Open Journal of Respiratory Diseases
Vol.3 No.4(2013), Article ID:38095,10 pages DOI:10.4236/ojrd.2013.34021
Multivariate Asthma Phenotypes in Adults: The Quebec City Case-Control Asthma Cohort
1Centre de Recherche de l’Institut Universitaire de Cardiologie et de Pneumologie de Québec, Québec City, Canada
2Department of Molecular Medicine, Laval University, Québec City, Canada
Email: *yohan.bosse@criucpq.ulaval.ca
Copyright © 2013 Émilie Lavoie-Charland et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received July 19, 2013; revised August 28, 2013; accepted September 30, 2013
Keywords: Asthma; Phenotypes; Mild-Moderate Asthma; Cluster Analysis; Factor Analysis
ABSTRACT
Background and Objectives: Asthma is a heterogeneous disease where patient severity can be classified according to various models based on numerous variables. Large collections of well-phenotyped subjects are needed to find distinct clusters of patients for personalized medicine and future genetic studies. The objective of this study is to describe the collection of the Quebec City Case-Control Asthma Cohort and to identify homogeneous subgroups of asthma patients based on clinical characteristics. Methods: This cohort is part of an ongoing project initiated in 2007 to elucidate the genetic basis of asthma. All subjects are randomly recruited at the same site following advertisements. Subjects are unrelated French Canadian white adults 18 years of age or older. Each participant underwent a spirometry, methacholine challenge, and allergy skin-prick tests. Blood was collected for DNA, cell counts and total serum IgE measurements. So far, 982 subjects have been recruited and classified as cases (n = 566) or controls (n = 416). We performed factor and cluster analyses on collected phenotypes from this set to identify subgroups of phenotypically similar asthmatic patients. Results: Factor analysis with 13 variables led to the selection of five factors: lung function, numbers of allergens, blood eosinophil percentage, smoking status and age. K-means cluster analysis on the reduced dataset produced four significantly different groups with the following characteristics: smoking history, low atopy and low lung function, high atopy, and young non-smoking with average atopy. Conclusions: The Quebec City Case-Control Asthma Cohort is a new resource for local and collaborative clinical and genetic research on asthma. This new collection reveals distinct multivariate phenotypes of adult asthma that are likely to be important for future genetic studies and targeted therapies.
1. Introduction
Large collections of participants with and without asthma are the essence of clinical and genetic research in asthma. Increasingly large-scale collaborative studies are performed to identify the genetic architecture of asthma [1, 2]. Despite these major efforts, only a fraction of the heritable components of asthma has been identified and much remains to be discovered. Pooling resources as accomplished by the European-based GABRIEL [1] and the US-based EVE [2] consortiums are essential to elucidate the genetics of asthma. However, asthma is a heterogeneous disease [3]. Pooling resources to increase sample size is required, but subgrouping phenotypes is likely to be as much important to understand the molecular basis of asthma.
Accurate description of asthma phenotypes is an ongoing challenge [3-6]. Cluster analysis aggregating phenotypes without a priori assumptions has become an optimal choice to define asthma [7]. Cluster analysis can offer clinicians and researchers a better insight on the multiple phenotypes present in asthma patients for targeted therapies and future genetic studies in more homogenous subgroups of patients. So far, heterogeneity of the disease has led to various subgroupings of asthma phenotypes in adults [5,6,8,9] and children [10,11]. Distinct adult airway limitation phenotypes were described [12], but adult asthma phenotypes still lack consensus for both severe and mild to moderate asthma.
Here we describe an ongoing effort to build a new case-control cohort to study the genetics of asthma in Quebec City. The main goal of the Quebec City CaseControl Asthma Cohort is to drive our local genetic research program on asthma, but also to serve a wide range of purposes from a replication cohort for others’ hypothesis-driven research to participation in large-scale genome-wide testing collaborative efforts nationally and internationally aiming to improve our molecular understanding of asthma. In this report, we focus on the phenotypes of participants enrolled in the Quebec City CaseControl Asthma Cohort. We aim to describe the collection and clinical characteristics of participants and to perform cluster analyses with this cohort of mild to moderate adult asthma. In addition, we want to establish subgrouping phenotypes of asthma that will be used in future clinical and genetic studies using the Quebec City CaseControl Asthma Cohort.
2. Methods
2.1. Study Subjects and Clinical Data
Asthmatic and non-asthmatic unrelated French Canadian (white) subjects 18 years and older are recruited from local advertisements in media or following a visit to the respiratory clinic to participate in other research projects. Patients with chronic obstructive pulmonary disease, systemic inflammatory diseases, and body mass index (BMI) above 40 kg/m2 are excluded. Personal health information, medication use, and clinical test measurements are collected in a local electronic database. A blood sample is collected and stored at the Institut universitaire de cardiologie et de pneumologie de Québec (IUCPQ) site of the Respiratory Health Network Tissue Bank of the Fonds de recherche du Québec-Santé (www.tissuebank.ca). The study protocol was approved by the Research Ethics Board of the IUCPQ. All participating subjects signed an informed consent approved by the REB. Subjects are de-identified using a code number for confidentiality. Access to data is protected using the data management structure approved by the REB.
2.1.1. Spirometry
All patients undergo pulmonary function testing using a spirometer (Medi-Soft Exp’air impression, Roxon meditech ltd, Montreal). Spirometry is carried out according to the American Thoracic Society guidelines [13,14]. Instructions to perform the forced expiratory maneuver are given to patients by an experienced research nurse and three reproducible measurements of forced expiratory volume in 1 sec (FEV1) and forced vital capacity (FVC) are obtained. Predicted lung function values are obtained from Knudson et al. [15].
2.1.2. Methacholine Challenge
Airway responsiveness to methacholine is measured using the 2-min tidal method [13]. Briefly, after baseline measurements of FEV1 and FVC, each subject inhaled saline (0.9%) followed by consecutive doubling concentrations of methacholine between 0.03 to 64 mg/mL. Methacholine aerosols are inhaled for 2 min at 5-min intervals. The FEV1 is measured 30 and 90 seconds after each inhalation or every two minutes until the FEV1 improved compared with the precedent. The test is stopped when the FEV1 falls by more than 20% compared to baseline or at a concentration of 64 mg/mL for negative responders. An acceptable-quality FEV1 needs to be obtained at each time point, otherwise, a second attempt is performed. We used the lowest FEV1 value after saline inhalation and the lowest FEV1 value after each dose to calculate the fall in FEV1. The reproducibility of the technique was previously reported [16]. The test result is expressed as the provocative dose of methacholine inducing a 20% fall in FEV1 (PC20) and airway hyperresponsiveness (AHR) is defined as PC20 < 8 mg/ml.
2.1.3. Asthma Status
Asthma diagnosis was confirmed by a physician (L.-P. Boulet and M. Laviolette) based on clinical symptoms, lung function and airway responsiveness. Asthma severity was determined according to the Canadian Asthma Guidelines [17].
2.1.4. Skin-Prick Tests
Skin-prick tests (SPT) with allergens were performed to measure the allergic status. A total of 25 inhalant allergens were tested including animal hair and danders (cat, dog, horse, and cattle), feather mix, house dust mix, house dust mites (dermatophagoides pteronyssinus and dermatophagoides farinae), trees (white ash, cottonwood, birch, american elm, boxelder maple, and oak), grass (grass mix, timothy, perennial ryegrass, ragweed, sagebrush-mugwort, English plantain, and cocklebur), and molds (Alternaria tenuis, Hormodendrum and Aspergillus fumigatus). Subjects are considered atopic if at least one allergen caused a wheal diameter of at least 3 mm at 10 min in the presence of a negative saline control and a positive histamine response [18].
2.1.5. Total Serum IgE
Total serum immunoglobulin E (IgE) was measured with enzyme immunofluorometry (E-170 Module for MODULAR® ANALYTICS EVO, Roche Diagnostics Ltd, Switzerland).
2.1.6. Blood Cell Counts
Complete blood counts were determined at the Hospital’s Clinical Laboratory by automated procedures. Measured parameters include total white count and counts for neutrophils, lymphocytes, monocytes, eosinophils, and basophils.
2.1.7. Blood Buffy-Coat
A blood sample is collected and the buffy coat is obtained after centrifugation. The buffy coat is kept at −80˚C prior to DNA extraction. The biological materials (i.e. buffy coat, DNA and serum) are kept frozen in our local biobank for future studies.
2.2. Statistical Analysis
Variables not normally distributed were log10 transformed before analysis. The differences between cases and controls were assessed by a two-tailed unpaired Student’s t-test for quantitative variables and with chi-square tests for dichotomous or categorical variables. All statistical analyses were performed using the R statistical software version 2.15.3.
2.2.1. Variable Reduction, Data Transformation, and Factor Analysis
181 variables were available for analysis. Initial reduction of the dataset led to 9 continuous and 29 categorical variables. Skin prick test results were grouped by type of allergen, by total allergen count and by atopy status, in order to reduce size of the final dataset. IgE levels and percent eosinophil were log10 transformed in order to normalize their distribution. PC20 scores were transformed into a dichotomous AHR variable as described above. In order to eliminate the correlation between variables, factor analysis (maximum likelihood) with orthogonal varimax rotation was applied. This standard method identifies independent groups of variables. Only asthmatic patients with complete information on the remaining 13 variables were included in the factor analysis. Factor analysis was repeated for different transformations of the allergy count. Factor analysis with the allergy transformation that explained the greatest proportion of the model’s variance was selected. Factors with eigenvalues greater than 1 were analysed and only variables with absolute value of loadings greater than 0.4 were considered significantly correlated to the factors.
2.2.2. Cluster Analysis
Cluster analyses were restricted to asthma cases with complete information for the variables selected from the factor analysis. Variables selected from the factor analysis were standardized prior to cluster analyses as recommended [19] and participants with missing data for these variables were excluded. Optimal number of clusters k was evaluated graphically by Ward’s agglomerative hierarchical clustering [6,9]. K-means clustering (Cluster package) separated the dataset into k clusters. Differences between clusters were evaluated by Kruskal-Wallis and chi-squared test (or exact fisher as appropriate) for continuous and categorical variables, respectively. Differences were deemed significant if the P-value was smaller than 0.05.
3. Results
In this initial report, we included data on 982 subjects, 566 asthmatic subjects and 416 controls recruited from October 2007 to March 2013. The recruitment rate for the full five years (i.e. 2008 to 2012) averages 190 subjects per year. The rate of missing values for the variables of interest is low: no missing values for anthropometric variables, 9 (0.9%) for spirometry, 68 (6.9%) for methacholine challenge, 43 (4.4%) for IgE levels, 18 (1.8%) for blood cell counts, and 4 (0.4%) for atopy. So far, more women (n = 623) than men (n = 359) were enrolled in the cohort.
Table 1 presents the clinical characteristics of cases and controls. No difference in age or in the proportion of men and women is observed between these two groups.
The mean body mass index (BMI) is higher in cases compared to controls, but this difference is not clinically relevant. Table 2 presents asthma medication for cases. Figure 1 shows age distributions according to asthma status. As observed in this figure, a greater proportion of young adults (20 to 30 years) was recruited.
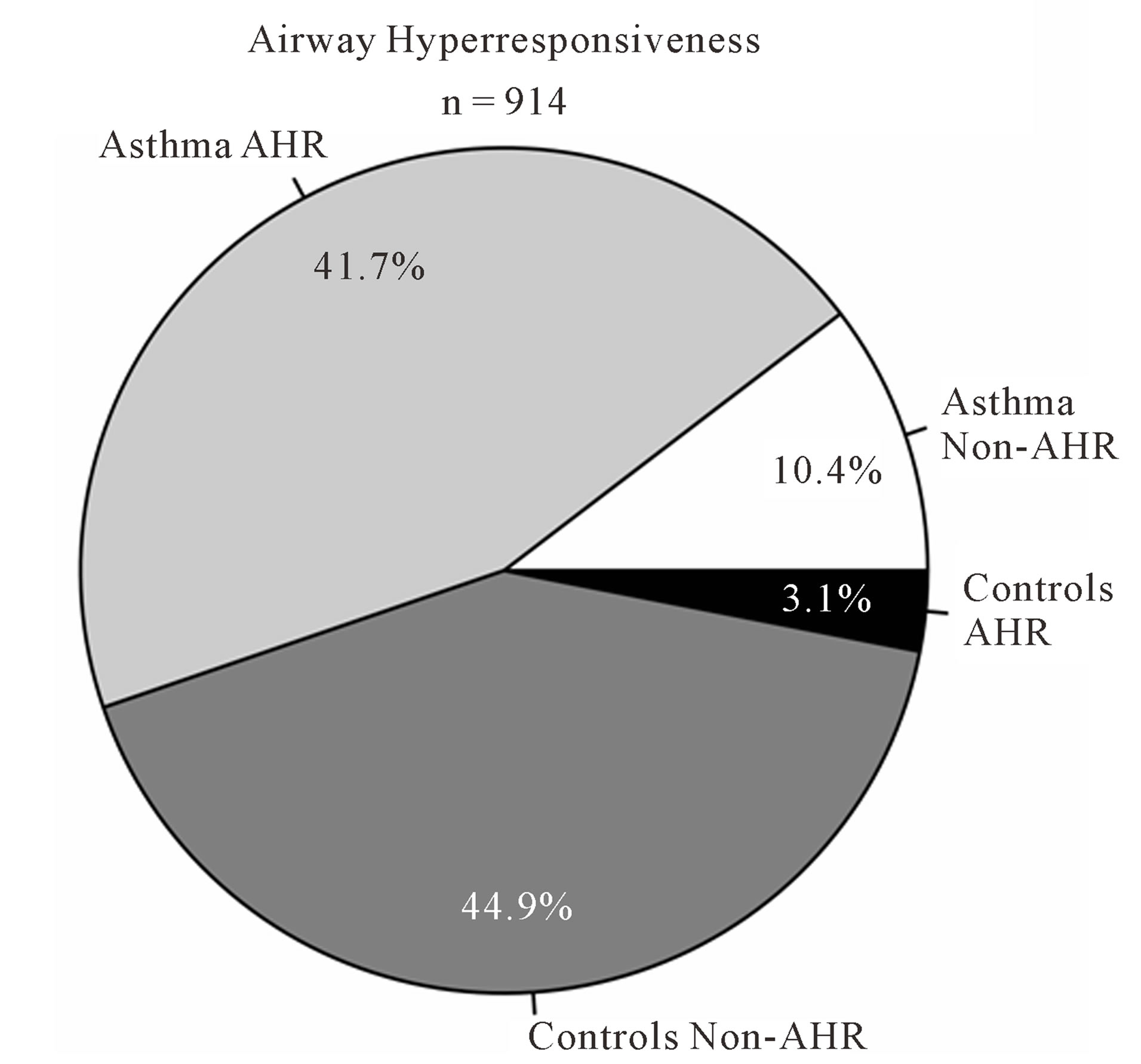
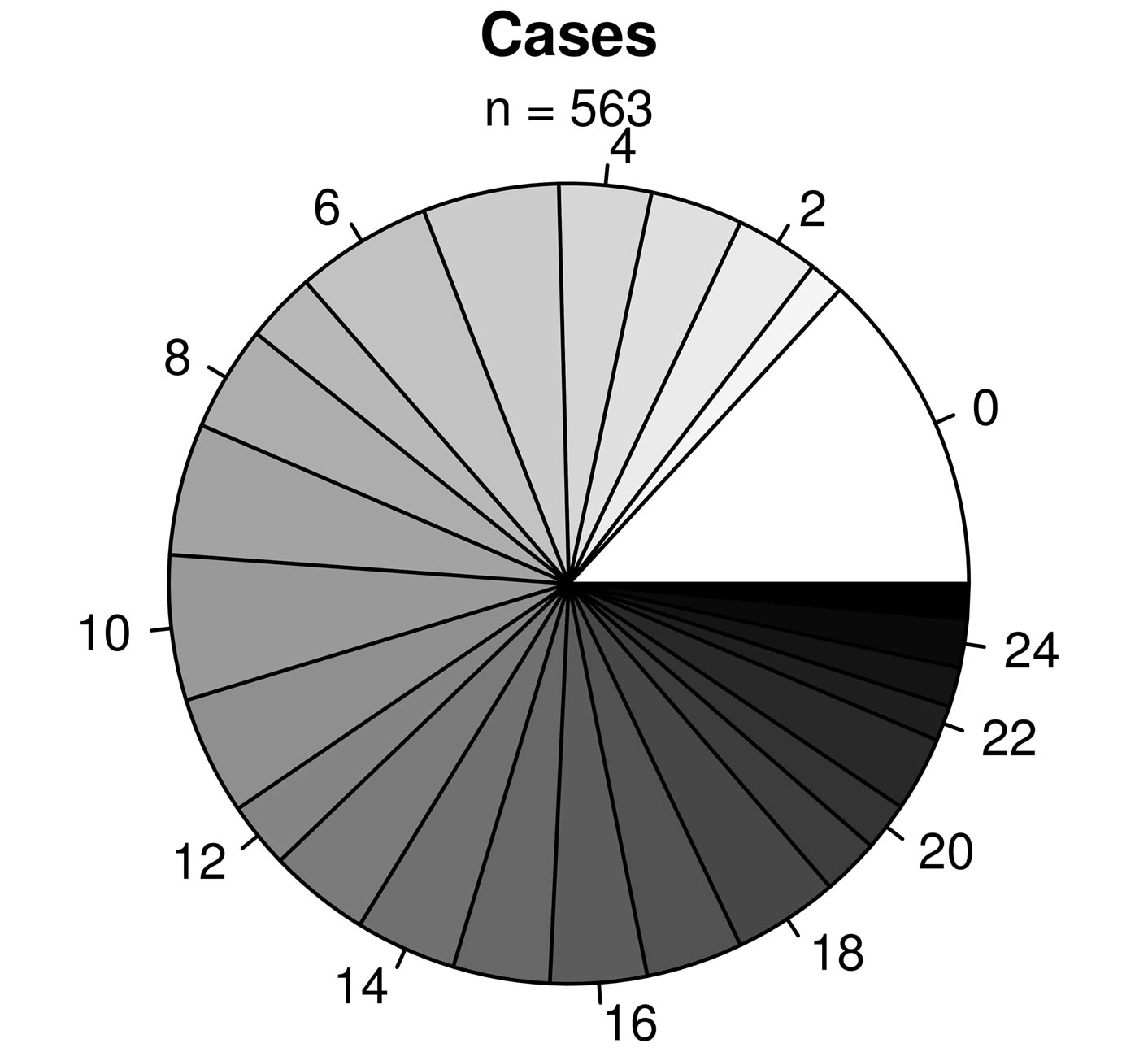
As expected, predicted lung function is lower in asthmatic subjects (Table 1). The proportion of AHR is also increased in cases compared to controls (Figure 2(a)). 81.9% of asthmatic subjects and 6.9% of controls had airway hyperresponsiveness at the time of testing. Allergies are also more frequent in cases compared to controls (Figure 2(b)); 88.1% and 59.7% of asthmatic subjects and controls were atopic, respectively. In addition, subjects with atopy and asthma have a greater number of positive responses to allergens compared to control subjects with atopy (Figure 3). The greater percentage of allergic response among subjects with asthma is also observed across all allergens evaluated (Figure 4). Blood eosinophil and total IgE levels are higher in asthmatics (Table 1). There is also a significant correlation between these two variables (r = 0.33, P ≤ 0.0001). The proportion of ex-smokers is higher in patients with asthma compared to controls, but this difference is not statistically significant (Table 1).
3.1. Cluster Analysis
377 asthmatics had complete information for the 13 variables selected for factor analysis. Factor analysis (Table 3) with total count of positive allergens as the allergy transformation led to the selection of 5 factors with eigenvalues >1 which explained 53.1% of the total variance in the dataset. Factors were representative of: smoking status, lung function, white blood cell counts, allergy, and age (at evaluation and at onset).
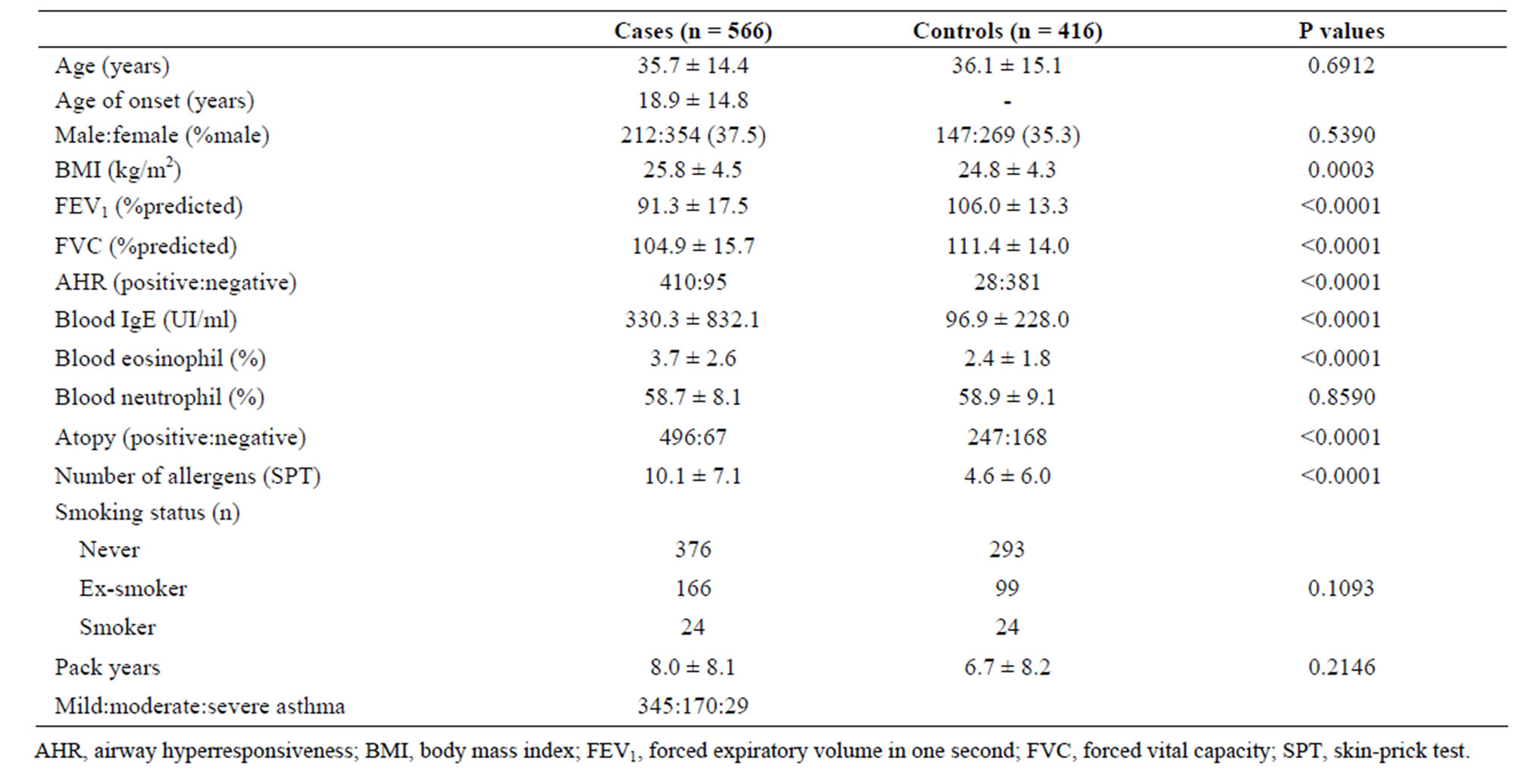
Table 1. Clinical characteristics of the Quebec City Case-Control Asthma Cohort.
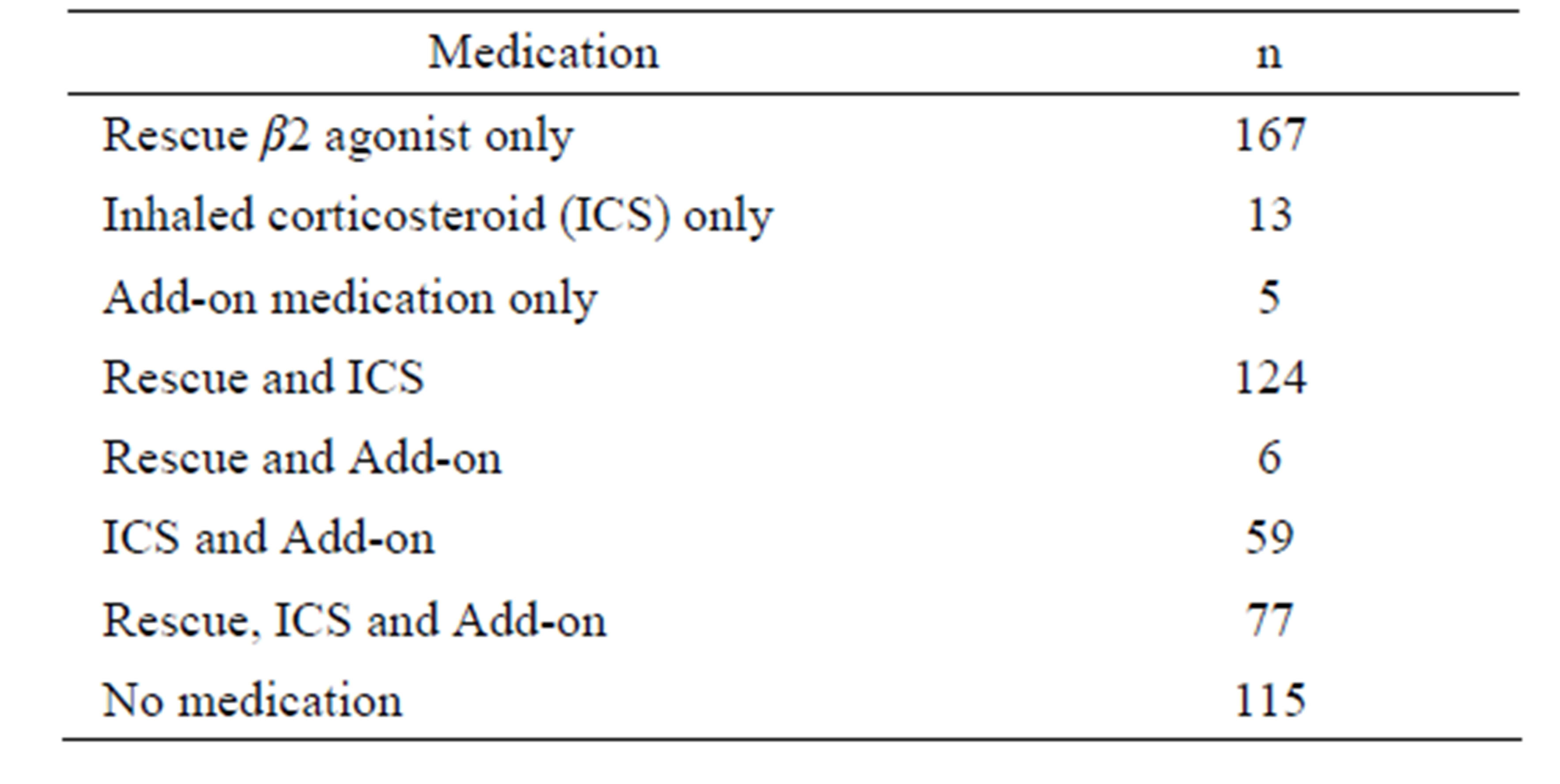
Table 2. Asthma medication.
For each factor, the variable most highly correlated to the factor (or containing fewer missing data) was selected for further analysis: smoking status, %predicted FEV1, log10% eosinophil, number of positive allergens, and age. All highly correlated variables are in bold in Table 3. We added 145 asthmatics with complete information for these five variables to the dataset for subsequent K-means analysis (n = 522). According to the dendrogram of the agglomerative hierarchical clustering, four clusters were required to separate the 522 subjects using the standardized variables described above.
K-means clustering (Table 4) was applied on participants. Clusters 1 to 4 contain 105, 104, 138, 175 subjects, respectively, and are further described below. As expected, clusters differ significantly for at least one of the five variables selected by factor analysis.
3.1.1. Smoking History
20% of participants were part of this smoking cluster. 78% of them were ex-smokers while the others were current-smokers. Age average was 35 and the majority of patients in this cluster were allergic to animal hair and danders. Blood eosinophil percentage (2.77%) was lower than the cohort average (3.71%). Most members of cluster 1 were diagnosed with mild asthma (71%).
3.1.2. Low Atopy & Poor Lung Function
This cluster of older patients (mean age of 57) included 20% of participants. 52% were ex-smokers while the others were never-smokers. Percent positive allergens were lower than the asthma cohort average for all types of allergens, but % eosinophil (4.13%) was slightly higher than average (3.71%). Mean percent predicted FEV1 was the lowest of all groups (73.9%). 21 out of the 23 severe asthmatic patients included in the analysis were in this cluster.
3.1.3. High Atopy
The third cluster included 26% of all cases and was composed of mostly never-smokers with age average of 31. This cluster had the highest percentage of positive allergens compared to other clusters and most patients in cluster 3 were positive to animal allergens. Mean %predicted FEV1 was lower than average and %eosinophil was the highest among clusters. 57% of participants in cluster 3 were diagnosed with mild asthma.
3.1.4. Young Non-Smoking & Average Atopy
34% of asthmatic participants were clustered in this non-smoking cluster with a mean age of 27. Average %
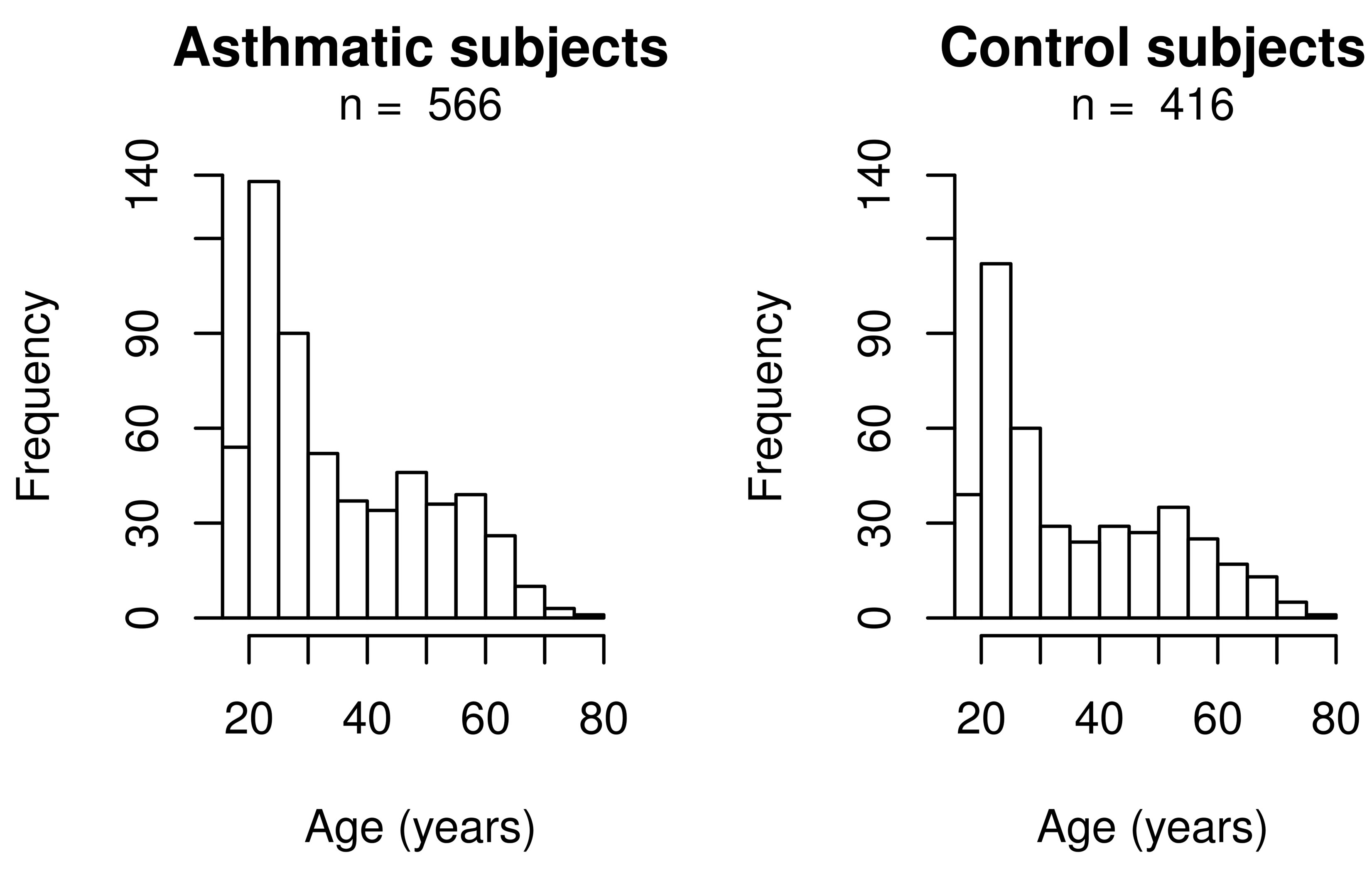
Figure 1. Age distribution according to asthma status.
 (a)
(a)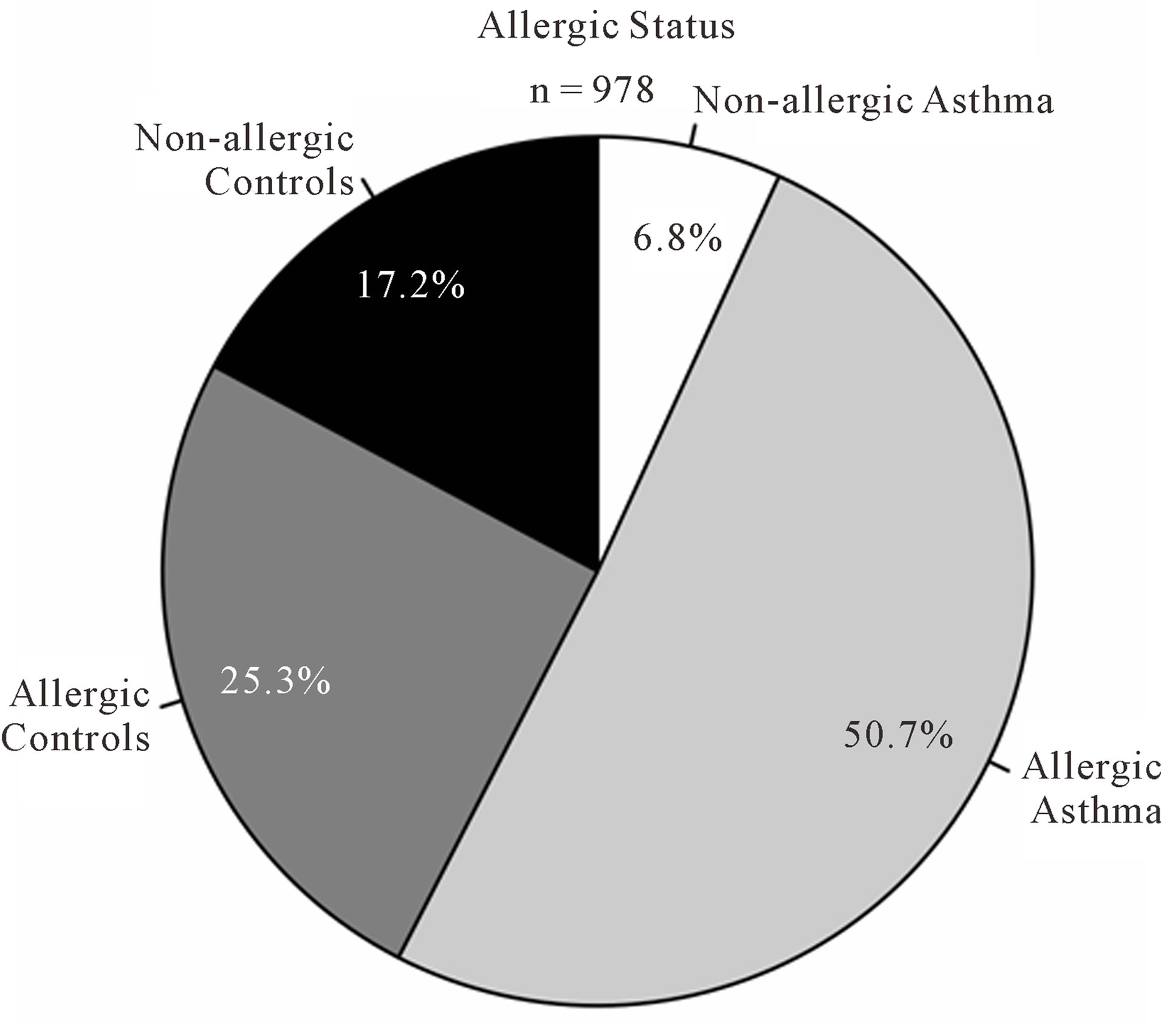 (b)
(b)
Figure 2. Atopy and airway hyperresponsiveness. (a) Proportions of subjects with airway hyperresponsiveness (AHR) in cases and controls; (b) Proportions of subjects with allergies in cases and controls.
 (a)
(a)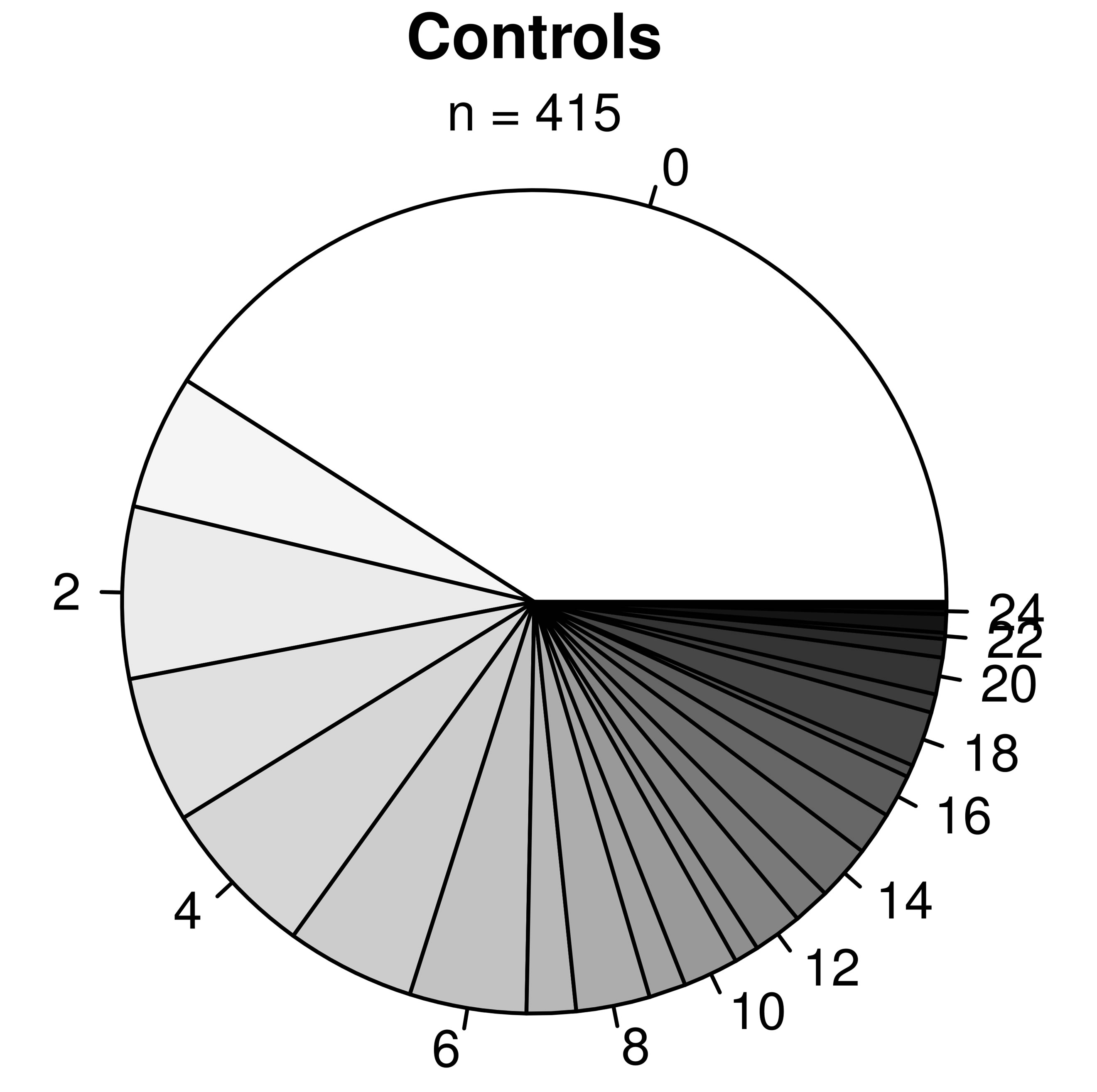 (b)
(b)
Figure 3. Proportions of subjects with x number of allergies for asthmatic (top panel) and non-asthmatic (bottom panel) subjects. The labels surrounding the pies indicate the number of allergies.
eosinophil was the lowest amongst the four groups. 78% of patients in cluster 4 were diagnosed with mild asthma.
4. Discussion
We describe for the first time the Quebec City CaseControl Asthma Cohort. Currently this cohort consists of 982 subjects including 566 asthmatic and 416 controls well-characterized for asthma and related phenotypes. All participants are white adults and more women are enrolled so far. One important objective of this cohort is to power our own clinical and genomic research program on asthma. The cohort is also planned for genetic replication purposes and participation in large-scale collaborating efforts to elucidate the genetic architecture of asthma. Pooling resources will be needed to achieve the later objective, but subgrouping asthma phenotypes will be as important. In this report, we defined four clusters of asthma patients based on demographic and clinical characteristics. Individuals within subgroups are more likely to share the same underlying molecular basis.
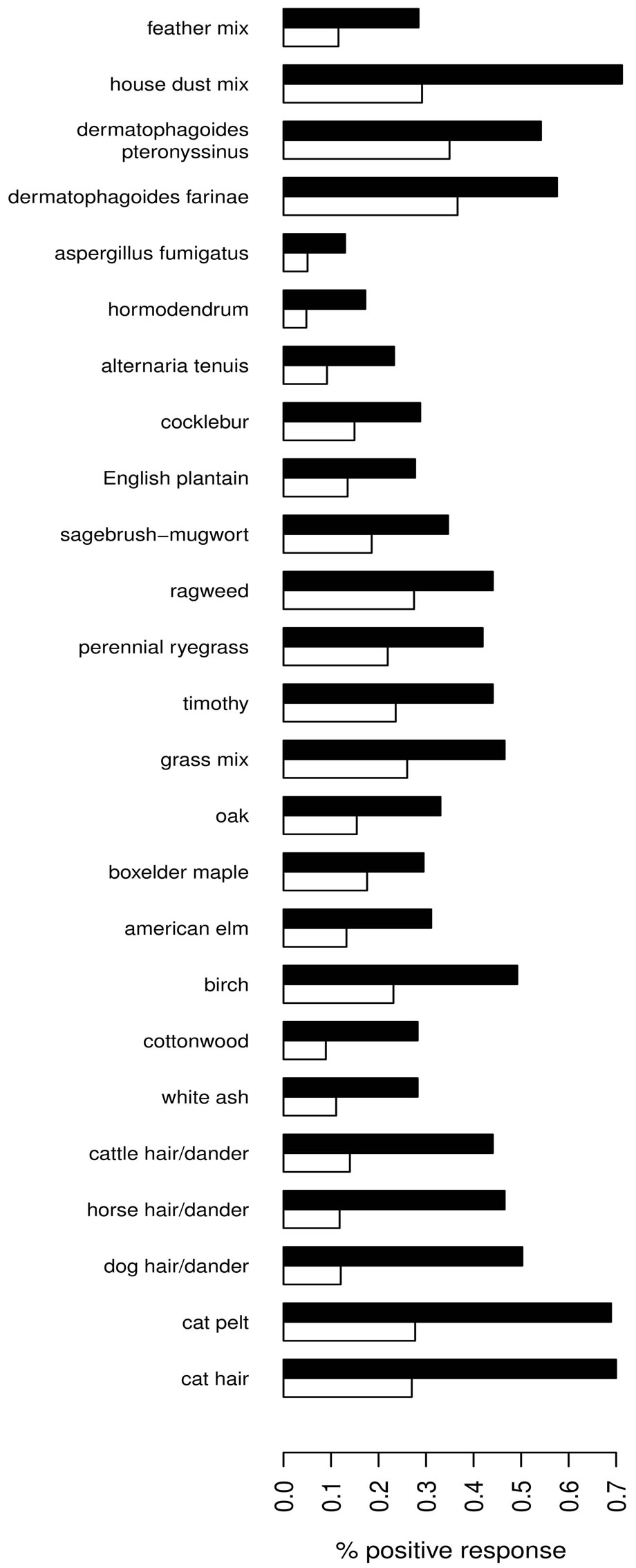
Figure 4. Percentage of positive response for tested allergens (y-axis) among subjects with (black) and without (white) asthma.
Optimal description and classification of participants based on clinical features are necessary to identify the genetic components of asthma. Accordingly, cluster analysis is an appropriate approach to clearly define our cohort and orient further studies. Clustering techniques are based on similarities (or differences) between observations, where individuals with high similarities (or small differences) for selected variables will have a greater probability of joining the same cluster. As performed previously [8,10,20], highly correlated variables were not included in the clustering, insuring that distinct asthma factors are included in the analyses.
This was achieved through factor analysis and factors obtained reflected known aspects of asthma diagnosis. Although our method was statistically-based, it did not limit the clinical significance of the selected variables. Atopy [6], sputum eosinophilic inflammation [6,21,22] and immunoglobulin E levels [21] have been described as factors of asthma, while age and smoking are both known to affect asthma diagnosis and treatment [23].
Selecting the appropriate numbers of clusters is a challenging aspect of cluster analysis. Although numerical and objective indexes exist, they are limited to particular types of variables, datasets or cluster shapes. Visual techniques through dendrogram analysis [6] are subjective, but may be the best approach when little is known on the dataset’s structure. In our study, numerical indexes were difficult to apply to our dataset since cluster shape was unknown and clustering was based on both continuous and categorical variables. In the past, hierarchical clustering has been used in a conclusive fashion to describe asthma phenotypes [10-12,24], which validates a hierarchical clustering determination of optimal numbers of clusters in asthma cohorts as described in this study.
This study has limitations. It is possible that other variables not measured in this study may better explain asthma even though factor analysis has provided us with five variables correlated to five independent factors. It would be insightful to compare different variables to determine which would be optimal to both represent factors of asthma and cluster phenotypes. Through our cluster analysis, we have shown that adult asthma can be characterized by 4 clusters based on 5 variables, but our results can only present a snapshot of a disease which evolves over time (e.g. immunoglobulin E levels and percent predicted FEV1 can vary within a day). Such variations imply that asthmatic participants may cluster differently on different visits. Longitudinal studies are needed to confirm the stability of clusters over time. It should be noted that mild to moderate asthmatics compose most of our cases. At this point, we cannot evaluate the impact of severe asthma on clustering. We are currently recruiting patients with severe asthma via the hospital’s asthma clinic.
It is important to note that asthma patients in this cohort are relatively young (with an average age of 36), and have a mild to moderate diagnosis of asthma and the majority are atopic (86%). These factors must be considered when our results are compared to previous clustering
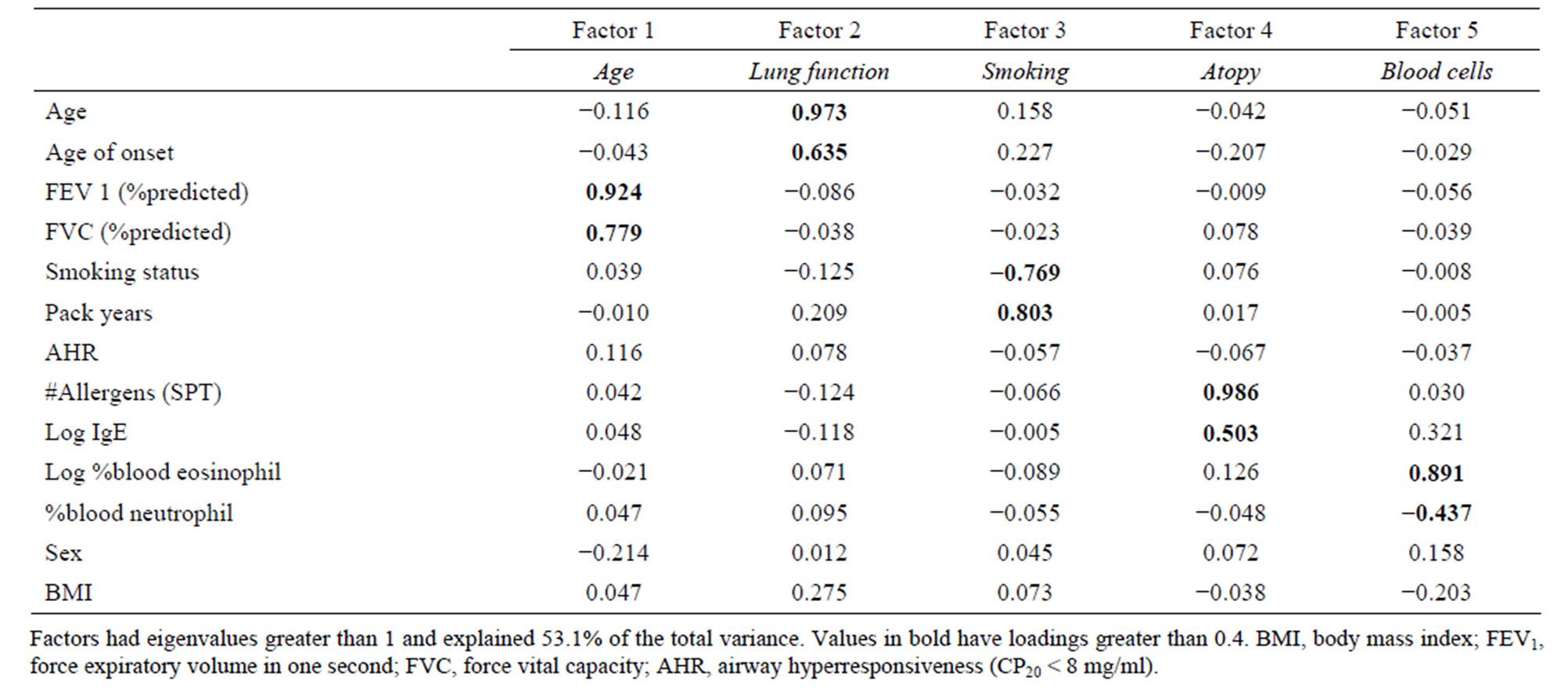
Table 3. Factor analysis with varimax rotation on cases.
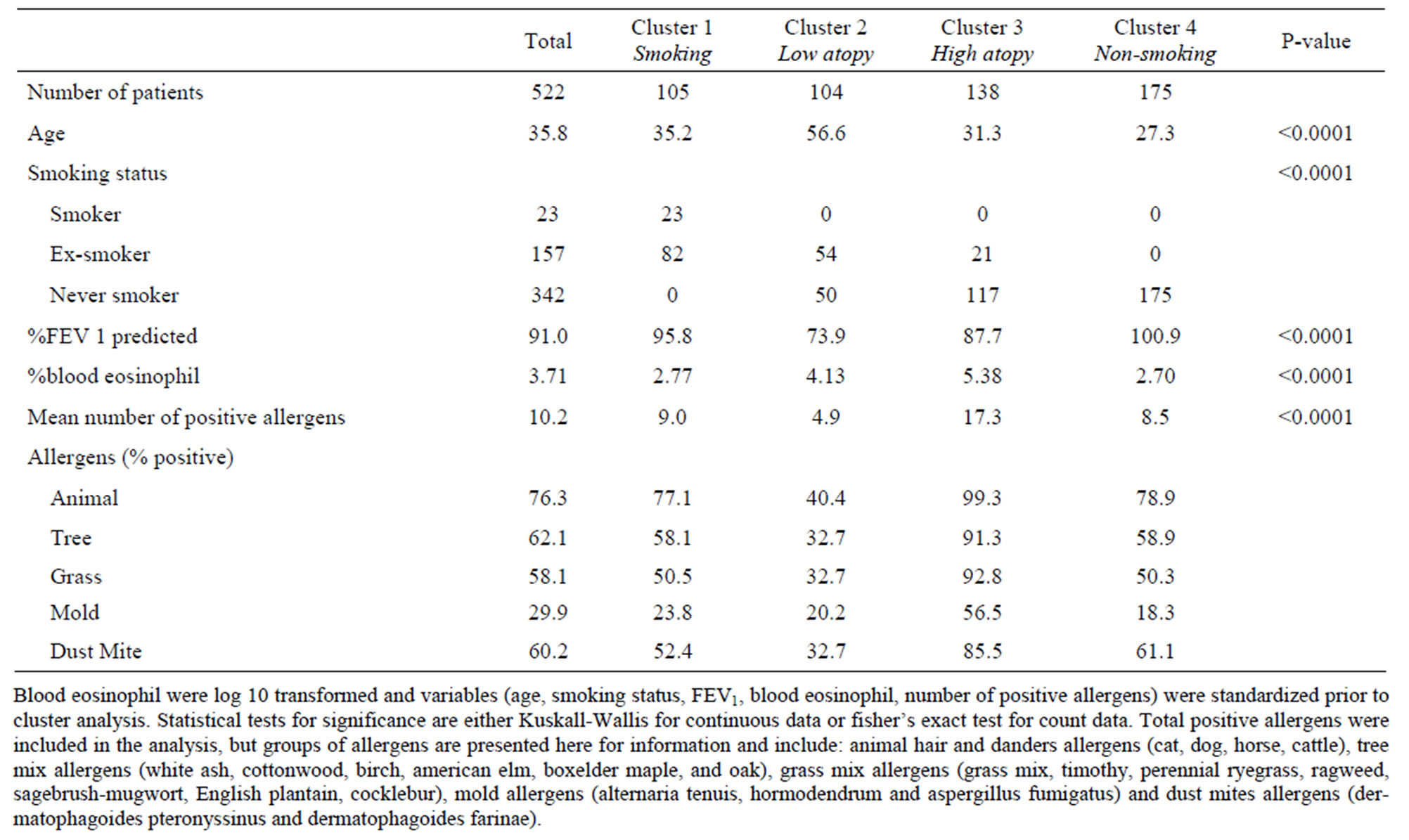
Table 4. K-means clustering on asthma participants.
analyses in asthma [5,6,12]. The current study may help define asthma in young allergic adults. Difficulties arise when comparing the findings of our cohort with other studies, due to low resemblance in variable selections. Trends in clustering are observed with the Severe Asthma Research Program (SARP) [10] (cohort age average of 37 years). The youngest group in SARP and our study is characterized with relatively high atopy, while the older groups are characterized by lower baseline lung function. A recent study on two independent Korean cohorts [5] presented clusters that are different compared to our study, even if variables in both analyses were similar (age, smoking status, atopy). Consensus between studies is needed to identify the best combination of variables to classify asthma patients in homogeneous and clinically relevant subgroups. Heterogeneity being a key component of asthma, phenotype alone may well be insufficient to properly describe the disease. Recent progresses in genetics of asthma, as discussed below, offer the possibility to include genetic and molecular data in multivariate phenotype of asthma and will need to be considered in future studies.
Understanding the genetic component of asthma is challenging [25]. Recent GWA studies on asthma have discovered susceptibility genes robustly associated with the disease [23,26-40]. However, this approach is powered to detect mostly common genetic variants, underestimating rare ones [41]. More comprehensive genetic studies are needed to identify the missing heritability of asthma including the rare variants [42] and structural variations [43]. In addition, targeted, exome, and wholegenome sequencing are increasingly applied to refine GWAS-nominated loci or to discover new genetic variants involved in complex diseases. Again, significant progress in this area is needed in the field of asthma. Technological advances and new genomic approaches offer great promise for improving health through personalized medicine [44,45]. However, none of the genomic approach described above are feasible without a large dataset of patients highly characterized for the disease under investigation. The most promising and upcoming genomic applications require increasingly large sample size and sufficient quantity of DNA. Existing cohorts to study the genetics of asthma have a finite amount of DNAs and more efforts are needed to collect new subjects and to expand existing cohorts for continuing progresses in the field. The ongoing collection of the Quebec City Case-Control Asthma Cohort aims at providing such resource in the field of asthma. The establishment of this resource is a major part of our genetic research program and will be the essence of the following studies that we are planning on the genetics of asthma. We hope that this cohort will contribute in multi-center efforts intent to understand the molecular basis of asthma and welcome collaborations for replicating genetic findings.
With the recent advances in genomic technologies, the bottleneck of genetic research has shifted from our capacity to measure DNA to the availability of a large number of patients well-characterized for the disease under study. This need was one of the primary rationales to initiate the Quebec City Case-Control Asthma Cohort. DNAs from this cohort will be studied using contemporary and upcoming genomic approaches to elucidate the genetic architecture of asthma. These include the study of rare and structural genetic variants and more comprehensive genomic analyses such as exome and whole-genome sequencing. Application of these molecular approaches to subgroups of phenotypically similar asthmatics described in this manuscript is required to fully understand the genetic susceptibility to asthma and to ensure pointof-care implementation of personalized medicine.
5. Acknowledgements
We thank the research subjects for participating in this study. The authors would like to recognize the contribution of devoted research nurses and staff that collected the Quebec City Case-Control Asthma Cohort including Johane Lepage, Joanne Milot, Hélène Villeneuve, Francine Deschesnes, Marie-Ève Boulay, and Hélène Turcotte. We are also grateful to the dedicated research staff at the Respiratory Health Network Tissue Bank for storing of biological materials and constantly updating the clinical database including Christine Racine, Sabrina Biardel, Marie-Christine Allard, and Geneviève Guèvremont. Finally, we are thankful for the support and help of Nathalie Gaudreault, Laura Sbarra and Cyndi Henry for extracting the DNA and preparing the samples for genetic/genomic applications.
The collection of the Quebec City Case-Control Asthma Cohort was supported by grants from the Chaire de pneumologie de la Fondation JD Bégin de l’Université Laval, the Fondation de l’Institut universitaire de cardiologie et de pneumologie de Québec, and the Respiratory Health Network of the FRQS. Y. Bossé is a research scholar from the Heart and Stroke Foundation of Canada.
REFERENCES
- M. F. Moffatt, I. G. Gut, F. Demenais, et al., “A LargeScale, Consortium-Based Genomewide Association Study of Asthma,” New England Journal of Medicine, Vol. 363, No. 13, 2010, pp. 1211-1221. http://dx.doi.org/10.1056/NEJMoa0906312
- D. G. Torgerson, E. J. Ampleford, G. Y. Chiu, et al., “Meta-Analysis of Genome-Wide Association Studies of Asthma in Ethnically Diverse North American Populations,” Nature Genetics, Vol. 43, No. 9, 2011, pp. 887- 892. http://dx.doi.org/10.1038/ng.888
- S. E. Wenzel, “Asthma: Defining of the Persistent Adult Phenotypes,” Lancet, Vol. 368, No. 9537, 2006, pp. 804-813. http://dx.doi.org/10.1016/S0140-6736(06)69290-8
- J. Kiley, R. Smith and P. Noel, “Asthma Phenotypes,” Current Opinion in Pulmonary Medicine, Vol. 13, No. 1, 2007, pp. 19-23.
- T. B. Kim, A. S. Jang, H. S. Kwon, et al., “Identification of Asthma Clusters in Two Independent Korean Adult Asthma Cohorts,” The European Respiratory Journal: Official Journal of the European Society for Clinical Respiratory Physiology, Vol. 41, No. 6, 2013, pp. 1308-1314.
- P. Haldar, I. D. Pavord, D. E. Shaw, et al., “Cluster Analysis and Clinical Asthma Phenotypes,” American Journal of Respiratory and Critical Care Medicine, Vol. 178, No. 3, 2008, pp. 218-224. http://dx.doi.org/10.1164/rccm.200711-1754OC
- P. Shirtcliffe, M. Weatherall, J. Travers and R. Beasley, “The Multiple Dimensions of Airways Disease: Targeting Treatment to Clinical Phenotypes,” Current Opinion in Pulmonary Medicine, Vol. 17, No. 2, 2011, pp. 72-78. http://dx.doi.org/10.1097/MCP.0b013e328341f181
- V. Siroux, X. Basagana, A. Boudier, et al., “Identifying Adult Asthma Phenotypes Using a Clustering Approach,” The European Respiratory Journal: Official Journal of the European Society for Clinical Respiratory Physiology, Vol. 38, No. 2, 2011, pp. 310-317. http://dx.doi.org/10.1183/09031936.00120810
- M. Amelink, S. B. de Nijs, J. C. de Groot, et al., “Three Phenotypes of Adult-Onset Asthma,” Allergy, Vol. 68, No. 5, 2013, pp. 674-680. http://dx.doi.org/10.1111/all.12136
- A. M. Fitzpatrick, W. G. Teague, D. A. Meyers, et al., “Heterogeneity of Severe Asthma in Childhood: Confirmation by Cluster Analysis of Children in the National Institutes of Health/National Heart, Lung, and Blood Institute Severe Asthma Research Program,” The Journal of Allergy and Clinical Immunology, Vol. 127, No. 2, 2011, pp. 382-389.
- J. Just, R. Gouvis-Echraghi, S. Rouve, et al., “Two Novel, Severe Asthma Phenotypes Identified during Childhood Using a Clustering Approach,” The European Respiratory Journal: Official Journal of the European Society for Clinical Respiratory Physiology, Vol. 40, No. 1, 2012, pp. 55-60. http://dx.doi.org/10.1183/09031936.00123411
- M. Weatherall, J. Travers, P. M. Shirtcliffe, et al., “Distinct Clinical Phenotypes of Airways Disease Defined by Cluster Analysis,” The European Respiratory Journal: Official Journal of the European Society for Clinical Respiratory Physiology, Vol. 34, No. 4, 2009, pp. 812-818.
- Society AT, “Standardization of Spirometry, 1994 Update. American Thoracic Society,” American Journal of Respiratory and Critical Care Medicine, Vol. 152, No. 3, 1995, pp. 1107-1136. http://dx.doi.org/10.1164/ajrccm.152.3.7663792
- R. Crapo, R. Casaburi, A. Coates, et al., “Guidelines for Methacholine and Exercise Challenge Testing—1999: This Official Statement of the American Thoracic Society Was Adopted by the ATS Board of Directors, July 1999,” American Journal of Respiratory and Critical Care Medicine, Vol. 161, No. 1, 2000, pp. 309-329.
- R. Knudson, M. Lebowitz, C. Holberg and B. Burrows, “Changes in the Normal Maximal Expiratory Flow-Volume Curve with Growth and Aging,” American Review of Respiratory Disease, Vol. 127, No. 6, 1983, pp. 725-734.
- E. Juniper, P. Frith, C. Dunnet, D. Cockcroft and F. Hargreave, “Reproducibility and Comparison of Response to Inhaled Histamine and Methacholine,” Thorax, Vol. 33, No. 6, 1978, pp. 705-710. http://dx.doi.org/10.1136/thx.33.6.705
- M. D. Lougheed, C. Lemiere, S. D. Dell, et al., “Canadian Thoracic Society Asthma Management Continuum— 2010 Consensus Summary for Children Six Years of Age and over, and Adults,” Canadian Respiratory Journal, Vol. 17, No. 1, 2010, pp. 15-24.
- Immunology EAoAaC, “Skin Tests Used in Type I Allergy Testing Position Paper. Sub-Committee on Skin Tests of the European Academy of Allergology and Clinical Immunology,” Allergy, Vol. 44, Suppl. 10, 1989, pp. 1-59.
- B. S. Everitt, S. Landau, M. Leese and D. Stahl, “Hierarchical Clustering,” Cluster Analysis, John Wiley & Sons, Ltd., pp. 71-110.
- W. C. Moore, D. A. Meyers, S. E. Wenzel, et al., “Identification of Asthma Phenotypes Using Cluster Analysis in the Severe Asthma Research Program,” American Journal of Respiratory and Critical Care Medicine, Vol. 181, No. 4, 2010, pp. 315-323. http://dx.doi.org/10.1164/rccm.200906-0896OC
- T. S. Lapperre, J. B. Snoeck-Stroband, M. M. Gosman, et al., “Dissociation of Lung Function and Airway Inflammation in Chronic Obstructive Pulmonary Disease,” American Journal of Respiratory and Critical Care Medicine, Vol. 170, No. 5, 2004, pp. 499-504. http://dx.doi.org/10.1164/rccm.200401-112OC
- E. R. Rosi, M. C. Grazzini, M. Duranti, R. Scano and G. Scano, “Sputum Analysis, Bronchial Hyperresponsiveness, and Airway Function in Asthma: Results of a Factor Anlaysis,” Journal of Allergy and Clinical Immunology, Vol. 103, No. 2, 1999, pp. 232-237. http://dx.doi.org/10.1016/S0091-6749(99)70496-3
- E. Noguchi, H. Sakamoto, T. Hirota, et al., “GenomeWide Association Study Identifies HLA-DP as a Susceptibility Gene for Pediatric Asthma in Asian Populations,” PLOS Genetics, Vol. 7, No. 7, 2011, Article ID: e1002170. http://dx.doi.org/10.1371/journal.pgen.1002170
- E. R. Sutherland, E. Goleva, T. S. King, et al., “Cluster Analysis of Obesity and Asthma Phenotypes,” PLoS One, Vol. 7, No. 5, 2012, Article ID: e36631. http://dx.doi.org/10.1371/journal.pone.0036631
- Y. Bosse and T. J. Hudson, “Toward a Comprehensive Set of Asthma Susceptibility Genes,” Annual Review of Medicine, Vol. 58, 2007, pp. 171-184. http://dx.doi.org/10.1146/annurev.med.58.071105.111738
- B. E. Himes, G. M. Hunninghake, J. W. Baurley, et al., “Genome-Wide Association Analysis Identifies PDE4D as an Asthma-Susceptibility Gene,” American Journal of Human Genetics, Vol. 84, No. 5, 2009, pp. 581-593. http://dx.doi.org/10.1016/j.ajhg.2009.04.006
- D. F. Gudbjartsson, U. S. Bjornsdottir, E. Halapi, et al., “Sequence Variants Affecting Eosinophil Numbers Associate with Asthma and Myocardial Infarction,” Nature Genetics, Vol. 41, No. 3, 2009, pp. 342-347. http://dx.doi.org/10.1038/ng.323
- C. Ober, Z. Tan, Y. Sun, et al., “Effect of Variation in CHI3L1 on Serum YKL-40 Level, Risk of Asthma, and Lung Function,” New England Journal of Medicine, Vol. 358, No. 16, 2008, pp. 1682-1691. http://dx.doi.org/10.1056/NEJMoa0708801
- S. Weidinger, C. Gieger, E. Rodriguez, et al., “GenomeWide Scan on Total Serum IgE Levels Identifies FCER1A as Novel Susceptibility Locus,” PLoS Genet, Vol. 4, No. 8, 2008, Article ID: e1000166. http://dx.doi.org/10.1371/journal.pgen.1000166
- S. Choudhry, M. Taub, R. Mei, et al., “Genome-Wide Screen for Asthma in Puerto Ricans: Evidence for Association with 5q23 Region,” Human Genetics, Vol. 123, No. 5, 2008, pp. 455-468. http://dx.doi.org/10.1007/s00439-008-0495-7
- A. T. DeWan, E. W. Triche, X. Xu, et al., “PDE11A Associations with Asthma: Results of a Genome-Wide Association Scan,” The Journal of Allergy and Clinical Immunology, Vol. 126, No. 4, 2010, pp. 871-873.
- D. B. Hancock, I. Romieu, M. Shi, et al., “Genome-Wide Association Study Implicates Chromosome 9q21.31 as a Susceptibility Locus for Asthma in Mexican Children,” PLoS Genet, Vol. 5, No. 8, 2009, Article ID: e1000623. http://dx.doi.org/10.1371/journal.pgen.1000623
- J. Hui, A. Oka, A. James, et al., “A Genome-Wide Association Scan for Asthma in a General Australian Population,” Human Genetics, Vol. 123, No. 3, 2008, pp. 297- 306. http://dx.doi.org/10.1007/s00439-008-0477-9
- S. H. Kim, B. Y. Cho, C. S. Park, et al., “Alpha-T-Catenin (CTNNA3) Gene Was Identified as a Risk Variant for Toluene Diisocyanate-Induced Asthma by Genome-Wide Association Analysis,” Clinical and Experimental Allergy: Journal of the British Society for Allergy and Clinical Immunology, Vol. 39, No. 2, 2009, pp. 203-212. http://dx.doi.org/10.1111/j.1365-2222.2008.03117.x
- R. A. Mathias, A. V. Grant, N. Rafaels, et al., “A Genome-Wide Association Study on African-Ancestry Populations for Asthma,” The Journal of Allergy and Clinical Immunology, Vol. 125, No. 2, 2010, pp. 336-346.
- X. Li, T. D. Howard, S. L. Zheng, et al., “Genome-Wide Association Study of Asthma Identifies RAD50-IL13 and HLA-DR/DQ Regions,” The Journal of Allergy and Clinical Immunology, Vol. 125, No. 2, 2010, pp. 328-335.
- M. F. Moffatt, M. Kabesch, L. Liang, et al., “Genetic Variants Regulating ORMDL3 Expression Contribute to the Risk of Childhood Asthma,” Nature, Vol. 448, No. 7152, 2007, pp. 470-473. http://dx.doi.org/10.1038/nature06014
- P. M. Sleiman, J. Flory, M. Imielinski, et al., “Variants of DENND1B Associated with Asthma in Children,” New England Journal of Medicine, Vol. 362, No. 1, 2010, pp. 36-44. http://dx.doi.org/10.1056/NEJMoa0901867
- T. Hirota, A. Takahashi, M. Kubo, et al., “Genome-Wide Association Study Identifies Three New Susceptibility Loci for Adult Asthma in the Japanese Population,” Nature Genetics, Vol. 43, No. 9, 2011, pp. 893-896. http://dx.doi.org/10.1038/ng.887
- M. A. Ferreira, M. C. Matheson, D. L. Duffy, et al., “Identification of IL6R and Chromosome 11q13.5 as Risk Loci for Asthma,” Lancet, Vol. 378, No. 9795, 2011, pp. 1006-1014. http://dx.doi.org/10.1016/S0140-6736(11)60874-X
- W. Y. Wang, B. J. Barratt, D. G. Clayton and J. A. Todd, “Genome-Wide Association Studies: Theoretical and Practical Concerns,” Nature Reviews, Vol. 6, No. 2, 2005, pp. 109-118. http://dx.doi.org/10.1038/nrg1522
- D. G. Torgerson, D. Capurso, R. A. Mathias, et al., “Resequencing Candidate Genes Implicates Rare Variants in Asthma Susceptibility,” American Journal of Human Genetics, Vol. 90, No. 2, 2012, pp. 273-281. http://dx.doi.org/10.1016/j.ajhg.2012.01.008
- M. A. Ferreira, A. F. McRae, S. E. Medland, et al., “Association between ORMDL3, IL1RL1 and a Deletion on Chromosome 17q21 with Asthma Risk in Australia,” European Journal of Human Genetics, Vol. 19, No. 4, 2011, pp. 458-464. http://dx.doi.org/10.1038/ejhg.2010.191
- E. D. Green and M. S. Guyer, “National Human Genome Research I. Charting a Course for Genomic Medicine from Base Pairs to Bedside,” Nature, Vol. 470, No. 7333, 2011, pp. 204-213. http://dx.doi.org/10.1038/nature09764
- M. A. Hamburg and F. S. Collins, “The Path to Personalized Medicine,” New England Journal of Medicine, Vol. 363, No. 4, 2010, pp. 301-304. http://dx.doi.org/10.1056/NEJMp1006304
NOTES
*Corresponding author.

