogonia [10]. NANOS2 can substitute NANOS3 during early PGC development but by contrast NANOS3 does not rescue the defects in NANOS2-null mice [13]. In addition, NANOS2 is known to suppress meiosis [14]. In human, NANOS3 is expressed highly in germ cell nuclei where the protein co-localizes with chromosomal DNA during mitosis/meiosis [15]. NANOS3 plays a role in the maintenance of PGC migration to the primary gonads [1,16]. NANOS3-null-mice are characterized by a complete absence of germ cells in both sexes [10]. NANOS3 interacts with PUMILIO2 and delays the cell cycle progression of spermatogonial cells [17].
Human NANOS1 forms a stable complex with PUMILIO2 through interaction of highly conserved domains [18]. PIMILIO2 interacts with the DEAD-box RNA helicase GEMIN3, a microRNA biogenesis factor. GEMIN3 has been shown to co-precipitate with NANOS 1 and PUMILIO2 in transfected mammalian cells. By double immunofluorescence staining, protein complexes composed of NANOS1, PUMILIO2 and GEMIN3 are localized to the cytoplasmic granules which are also stained for DDX4, a marker protein of chromatoid body (CB) [19]. Moreover, PUMILIO2 and NANOS1 interact with SNARE-associated component SNAPIN to form a complex in human spermatogenic cells [20]. Nanos1 forms a complex with PUMILIO2 and functions to regulate the translation of selective mRNAs, specifically via association with the 3’-UTR of its mRNA targets. Thus, the roles of NANOS in mammalian spermatogenic cells are gradually being elucidated. Despite of the colocalization of NANOS1 with DDX4 in the CB, the interaction of the two proteins has not been shown in other nuage compartments such as intermitochondrial cement (IMC) and irregularly-shaped perinuclear granules (ISPG). Moreover, it has been reported that several nuage-resident proteins are localized to the non-nuage structures, including mitochondria-associated granules (MAG), granulated body (GB), reticulated body (RB) and ribosome aggregates (RA) [21,22]. It is unclear whether NANOS1 is localized to the nuage or non-nuage compartments. In the present work, we investigated the subcellular localization of NANOS1 in rat testis using immunofluorescence and immunoelectron microscopy. Our data show that NANOS1 is co-localized with DDX4 in all nuage compartments and non-nuage structures.
2. Materials and Methods
2.1. Animals
Male guinea pigs (320 - 450 g) and Male Wistar rats (180 - 220 g) were obtained from Kyudo Co. (Tosu, Japan) and fed appropriate standard diets and water ad libitum. The animal experiments were performed in accordance with the guidance for animal experiments issued by Nagasaki International University.
2.2. Antibodies
Polypeptide (MEAFPWAPRSPRRARAPAP) consisting of 19 amino acids at the N-terminus of mouse NANOS1 was synthesized and at the C-terminus cysteine was added to facilitate chemical binding to the carrier protein, The polypeptide (1mg) was conjugated with egg albumin (50 mg) by m-maleimidobenzoyl-N-hydrosuccinimide ester. The conjugate was emulsified with Freund’s complete adjuvant. Guinea pigs were intracutaneously injected with 50 µg of peptide. The injections were carried out 4 times with 2-weeks interval, followed by collection of blood samples 2 weeks after the last injection. The specific antibody was affinity-purified by the peptidecoupled column. Rabbit antibody against mouse DDX4 was prepared as previously [23]. Horseradish peroxidase (HRP)-labeled swine anti-guinea pig IgG was obtained from DAKO Japan (Tokyo). Rabbit anti-mitochondrial isocitrate dehydrogenase (ICD1) was described previously [24]. Rabbit anti-histone H2B, rabbit anti-protein disulfide isomerase (PDI), and rabbit anti-tubulin were obtained from Cell Signaling Technology (Boston, MA), Sigma-Aldrich Japan (Tokyo), and Thermo Fisher Scientific (Fremont, CA), respectively. Alexa 568® or Alexa 488® conjugated goat anti-rabbit IgG or goat anti-rat IgG were obtained from Molecular Probes (Eugene, OR). HRP-labeled goat antibody to rabbit IgG was purchased from DAKO Japan (Tokyo) and Invitrogen Japan, respectively. Protein A-gold probe (15 nm gold) was prepared by the method of de Roe et al. [25].
2.3. Biochemical Analysis
2.3.1. Western Blotting
Rat testes were homogenized in 5 mM MOPS-KOH buffer (pH 7.4) containing 0.25 M sucrose, 1 mM ethylenediaminetetraacetic acid, 1 mM phenylmethylsulfonyl fluoride and a cocktail of protease inhibitors (1 µg/ml), including leupeptin, pepstatin, aprotinin, and antipain (medium A), using a Potter-Elvehjem homogenizer. Ten percent homogenate (w/v) was centrifuged at 800 × g for 10 min. The resulting supernatant was centrifuged at 10,000 × g for 20 min and the pellet (mitochondrial fraction) was suspended in a small volume of medium A. The supernatant was centrifuged at 100,000 × g for 60 min in a Beckman ultracentrifuge using a SW 41 swing rotor. The resulting pellets were suspended in medium A and used as the microsomal fraction, while the supernatant was used as the cytosol fraction. The cell fractions were stored at −70˚C. Protein concentrations were determined by the bicinchoninic acid method (Pierce Chemical, Rockford, IL) using bovine serum albumin (BSA) as a standard. The protein concentrations of the fractions were adjusted to 1 mg/ml, mixed with one volume of sample buffer for SDS-PAGE, and heated in boiling water for 5 min. Ten micrograms of each sample were analyzed by western blotting. The molecular mass of NANOS1 was estimated by prestained protein maker (Nippon Genetics Europe, Dueren, Germany).
2.3.2. Dot Blot Analysis of NANOS1 Content in Cell Fractions of Mouse Testis
The mitochondrial, microsomal, and cytosolic cell fractions were isolated from rat testes homogenate by differential centrifugation as described above. Nuclei were isolated by the method of Rickwood and Ford [26].
Briefly, the crude nuclear fraction (800 g pellet) was suspended in medium A containing 1% Triton X-100 and centrifuged at 800 g for 10 min. The resulting pellet was suspended in 2 ml of medium A, mixed with 72.5% (wt/vol) metrizamide (Sigma-Aldrich) in medium A without 0.25 M sucrose and centrifuged at 10,000 g for 20 min at 5˚C The nuclei-rich pellicles at the surface of the metrizamide solution were collected, suspended in a 10-times volume of medium A, and then centrifuged at 6000 g for 10 min. The pellet was suspended in 1.6 ml of medium A. Each fraction was diluted by medium A. The diluted fractions (90 μl) were mixed with 90 µl of SDS-PAGE sample buffer and 20 μl of 0.3 M iodoacetamide, treated in boiling water for 5 min, and centrifuged at 10,000 g for 10 min. The supernatant fractions were diluted 1000-fold and 1 - 5 µl was loaded onto polyvinylidene difluoride (PVDF) membranes for immunoblotting. NANOS1 was visualized by combination of HRP-labeled swine anti-guinea pig IgG and 3,3’-diaminobenzidine (DAB) reaction. Internal standard proteins, histone H2A (H2A, nuclei), mitochondrial isocitrate dehydrogenase (ICD1, mitochondria), protein disulfide isomerase (PDI, microsomes), and α-tubulin (α-Tub, cytosol) were visualized using a combination of rabbit antibodies against each protein, HRP-labeled goat antirabbit IgG, and DAB reaction. The staining intensity was measured by densitometer. The total amount of NANOS1 in each cell fraction was calculated as follows: the densitometric values obtained were multiplied by the final volume of each fraction. The data were obtained from three measurements and the average values and stand ard deviations were plotted.
2.4. Immunocytochemical Analysis
2.4.1. Immunofluorescence Staining of Testis Tissue Sections
Rat testes were embedded in Tissue-Tek (SAKURA Fintec, Tokyo, Japan) and frozen in isopentane cooled by liquid nitrogen. Frozen sections (6 μm thickness) were cut with a Leitz cryostat (Leica Instruments, Nussloch, Germany) and picked up on silicone-coated glass slides. Sections were fixed in a fixative consisting of 4% paraformaldehyde, 0.01% CaCl2, and 0.1 M HEPES-KOH buffer (pH 7.4) for 15 min at room temperature (RT). Sections were treated with 0.2% Triton X-100 and 0.2% Saponin for 15 min and then incubated in blocking solution containing 2% fish gelatin and 15 mM NaN3 in PBS for 30 min. The sections were subsequently incubated with guinea pig anti-NANOS1 antibody (1 μg/ml) overnight at 4˚C. After washing in PBS, they were incubated with Alexa 488 or 568-labeled goat anti-guinea pig IgG solutions containing 3 μM DAPI (Hoechst, Tokyo) for 60 min at RT. For the immunofluorescence control, preimmune guinea pig sera were used for the primary reaction, followed by Alexa 488-labeled or 568-labeled secondary antibodies. For double staining, sections were incubated separately with mixtures of rabbit anti-DDX4/guinea pig anti-NANOS1, followed by incubation with Alexa 488- or 568-labeled secondary antibodies. The sections were examined with a Nikon Eclipse E600 fluorescence microscope (Nikon, Tokyo).
The images were merged using Adobe Photoshop 7.0 to visualize cell contours. The stage of seminiferous tubules was judged from the size and shape of spermatocytes and spermatids stained by DAPI with reference to the stages of the cycle illustrated by Russell and coworkers [27].
2.4.2. Immunoelectron Microscopic Staining of Rat Testis
Rat testes were dissected out, cut into small tissue blocks, and fixed in 4% paraformaldehyde + 0.2% glutaraldehyde in HEPES-KOH buffer (pH 7.4) for 1h at 4˚C. The tissue blocks were dehydrated in graded ethanol series at −20˚C and embedded in LR White, which was polymerized under UV light at −20˚C. Thin sections of LR White-embedded testis tissues were cut with a diamond knife equipped with a Reichert Ultracut R, mounted on nickel grids, and incubated with affinity purified guinea pig anti-NANOS1 (1 µg/ml) overnight at 4˚C. Preimmune guinea pig sera were used instead of the primary specific antibody for the control experiments. Sections were contrasted and then examined with a Hitachi H7650 electron microscope (Hitachi, Tokyo, Japan) at acceleration voltage of 80 kV. The stage of seminiferous tubules and step of spermatids were judged according to stages of the cycle illustrated by Russell et al. [27].
3. Results
3.1. Western Blotting
Our peptide antibody reacted with two protein bands with molecular masses of 29 and 40 kDa, respectively. The former band was detected in all cell fractions, whereas the latter was present only in the mitochondrial and cytosolic fractions (Figure 1). Dot blot analysis of NANOS1 contents in cell fractions showed that approximately 61%, 27%, 8%, and 4% of the protein was detected in the cytosolic, mitochondrial, nuclear, and microsomal fractions, respectively. Marker proteins, H2A, ICD1, PDI, and α-Tub, were detected prominently in the nuclear, mitochondrial, microsomal, and cytosol fractions, respectively (Figure 1(b), Dot blot).
3.2. Immunofluorescence Staining of NANOS1 Protein (NANOS1) in Rat Testis
Two staining patterns were noted in spermatogenic cells after immunofluorescence staining of NANOS1, one being diffusive in the cytoplasm and the other with a discrete granular distribution. The former staining pattern was observed in spermatocytes and spermatids at all steps with various staining intensities Figures 2(a), (d) and (g). The latter was seen in early to late spermatids and the size and shape of stained granules varied depending on their differentiation steps. In spermatids at steps 1 to 8, irregularly shaped granules were strongly stained, which were also positive for DDX4, a marker for chromatoid bodies (CB) (Figures 2(a)-(c)). Small granules located in the cytoplasmic lobe of spermatids at steps 10-13 were also stained for NANOS1 but not for DDX4 (Figures 2(d)-(i)). In pachytene spermatocytes, irregularly-shaped perinuclear granules (ISPG) were stained weakly for NANOS1 but strongly for DDX4 (Figures 2(d)-(i)). Frequently, the granules stained only either for NANOS1 or DDX4 were noted (Figures 2(d)-(i)). The nuclei of spermatocytes were almost negative for NANOS1 but those of elongated spermatids at steps 9-10 were slightly
 (a)
(a) (b)
(b)
Figure 1. (a) Western blotting analysis of cell fractions of rat testis. Lane 1: molecular weight marker, lane 2: homogenate, lane 3: nuclear fraction, lane 4: mitochondrial fraction, lane 5: microsomal fraction, lane 6: cytosol fraction. A single band with a molecular mass of 29 kDa is seen (arrows); (b) Dot blotting analysis of cell fractions isolated from rat testis homogenate. N: nuclear fraction, Mt: mitochondrial fraction, Mc: microsomal fraction, Cy: cytosol fraction. Dot blot shows marker protein of each cell fraction. H2A: histone H2A, ICD1: mitochondrial isocitrate dehydrogenase, PDI: protein disulfide isomerase, α-Tub: α- tubulin.
stained (Figure 2(d)). Areas surrounding clusters of elongated spermatid heads were stained for NANOS1 (Figure 2(a)). Flagella of spermatids at steps 12-19 were weakly stained (Figure 2(g)). At higher magnification, staining of CB for NANOS1 and DDX4 was almost completely overlapped in early spermatid (Figure 3(a)) but partly stuck out each other in spermatids at steps 5 and 6 (Figure 3(b)). ISPG in pachytene spermatocytes at stage IX-XII were stained partially for NANOS1 and DDX4 (Figure 3(c)). All the staining for NANOS1 and DDX4 was not observed in the control sections incubated in preimmune sera, followed by the secondary antibodies conjugated with Alexa 568® or Alexa 488® (not shown).
3.3. Immunoelectron Microscopic Localization of NANOS1 in Nuage Compartments
NANOS1 signals detected in all nuage compartments as classified by Russell and Frank [28], including 70 - 90 nm particles, cluster of 60 - 90 nm particles, intermitochondrial cement (IMC), chromatoid body (CB) and satellite body (SB) with various labeling intensities. The
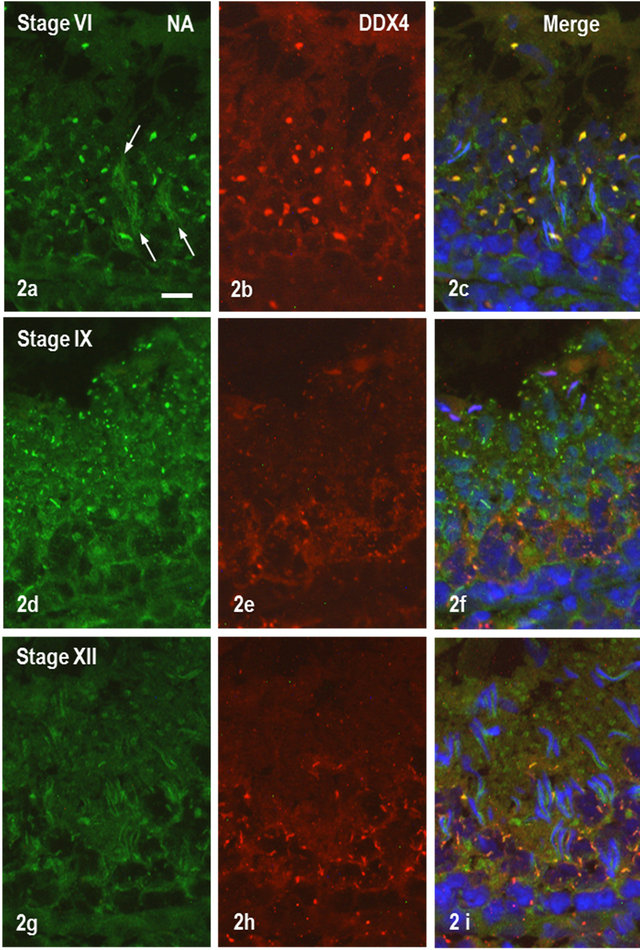
Figure 2. IF staining of rat testis. (a)-(c). Seminiferous tubule at stage IV. (a) Staining for NANOS1; (b) Staining for DDX4; (c) Merged (a) and (b) and staining with DAPI for nuclei. Relatively large granules are stained for both NANOS1 and DDX4 but staining around clusters of spermatid heads is for solely for NANOS1. Bar = 50 μm; (d)-(f) Seminiferous tubule at stage IX. (d) Staining for NANOS1; (e) DDX staining; (f) Merged (d) and (e) and stained with DAPI. Note that NANOS1-stained granules in step 10 spermatids are almost negative for DDX4 staining, whereas DDX4-positive small granules in pachytene spermatocytes are weakly stained for NANOS1. (g)-(i) Tubule at stage XII. (g) Staining for NANOS1; (h) Staining for DDX4; (i) Merged (g) and (h) and stained with DAPI. Granules in step 12 spermatids some in pachytene spermatocytes are stained only for NANOS1, whereas irregularly shaped perinuclear granules are stained for both NANOS1 and DDX4 with diverse intensities.
staining intensity of NANOS1 in 70 - 90 nm particles was very weak, as compared with the staining for DDX4 (data not shown).The clusters of 60 - 90 nm particles corresponding to ISPG identified by immunofluorescence staining were intermediately stained for NANOS1 (Figure 4(a)). The labeling intensity for NANOS1 was weaker than that for DDX4 (Figure 4(a), inset). Many of gold-labeled signals were found to be associated with the surface of the dense material of the cluster (Figure 4(a)).

Figure 3. High power view of double IF staining for NANOS1 (green) and DDX4 (red). (a) Granules in step 4 spermatids are stained for both proteins. (a) Stage IV. Large granules of spermatid at step 4 are stained for both proteins (arrows). Many small granules in pachytene spermatocytes (P) are stained for DDX4 and small numbers of granules are stained for NANOS1; (b) Stage VI. Spermatids are shown. Irregularly-shaped granules are stained for both proteins with various staining intensities (arrows); (c) Stage XI. Pachytene spermatocytes are shown. NANOS1 is stained in numerous small granules whereas DDX4 is in ISPG, some of which are positive for NANOS1. Bar = 10 mm.
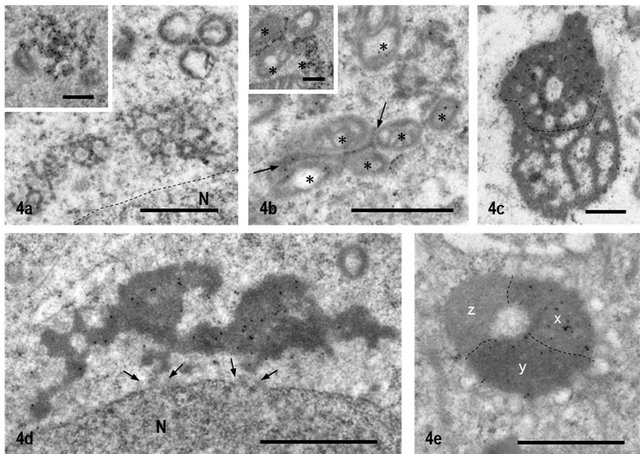
Figure 4. Immunoelectron Microscopic staining for NANOS1 and DDX4. (a) Cluster of 60-90nm particles corresponding to ISPG by IF staining is weakly labeled for NANOS1, whereas the same structure is stained strongly for DDX4 (Inset). Dashed line shows nuclear membrane. N: nucleus; (b) IMC of pachytene spermatocyte at stage IX. It is weakly labeled for NANOS1 (arrows) but very strongly labeled for DDX4 (Inset). *: mitochondria; (c) Typical CB in step 5 spermatid. A half upper dashed line is darker than lower half and labeled stronger than lower part; (d) CB of step 1 spermatid stained for NANOS1. It locates closely to the nucleus. Between CB and nuclear membrane vague particles are recognized (arrows). N: nucleus; (e) Late CB in step 9 spermatid. Three different areas (x, y and z) which are bounded with dashed lines are seen. Labeling intensity is highest in the area x, decreases in the y and no label in the z. Bar = 1 μm for all pictures and 0.5 μm for inset.
NANOS1 labeling in IMC was also low, compared with DDX4 labeling (Figure 4(b)). Frequently, the structure of IMC was connected to the cluster of 60 - 90 nm particles. There were two types of CBs, one having homogeneous matrix and the other being heterogeneous. Both types reacted positively for NANOS1 with various staining intensities. The former type of CB was observed generally in early round spermatids, located closely to the nuclear envelope and seemed to communicate with the nucleus (Figure 4(d)). The latter type of CB was seen in spermatids at steps 5-10 and had the areas stained heterogeneously from negative to positive (Figure 4(c)). This heterogeneous labeling pattern increased as differentiation of spermatids progressed. In the CBs of late elongated spermatids, even the completely unlabeled area appeared (Figure 4(e)). SB appeared in pachytene spermatocytes at stages IX-XI and was relatively heavily labeled for NANOS1 (Figure 5(a)). Gold-labeled signals were associated with the dense cord-like structure. In the meiotic cells, NANOS1 was localized in small particles, which were scattered in the cytoplasm or closely associated with mitochondria (Figure 5(b)). All the gold signals were not observed in the immunoelectron microscopic control sections incubated with preimmune sera, followed by incubation with protein A-gold probe (Figures 6(a)-(e)).
3.4. Immunoelectron Microscopic Localization of NANOS1 in Non-Nuage Structures
Next we focused on non-nuage structures, in (MAG), granulated body (GB), reticulated body (RB), ribosome aggregate (RA), head cap of late spermatids, manchette, and dense fiber of flagella.
MAG was 1 - 2 µm in diameter, composed of a fine granular material, and appeared in the elongated cytop-

Figure 5. (a) SB seen in pachytene spermatocyte at stage IX. Gold particles showing NANOS1 are associated closely with dense reticular network; (b) Dividing spermatocyte. NANOS1 signals are seen in small dense particles which located closely to mitochondria (*) (arrows). Ch: chromosome. Bar = 0.5 mm.
Figure 6. Nuage structures in immunoelectron microscopic control sections incubated with preimmune sera, followed by protein A-gold probe. (a) ISPG; (b) IMC; (c) CB; (d) SB. and (e) MAG appeared in dividing spermatocyte. No gold labeling is observed. Bar = 0.5 μm.
lasmic lobes of steps 9-17 spermatids. Its marked feature was the close association with mitochondria [29]. MAG was relatively strongly labeled for NANOS1 (Figure 7(a)) GB appeared in the cytoplasm lobes of step 14 spermatids, was most abundant in step 17 spermatids, and decreased in number during subsequent stages of sper matogenesis. The dense matrix of GB was labeled for NANOS1 but not less dense particles closely associated with GB (Figure 7(b)). RB first appeared in the cytoplasm of step 14 spermatids and completely disappeared in step 18 spermatids. It was characterized by several dense, anastomosed cords with a width of 80 - 100 nm. Gold signals showing NANOS1 were closely associated with the surface of RB but not with the densecord (Figure 7(c)). RA was observed in the residual body of step 19 spermatids and was composed of massive ribosome particles and other unknown materials. NANOS1 signals were scattered in RA but not in the cytoplasm surrounding it (Figure 7(d)). In spermatids after step 15, NANOS1 signals were noted in the ventral head cap but the dorsal head cap was weakly or not labeled (Figure 7(e)). The acrosome itself was almost negative. In the flagellum of spermatids at steps 15-17, outer dense fibers of the middle piece and principal piece were stained for NANOS1 but not microtubules of the end piece (Figures 7(f) and (h)). In immunoelectron microscopic control sections incubated with preimmune sera, followed by protein A-gold probe, gold labeling in aforementioned structures was abolished (Figures 8(a)- (h)).
3.5. NANOS1 Expression in Spermatogenic Cells during Spermatogenesis in Rat Testis
Figure 9 summarizes the results of this experiment. Data from immunofluorescence and immunoelectron microscopy were combined and the expression of NANOS1 in nuage and non-nuage compartments was evaluated by staining intensity. In spermatocytes, weak cytosolic staining for NANOS1 was observed during all stages. NANOS1 appeared in ISPG and IMC of pachytene spermatocytes at stage V, the strongest staining seen in stages IX-X, decreased gradually in subsequent stages. NANOS1 started to appear in SB of pachytene spermatocytes at stage VIII, and then it rapidly increased, reached the peak level at stage IX, then started to decrease, and disappeared in stage XI. NANOS1 in CB appeared in round spermatids at step 1, increased rapidly to reach the peak level at steps 2-3. This state was maintained until step 8, following the signals decreased gradually and eventually disappeared in step 11. As described above, NANOS1 was localized in four kinds of non-nuage structures, MAG, RB, GB and RA located in the elongated cytoplasmic lobe of late spermatids. MAG was detected in spermatids at steps 10-17 while
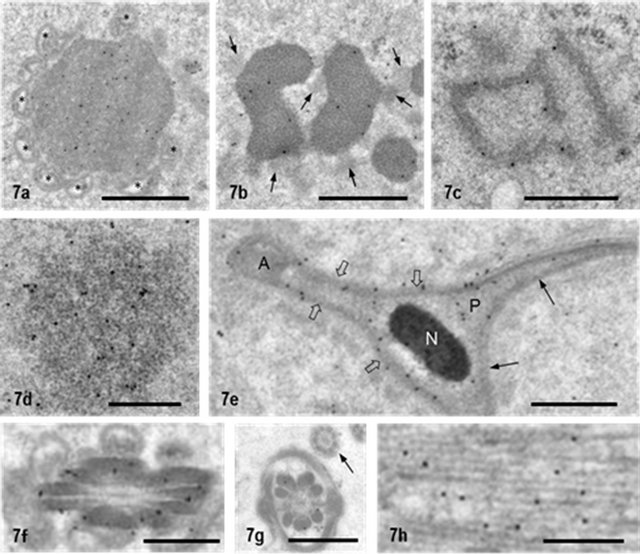
Figure 7. NANOS1 localization in non-nuage compartments. (a) MAG seen in elongated cytoplasmic lobe of step 15 spermatid. It is surrounded by many mitochondria (*). NANOS1 signals are present in the matrix; (b) GB in spermatids at step 15. Small less dense particles attach to it (arrows). Gold particles showing NANOS1 are in the fine granular matrix; (c) RB in spermatid at step 15. NANOS1 signals are associated with rod-like structures; (d) RA in the residual body of step-19 spermatid; (e) Cross-section of sickle-shaped nucleus of step 17 spermatid. Head cap is labeled for NANOS1 (open arrows) but the cap at dorsal side is not (arrows). N: nucleus; (f) Cross section of flagellum of step16 spermatid. Outer dense fibers are labeled for NANOS1; (g) Cross-section of flagellum of step16 spermatid. Outer dense fibers are labeled for NANOS1 but microtubules in the end piece are not (arrow); (h) Manchette of step 10 spermatid. Gold particles are associated with microtubules. Bar = 1 μm for (a) and (b). Bar = 0.5 μm for the other pictures.

Figure 8. Non-nuage structures in Immuno-electron microscopy control sections. (a) MAG; (b) GB; (c) RB; (d) RA; (e) Head cap; (f) Cross section of flagellum at the middle piece; (g) Cross section of flagellum at the principal piece; (h) Manchette of step 10 spermatid. Bar = 0.5 μm for all.
NANOS1 was detected in all steps. RB appeared in late spermatids at steps 14-16 but the NANOS1 signals were seen in RB only at steps 15 and 16. GB occurred in spermatids at steps 15-16 and the signals for NANOS1 were associated with GB in all steps. RA was observed in the elongated cytoplasmic lobe of the step-18 spermatids
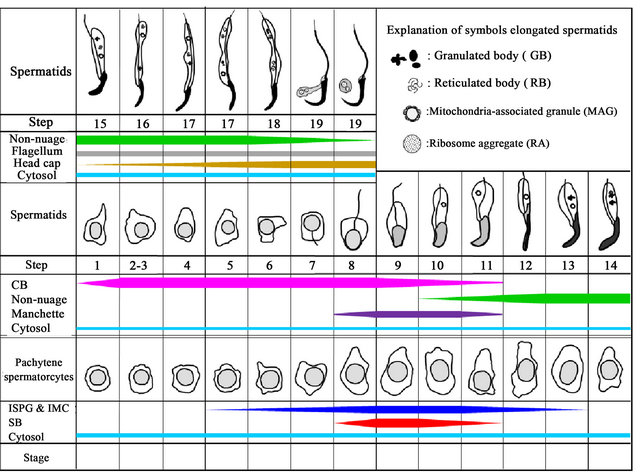
Figure 9. Expression of NANOS1 in nuage and non-nuage structures during spermatogenesis of rat testis. The width of colored bars indicates the number and size of nuage and non-nuage granules and cytosol staining, which are estimated arbitrarily. The position of colored bars indicates the stage (spermatocytes) and step (spermatids) where the staining of the granules and cytosol appears and disappears. ISPG and IMC are indicated by blue color, SB by red, CB by pink, non-nuage including GB, RB, MAG and RA, by green, cytosol by cyan, outer dense fiber by gray, manchette by violet, and head cap by ocher. Illustration showing developing cells is modified from that of Russell et al. [27].
and in the residual bodies of spermatids at step 19 and the NANOS1 signals were found to be stronger in step 19 than in step 18. Manchette appeared in spermatids at step 7 and continued to be observed until step 14. However, the signals for NANOS1 were detected in manchette in spermatids at steps 8-11. Flagella started to form in spermatids at step 7 and the NANOS1 signals appeared first in the dense fiber of spermatids at step 12. Afterward, the signals were consistently observed until step 19. The NANOS1 signals in the head cap appeared in spermatids at step 15, gradually increased and reached the peak level at step 19. Weak cytosolic staining of NANOS1 in spermatids was seen in all steps.
4. Discussion
4.1. Subcellular Distribution of NANOS1 in Rat Testis Cell Fractions
Our peptide antibody against NANOS1 reacted with a main 29 KDa band and an additional 40 KDa band on by Western blotting analysis. The molecular mass of the main band is almost consistent with that calculated from amino acid sequence reported in SWISS-PROT database, indicating that the antibody reacts with NANOS1. Although the latter band was observed in the mitochondrial and cytosol fractions but not in the nuclear and microsomal fractions, it is unclear whether it shows the band larger molecular species of NANOS1 or the other protein. The larger molecules of NANOS1 were reported [15,30]. Dot blot analysis showed that approximately 61% of NANOS1 distributed in the cytosol fraction and about 27% in the mitochondrial fraction. The results are almost consistent with immunofluorescence staining. Immunoelectron microscopic observations show no labeling in the mitochondria, so the dot blot value for the mitochondria indicates NANOS1 containing IMC, CB and cluster of 60 - 90 nm particles, which are co-precipitated with the mitochondria after differential centrifugation.
4.2. Immunofluorescence Localization of NANOS1 in Rat Testis
Our immunofluorescence staining results showed that NANOS1 is distributed in the cytosol and granular components of spermatogenic cells, but is absent in the nucleus. Double staining for NANOS1 andDDX4, a marker for nuage, showed that the granular components were mainly nuage compartments in which CB and SB were major site and ISPG and IMC minor sites. The distribution of NANOS1 in CB has previously been reported [19]. The present study demonstrates that NANOS1 is present in all nuage compartments. Our double immunofluorescence staining results suggest that there is a dynamic development process of nuage compartments, in which the NANOS1 negative nuage compartments continue to fuse with NANOS1-positive ones. Moreover, cytosolic NANOS1 might be incorporated into the nuage compartments. On the other hand, the double staining results also revealed that there appear to have many NANOS1-containing granules that are not stained for DDX4 in spermatogenic cells. Most of them are distributed in the elongated cytoplasmic lobe of late spermatids. Immunoelectron microscopic observations reveal that these DDX4-negative granules are non-nuage structures.
4.3. Immunoelectron Microscopic Localization of NANOS1 in Nuage and Non-Nuage Compartments
In the present study, our data showed that NANOS1 is localized to five kinds of nuage compartments as described by Russell and Frank [28]. The NANOS1 signals were weaker than those observed for other nuage proteins such as DDX4 [23], DDX25 [21] and BRU-NOL2 [22]. As shown by immunofluorescence staining and dot blotting analysis, NANOS1 was abundantly present in the cytosol fraction. It is possible that NA-NOS1 is synthesized in the cytosol and transported to the nuage compartments. It has been shown that human NANOS1 interacts with PUMILIO2 through highly conserved domains to form a stable complex [18]. Human PIMILIO2 has been shown to interact with the DEAD-box RNA helicase GEMIN3, a microRNA biogenesis factor, which is co-precipitated with NANOS1 and PU-MILIO2 in transfected mammalian cells [19]. By double immunofluorescence staining, the complexes of NA-NOS1, PUMILIO2 and GEMIN3 have been localized to CB [19]. Thus, it is suggested that CB is the main subcellular site where NANOS1 forms complexes with PUMILIO2 and GEMIN3 in the spermatids. The present results showed that NANOS1 is also localized to other nuage compartments such as 70 - 90 nm particles, cluster of 60 - 90 nm particles, IMC and SB, which occurred mainly in pachytene spermatocytes. Thus, NANOS1-PUMILIO2-GEMIN3 complexes might be one of the building components of these nuage compartments. The staining heterogeneity within the CB, in which the NANOS1 staining of one area was stronger than the other, was noticeable in spermatids at steps 8-11. It is likely that the observed staining heterogeneity may represent degradation levels of CB contents as suggested previously [31].
The present study showed that NANOS1 is also localized to the non-nuage structures, such as MAG, GB, RB, RA, Head cap of the step-17 spermatids, and outer dense fibers of flagella. Although the functions of MAG, GB, RB, and RA remain unknown, our previous studies have shown that DDX25 and BRUNOL2 are present in these structures [21,23]. The existence of nuage proteins such as BRUNOL2, DDX25 and NANOS1 in these structures suggests that these proteins may have biological functions there. However, it is not clear whether they perform their primary functions, such as the regulation of translation and mRNA silencing by BRUNOL2 [32], the participation of DDX25 in the nuclear export of RNAs [33], and the involvement of NANOS1 in microRNA biogenesis together with PUMILIO and GEMIN3 [19].
In conclusion, NANOS1 is a protein component of all nuage compartments, including CB, IMC, ISPG, SB and cluster of 60 - 90 nm particles, as well as several nonnuage compartments such as MAG, GB and RB. NANOS1 may exert its biological function in the nuage compartments via formation of protein complexes with PUMILIO and GEMIN3. How NANOS1 performs its function in the non-nuage structures is unclear.
5. Acknowledgements
This work was supported by the University research fund, in part by a grant-in-aid (17570158) from the Ministry of Education, Science, Culture and Sport, and by Science Research Promotion Fund from the Promotion and Mutual Aid Corporation for Private Schools of Japan.
REFERENCES
- S. Kobayashi, M. Yamada, M. Asaoka and T. Kitamura, “Essential Role of the Posterior Morphogen Nanos for Germline Development in Drosophila,” Nature, Vol. 380, No. 6576, 1996, pp. 708-711. doi:10.1038/380708a0
- K. Subramaniam and G. Seydoux, “Nos-1 and Nos-2, Two Genes Related to Drosophila Nanos, Regulate Primordial Germ Cell Development and Survival in Caenorhabditis elegans,” Development, Vol. 126, No. 21, 1999, pp. 4861-4871.
- Z. Wang and H. Lin, “Nanos Maintains Germline Cell Self-Renewal by Preventing Differentiation,” Science, Vol. 303, No. 5666, 2004, pp. 2016-2019. doi:10.1126/science.1093983
- D. Curtis, J. Apfeld and L. Lehmann, “Nanos Is an Evolutionarily Conserved Organizer of Anterior-Posterior Polarity,” Development, Vol. 121, No. 6, 1995, pp. 1899- 1910.
- R. Lehmann and C. Nüsslein-Volhard, “The Maternal Gene Nanos Has a Central Role in Posterior Pattern Formation of the Drosophila embryo,” Development, Vol. 112, No. 3, 1991, pp. 679-691.
- C. Wang, K. K. Dickinson and R. Lehman, “Genetics of nanos Localization in Drosophila,” Developmental Dynamics, Vol. 199, No. 2, 1999, pp. 103-115. doi:10.1002/aja.1001990204
- Y. Wang, R. M. Zayas T. Guo and P. A. Newmark, “Nanos Function Is Essential for Development and Regeneration of Planarian Germ Cells,” Proceedings of National Academy of Sciences of the United States of America, Vol. 104, No. 14, 2007, pp. 5901-5906. doi:10.1073/pnas.0609708104
- G. Deshpande, G. Calhoun, J. L. Yanowitzand and P. D. Schedl, “Novel Functions of nanos in Down Regulating Mitosis and Transcription during the Development of the Drosophila Germline,” Cell, Vol. 99, No. 3, 1999, pp. 271-181. doi:10.1016/S0092-8674(00)81658-X
- Y. Hayashi, M. Hayashi and S. Kobayashi, “Nanos Suppresses Somatic Cell Fate in Drosophila Germ Line,” Proceedings of National Academy of Sciences of the United States of America, Vol. 101, No. 28, 2004, pp. 10338-10342. doi:10.1073/pnas.0401647101
- M.Tsuda, Y. Sasaoka, M. Kiso, K. Abe, S. Haraguchi, S. Kobayashi and Y. Saga, “Conserved Role of nanos Proteins in Germ Cell Development,” Science, Vol. 301, No. 5637, 2003, pp. 1239-1241. doi:10.1126/science.1085222
- K. Sato, Y. Hayashi, Y. Ninomiya, S. Shigenobu, K. Arita, M. Mukai and S. Kobayashi, “Maternal Nanos Represses hid/skl-Dependent Apoptosis to Maintain the Germ Line in Drosophila Embryos,” Proceedings of National Academy of Sciences of the United States of America, Vol. 104, No. 18, 2007, pp. 7455-7460. doi:10.1073/pnas.0610052104
- S. Haraguchi, M. Tsuda, S. Kitajima, Y. Sasaoka, A. Nomura-Kitabayashi, K. Kurokawa and Y. Saga, “nanos1: A Mouse nanos Gene Expressed in the Central Nervous System Is Dispensable for Normal Development,” Mechanism of Development, Vol. 120, No. 6, 2003, pp. 721-731. doi:10.1016/S0925-4773(03)00043-1
- A. Suzuki, M. Tsuda and Y. Saga, “Functional Redundancy among Nanos Proteins and a Distinct Role of Nanos2 during Male Germ Cell Development,” Development, Vol. 134, No. 1, 2007, pp. 77-83. doi:10.1242/dev.02697
- A. Suzuki and Y. Saga, “Nanos2 Suppresses Meiosis and Promotes Male Germ Cell Differentiation,” Genes & Development, Vol. 22, No. 4, 2008, pp. 430-435. doi:10.1101/gad.1612708
- V. T. A. Julaton and R. A. R. Pera, “NANOS3 Function in Human Germ Cell Development,” Human Molecular Genetics, Vol. 20, No. 11, 2011, pp. 2238-2250. doi:10.1093/hmg/ddr114
- A. Forbes and R. Lehmann, “Nanos and Pumilio Have critical Roles in the Development and Function of Drosophila Germline Stem Cells,” Development, Vol. 125, 1998, pp. 679-690.
- F. Lolicato, R. Marino, M. P. Paronetto, M. Pellegrini, S. Dolci, R. Geremia and P. Grimaldi, “Potential Role Nanos3 in Maintaining the Undifferentiated Spermatogonia Population,” Developmental Biology, Vol. 313, No. 2, 2008, pp. 725-738. doi:10.1016/j.ydbio.2007.11.011
- J. Jaruzelska, M. Kotecki, K. Kusz, A. Spik, M. Firpo and R. A. R. Pera, “Conservation of a Pumilio-Nanos Complex from Drosophila Germ Plasm to Human Germ Cells,” Development Genes and Evolution, Vol. 213, No. 3, 2003, pp. 120-126.
- B. Ginter-Matuszewska, K. Kusz, A. Spik, D. Grzeszkowiak, A. Rembiszewska, J. Kupryjanczyk and J. Jaruzelska, “NANOS1 and PUMILIO2 Bind microRNA Biogenesis Factor GEMIN3, within Chromatoid Body in Human Germ Cells,” Histochemistry and Cell Biology, Vol. 136, No. 3, 2011, pp. 279-287. doi:10.1007/s00418-011-0842-y
- B. Ginter-Matuszewska, A. Spik, A. Rembiszewska, C. Koyias, J. Kupryjańczyk and J. Jaruzelska, “The SNAREAssociated Component SNAPIN Binds PUMILIO2 and NANOS1 Proteins in Human Male Germ cells,” Molecular Human Reproduction, Vol. 15, No. 3, 2009, pp. 173-179. doi:10.1093/molehr/gap004
- Y. Onohara and S. Yokota, “Expression of DDX25 in Nuage Components of Mammalian Spermatogenic Cells: Immunofluorescence and Immunoelectron Microscopic Study,” Histochemistry and Cell Biology, Vol. 137, No. 1, 2012, pp. 37-51. doi:10.1007/s00418-011-0875-2
- H. Yonetamari, Y. Onohara and S. Yokota, “Localization of BRUNOL2 in Rat Spermatogenic Cells Revealed by Immunofluorescence and Immunoelectron Microscopic Techniques,” Open Journal of Cell Biology, Vol. 2, No. 2, 2012, pp. 11-20. doi:10.4236/ojcb.2012.22002
- Y. Onohara, T. Fujiwara, T. Yasukochi, M. Himeno and S. Yokota, “Localization of Mouse Vasa Homolog Protein in Chromatoid Body and Related Nuage Structures of Mammalian Spermatogenic Cells during Spermatogenesis,” Histochemistry and Cell Biology, Vol. 133, No. 6, 2010, pp. 627-639. doi:10.1007/s00418-010-0699-5
- C. M. Haraguchi, T. Mabuchi and S. Yokota, “Localization of a Mitochondrial Type of NADP-Dependent Isocitrate Dehydrogenase in Kidney and Heart of Rat: An Immunocytochemical and Biochemical Study,” Journal of Histochemistry and Cytochemistry, Vol. 51, No. 2, 2003, pp. 215-226. doi:10.1177/002215540305100210
- C. de Roe, P. J. Courtoy and P. Baudhuin, “A Model of Protein-Colloidal Gold Interactions,” Journal of Histochemistry and Cytochemistry, Vol. 35, No. 11, 1987, pp. 1191-1198. doi:10.1177/35.11.3655323
- D. Rickwood and T. C. Ford, “Preparation and Fractionation of Nuclei, Nucleoli and Deoxyribonucleoproteins,” In: D. Rickwood Ed., Iodinated Density Gradient Media, Practical Approach, IRL Press, Oxford, Washington DC, 1983, pp. 69-89.
- L. D. Russell, R. A. Ettlin, A. S. P. Hikimand and E. D. Clegg, “Histological and Histopathological Evaluation of Testis,” Cache River Press, Clearwater, 1990.
- L. Russell and B. Frank, “Ultrastructural Characterization of Nuage in Spermatocytes of the Rat Testis,” Anatomical Record, Vol. 190, No. 1, 1978, pp. 79-97. doi:10.1002/ar.1091900108
- Y. Clermont, R. Oko and L. Hermo, “Immunocytochemical Localization of Proteins Utilized in the Formation of Outer Dense Fibers and Fibrous Sheath in Rat Spermatids: An Electron Microscope Study,” Anatomical Record, Vol. 227, No. 4, 1990, pp. 447-457. doi:10.1002/ar.1092270408
- K. M. Kusz, L. Tomczyk, M. Sajek, A. Spik, A. LatosBielenska, P. Jedrzejczak, L. Pawelczyk and J. Jaruzelska, “The Highly Conserved NANOS2 Protein: Testis-Specific Expression and Significance for the Human Male Reproductionm,” Molecular Human Reproduction, Vol. 15, No. 3, 2009, pp. 165-171. doi:10.1093/molehr/gap003
- C. M. Haraguchi, T. Mabuchi, S. Hirata, T. Shoda, K. Hoshi, K. Akasaki and S. Yokota, “Chromatoid Bodies: Aggresome-Like Characteristics and Degradation Sites for Organelles of Spermiogeniccells,” Journal of Histochemistry and Cytochemistry, Vol. 53, No. 4, 2000, pp. 455-465. doi:10.1369/jhc.4A6520.2005
- C. Barreau, L. Paillard, A. Méreau and H. B. Osborne, “Mammalian CELF/Bruno-Like RNA-Binding Proteins: Molecular Characteristics and Biological Functions,” Biochemie, Vol. 88, No. 5, 2006, pp. 515-525. doi:10.1016/j.biochi.2005.10.011
- Y. Sheng, C.-H. Tsai-Morris, R. Gutti, Y. Maeda and M. L. Dufau, “Gonadotropin-Regulated Testicular RNA Helicase (GRTH/Ddx25) Is a Transport Protein Involved in Gene-Specific mRNA Export and Protein Translation during Spermatogenesis,” Journal of Biological Chemistry, Vol. 281, No. 46, 2006, pp. 35048-35056. doi:10.1074/jbc.M605086200

