Advances in Infectious Diseases
Vol.4 No.1(2014), Article ID:43310,7 pages DOI:10.4236/aid.2014.41001
Characteristics, Treatment, and Outcomes Associated with Clostridium difficile Associated Diarrhea in a Veterans Affairs Medical Center
David B. Huang1*, Han Lee2, Tom Chiang1
1Division of Infectious Diseases, Department of Medicine, Rutgers New Jersey Medical School, Newark, USA
2Department of Internal Medicine, Yale University School of Medicine, New Haven, USA
Email: *dhuang82@hotmail.com
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/


Received 16 November 2013; revised 16 December 2013; accepted 23 December 2013
ABSTRACT
We conducted a study to assess the characteristics, treatment and outcomes associated with Clostridium difficile associated diarrhea in a Veterans Affairs Medical Center. Fifty-eight consecutive individual cases of C. difficile infection in 2013 were observed within the Veterans Affairs New Jersey Health Care System (VA NJHCS). We molecularly typed all 58 individual strains and identified the associated characteristics, treatment and outcomes. Forty-four out of 58 specimens (76%) which were probed had characteristics of the epidemic strain BI/NAP1/027 making this virulent strain to be the predominate strain at the VA NJHCS. All C. difficile BI/NAP1/027 strains were resistant to fluoroquinolones and sensitive to fidaxomicin, metronidazole and vancomycin. Fidaxomicin had the most potent in vitro activity (MIC90 = 0.5 µg/ml) against the BI/NAP1/027 strain. Twenty-six of 44 patients (59%) with the virulent strain were from a long-term care facility (LTCF). Patients possessing the virulent strain from the LTCF had a mean APACHE II score of 14.1 and a predicted death rate of 21.9%. Two-third of patients were treated with metronidazole alone (mean APACHE II scores 9.6), and one-third required oral vancomycin and metronidazole (mean APACHE II scores 14.1). There were no C. difficile infection related deaths. C. difficile BI/NAP1/027, an epidemic strain, is the endemic strain at the VA NJHCS, but no increased mortality was seen with infection with this strain.
Keywords:Clostridium difficile; BI/NAP1/027; Treatment; Outcomes

1. Introduction
Clostridium difficile infection, caused by anaerobic Gram-positive bacteria, is a potential serious and life-- threatening cause of nosocomial diarrhea [1] [2] . C. difficile infection is the most frequent cause of healthcareassociated infectious diarrhea in hospitals and long-term health care facilities (LTCF) in industrialized countries. In the United States, C. difficile affects more than 300,000 hospitalized patients, approximately 14,000 people die each year, and it is estimated that C. difficile infection causes $3.2 billion in annual health care costs [3] . Prior antibiotic usage, especially broad-spectrum antibiotics (e.g., fluoroquinolones, cephalosporins, carbapenems and clindamycin), is a risk factor for developing C. difficile infection [4] -[6] . Broad-spectrum antibiotic usage eradicates the normal gut flora causing overpopulation with C. difficile and subsequent release of toxins that can cause diarrhea (~3 to 15 bowel movements per day), pseudomembranous colitis and toxic megacolon, and death.
In the last decade, there has been an increase of C. difficile infections across North America [7] -[10] . In 2005, a new toxic strain of C. difficile, resistant to fluoroquinolone antibiotics, was identified as a cause of geographically dispersed outbreaks accounting for the increase in C. difficile infections [8] [11] . Molecular analysis identified the epidemic strain of C. difficile as BI by restriction endonuclease analysis, North American pulse-fieldtype (NAP1) by pulse-field gel electrophoresis, and ribotype 027 by polymerase chain reaction [9] [12] [13] . This strain is referred to C. difficile BI/NAP1/027. This strain carries an 18-bp deletion of the tcdC suppressor gene which is thought to be a negative regulator of toxin A and B production [9] . The BI/NAP1/027 strain produces significantly greater quantities of toxins A and B, and it is more resistant to fluoroquinolones. It can cause more refractory disease. It is more likely to relapse (20% - 30%) and is reported to be associated with higher rates of morbidity and mortality [8] [9] [12] [14] -[16] . Also associated with this strain is a binary toxin gene which is thought to be associated with the virulence of this strain of C. difficile [8] [9] [12] [14] -[16] . This epidemic strain of C. difficile has been linked with more severe symptoms and disease progression of C. difficile infection such as colitis, toxic megacolon, increased intensive care unit admissions, prolonged hospitalizations and death [7] [8] [12] [14] [15] [17] .
The outbreaks described in Quebec and the United States have been found to be largely due to the same virulent strain of C. difficile [1] [7] -[9] [12] [13] [15] [16] . Over the past several years, there has been an increase in rates of C. difficile infections within the Veterans Affairs New Jersey Health Care System (VA NJHCS) similar to what is being seen throughout Northern America.
We followed a prospective cohort of 58 consecutive individual cases of C. difficile infection in 2013 within the VA NJHCS and then identified characteristics, treatment and outcomes of these infections.
2. Materials and Methods
2.1. Surveillance Definitions, Patient Characteristics and Outcome Measures
This study was Institutional Review Board (IRB)-approved. C. difficile-associated diarrhea (CDAD) was defined by the presence of diarrhea and a positive assay for C. difficile toxin A, toxin B, or both; by the sudden onset of diarrhea with no alternative explanation and a diagnosis of pseudomembranous colitis on the basis of endoscopy; or by histologic evidence of the condition. A case was considered nosocomial if symptoms started 72 hours or more after a patient was admitted or if CDAD was diagnosed within one month after a previous admission. An episode was considered new if it occurred more than eight weeks after a previous diagnosis of CDAD.
Acute Physiology and Chronic Health Evaluation (APACHE) II scores were recorded on the day of the diarrheal illness and stool collection. APACHE II is a severity of disease classification system consisting of an integer score from 0 to 71 based on 12 routine physiological measurements; higher scores correspond to more severe disease and a higher risk of death. Predicted death rates were obtained from APACHE II scores.
2.2. Detection of C. difficile Toxin, Binary Toxin Genes, and Partial Deletions of the tcdC Gene
Isolation and identification were performed as previously described [8] . Fifty-eight consecutive individual stool samples that tested positive for C. difficile underwent susceptibility testing. Testing of C. difficile isolates for susceptibility to fidaxomicin, rifaximin, rifampin, ciprofloxacin, gatifloxacin, levofloxacin, clarithromycin, clindamycin, ceftriaxone, meropenem, piperacillin-tazobactam, metronidazole, and vancomycin was performed according to the Clinical Laboratory Standards Institute agar dilution method for susceptibility testing of anaerobes [18] .
PFGE of C. difficile isolates was performed according to the method described by Fawley and Wilcox [19] . The relatedness of the various isolates was determined according to the criteria of Tenover [20] and with use of Molecular Analyst software (Bio-Rad).
Analyses for binary toxin genes (cdtA and cdtB) and partial tcdC deletions were performed as described elsewhere [20] . C. difficile (CIP 107932) was used as the positive control for binary toxin genes and an 18-bp tcdC deletion, and C. difficile (ATCC 43255) served as the negative control.
2.3. Statistical Analyses
Descriptive data were processed using MS Excel (Microsoft, Redmond, Washington) and STATA version 8.2 (College Station, Texas). Statistical analyses were performed using Chi-square or Fisher’s exact test for categorical data and ANOVA for continuous variables. Data were examined in a two-tailed fashion to estimate the P value. P values of <0.05 were considered significant.
3. Results
3.1. Demographics
In 2013, 58 consecutive cases of CDAD positive cases were reviewed (Table 1). Thirty-two (55%) patients were from the hospital and 26 (45%) were from the LTCF. The average age of patients from the hospital versus the LTCF was 63 and 69 years, respectively. Eighty-six percent of the patients were male. Among patients from the hospital, the average number of inpatient days prior to diagnosis was 11. All 58 patients with infections due to C. difficile were molecularly probed.
3.2. Antibiotic Usage
Of the 58 patients, 56 had a prior history of antibiotic usage in the past 90 days (Table 2). These patients received antibiotics either as an inpatient or after discharge from the hospital within 90 days of being C. difficile positive. Fifty-four percent of patients received a β-lactam, 46% received a fluoroquinolone, and 7% received a macrolide.
3.3. Molecular Typing
Forty-four of the 58 specimens (76%) probed had characteristics of the BI/NAP1/027 strain (possessing the binary toxin genes, ribotype 27, and an 18 base pair deletion of the tcdC suppressor region) making the virulent strain previously seen in Quebec the predominate strain at the VA NJHCS. Twenty-six of the 44 patients (59%) with the virulent strain were from the LCTF.
3.4. Antimicrobial Susceptibility
All isolates were susceptible to fidaxomicin, rifaximin, rifampin, metronidazole and vancomycin, with minimal 90 percent inhibitory concentrations (MIC90) of 0.5, 4.0, 4.0, 0.5 and 1.0 μg per milliliter, respectively. All BI/NAP1/027 strains (n = 44) were resistant to ciprofloxacin, moxifloxacin, gatifloxacin, and levofloxacin (minimal inhibitory concentrations of at least 32 μg per milliliter) and susceptible to clindamycin (Table 3).
3.5. APACHE II Scores and Predicted Death Rates (PDR)
The mean APACHE II score for the BI/NAP1/027 strain was 13.2 with a PDR of 19%. For the strain carrying the 36 base pair deletion at the tcdC region and the binary toxin the mean APACHE II scores was 7.5 with a PDR of 8.6%. For the non virulent non ribotype 027 which lacked the binary toxin and had no deletions in the tcdC region the mean APACHE II score was 11 and PDR was 13.4% (BI/NAP1/027 vs. non ribotype 027, P < 0.05). Patients possessing the virulent strain from the LTCF had a mean APACHE II score of 14.1 and a PDR 21.9%. While patients from the hospital with the virulent strain had a mean APACHE II score of 11 and a PDR of 15.1% (LTCF vs. Hospital, P < 0.05).

Table 1. Demographic characteristics of 58 consecutive cases of Clostridium difficile associated diarrhea.
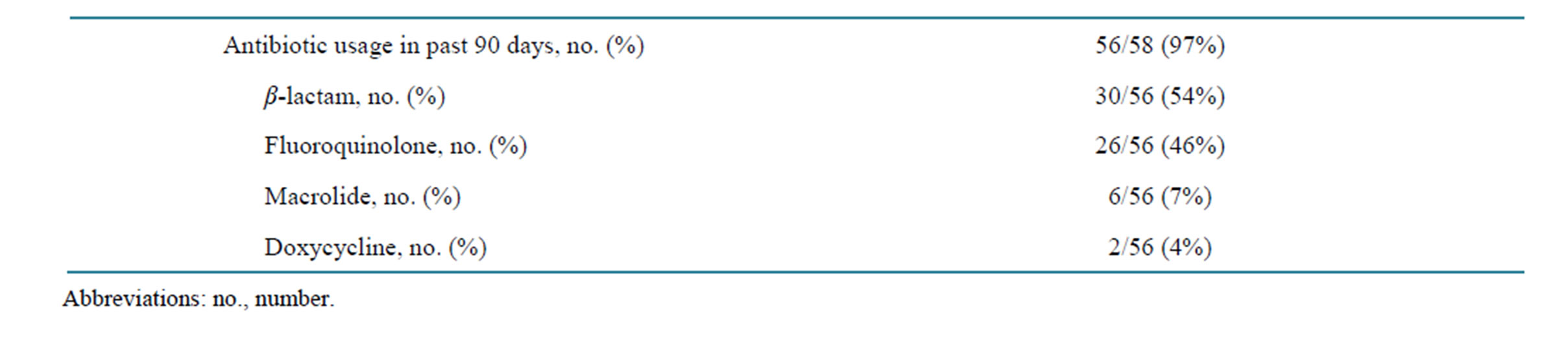
Table 2. Antibiotic usage of the 58 consecutive cases of Clostridium difficile associated diarrhea.
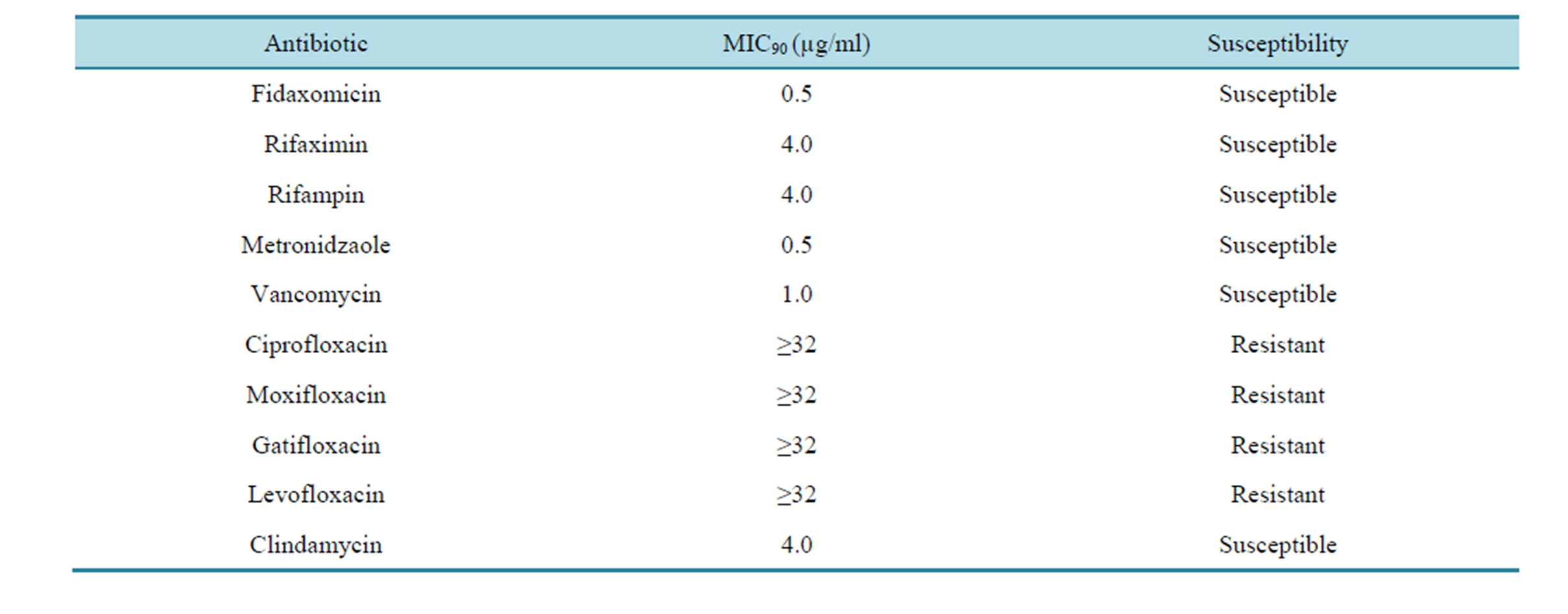
Table 3. Antibiotic MIC90 of C. difficile strain BI/NAP1/027 that were molecularly typed.
3.6. Treatments and Outcomes
Thirty-eight patients were treated with metronidazole alone (mean APACHE II scores 9.6) and 20 patients required oral vancomycin and metronidazole (mean APACHE II scores 14.1) due to inadequate response from metronidazole alone (metronidazole vs. vancomycin and metronidazole, P < 0.05). Six patients infected with BI/NAP1/027 developed complications with four developing colitis and two requiring a colectomy secondary to toxic megacolon. None of the 14 patients infected with non BI/NAP1/027 strain developed any complications (BI/NAP1/027 vs. non BI/NAP1/027, P < 0.05). There were no C. difficile infection related deaths.
4. Discussion
Our results show that the predominant strain in the VA NJHCS is the BI/NAP1/027, toxinotype III strain. This strain is the epidemic strain linked with the rise of CDAD associated morbidity throughout North America [9] and Quebec [8] . A 2007 Department of Health and Senior Services study which did a retrospective survey of 58 New Jersey hospitals found that there was an increase in CDAD cases per 1000 admissions from 3.3 in 2001 to 7.7 in 2004 showing greater than a 2 fold increase in this time period [21] . This may be associated to the epidemic strain though no molecular studies were conducted in the survey.
Antibiotic usage, including but not limited to clindamycin, β-lactams, β-lactamase inhibitors and fluoroquinolones, is known to be associated with increased risk for acquisition of C. difficile infection [22] -[24] . In our patient population, the predominant antibiotic received was a β-lactam antibiotic (54%). This practice of increased β-lactam usage was noted in a prior C. difficile infection study conducted in our VA in 2001 [25] . The epidemic strain has been found to be resistant to fluoroquinolones [1] [8] [9] [12] [26] and fluoroquinolone usage has been shown to be a strong risk factor for C. difficile infection [1] [8] [24] [26] -[28] . Forty six percent of our patients received fluoroquinolones. A study from the Atlanta, Georgia and the Hines, Illinois VA Medical center found increased rates of C. difficile infection in the VA health care system with the use of fluoroquinolones [9] [29] [30] .
The toxinotype III, ribotype 027 epidemic strain is the predominant strain of C. difficile in our Veterans Affairs (VA) patient population. Fifty five percent of our patients with the virulent strain were from the hospital and 45% were from LTCF. Four patients with BI/NAP1/027 strains were from nursing homes and had no recent hospitalizations. This finding is concerning and reflects a need for better infection control policies within our LTCF.
Comparing the three toxinotypes, the BI/NAP1/027 strain predictably had a high mean APACHE II score (13.2) and Predicted Death rate (PDR) (19%). Mean APACHE II scores and PDR were higher for LTCF patients with the BI/NAP1/027 strain [mean APACHE II score (14.1)/PDR (21.9%)] than those from the hospital with the same strain [mean APACHE II score (11.0)/PDR (15.1%)] (LTCF vs. Hospital, P < 0.05). The higher mean APACHE II scores and PDR among LTCF patients compared to hospital patients is likely because LTCF residents have increased underlying morbidities, as indicated by the higher Charlson Comorbidity Index, when they develop CDAD [8] [9] .
The binary toxin has been present in the epidemic strain though its exact role is not well defined [8] [9] [12] [14] -[16] . A 2005 study found a close though not statistically significant association between the presence of the binary toxin gene and more severe cases of C. difficile associated diarrhea [15] . The BI/NAP1/027 strain also has been associated with 18 base pair deletions in the tcdC region thought to be a negative regulator of toxin A and B production [9] and a single nucleotide deletion of position 117 which causes a frame shift mutation of the tcdC gene truncating it [31] [32] . Spigaglia et al. showed that variations in the tcdC may affect toxin production differently [33] . A study from the Netherlands had a C. difficile strain which had a 44 base pair deletion of the tcdC region suggesting that different mutations of the tcdC region may affect functionality and toxin production [34] .
We demonstrated that the epidemic ribotype 027/toxinotype III strain is the predominate strain within our VA population. However, infection with this strain did not result in increased mortality. All of these strains were fluoroquinolone resistant and sensitive to metronidazole and vancomycin. In this study, fidaxomicin, the most recently FDA approved treatment for C. difficile infection had the most potent in vitro activity against the BI/NAP1/027 strain. There were no BI/NAP1/027 strains that had a fidaxomicin or metronidazole MIC value of ˃ 0.5 μg/ml in this study. The MIC values for fidaxomicin were not affected by the gene profiles of the BI/NAP1/027 strain. In this study, another RNA polymerase inhibitor, rifaximin, which has been used for C. difficile infection was found to have higher MIC90 values for the BI/NAP1/027 strains. Despite fidaxomicin and rifaximin both being RNA polymerase inhibitors, no evidence were found for cross resistance of rifaximin and fidaxomicin. This is not unexpected given that these two transcriptional inhibitors interact with different regions of RNA polymerase. As expected, all BI/NAP1/027 strains were resistant to moxifloxacin, ciprofloxacin and levofloxacin.
5. Conclusion
In conclusion, the epidemic strain of C. difficile BI/NAP1/027 is now endemic at the VA NJHCS and its associated LTCF. Patients from the LTCF with BI/NAP1/027 had increased APACHE II scores compared to those from the hospital. This strain is susceptible to fidaxomicin, metronidazole and vancomycin. Of concern was that four patients with the epidemic strain from our LTCF had no recent hospitalizations. With such high morbidity, strict infection control measures and antibiotic restrictions especially the β-lactam class may reduce further spread of this virulent strain in this VA population.
Conflict of Interests and Financial Disclosures
None of the authors have any conflict of interests or financial disclosures to report.
Acknowledgements
None of the authors have any acknowledgements to report.
References
- Kazakova, S.V., Ware, K., Baughman, B., Bilukha, O., Paradis, A. and Sears, S. (2006) A Hospital Outbreak of Diarrhea Due to an Emerging Epidemic Strain of Clostridium difficile. Archives of Internal Medicine, 166, 2518-2524. http://dx.doi.org/10.1001/archinte.166.22.2518
- Johnson, S. and Gerding, D.N. (1998) Clostridium difficile-Associated Diarrhea. Clinical Infectious Diseases, 26, 1027-1034. http://dx.doi.org/10.1086/520276
- Treating Disease with Microbes: Bugs in the System. The Economist 2012.
- Baxter, R., Ray, G.T. and Fireman, B.H. (2008) Case-Control Study of Antibiotic Use and Subsequent Clostridium difficile-Associated Diarrhea in Hospitalized Patients. Infection Control and Hospital Epidemiology, 29, 44-50. http://dx.doi.org/10.1086/524320
- Gifford, A.H. and Kirkland, K.B. (2006) Risk Factors for Clostridium difficile-Associated Diarrhea on an Adult Hematology-Oncology Ward. European Journal of Clinical Microbiology and Infectious Diseases, 25, 751-755. http://dx.doi.org/10.1007/s10096-006-0220-1
- Palmore, T.N., Sohn, S., Malak, S.F., Eagan, J. and Sepkowitz, K.A. (2005) Risk Factors for Acquisition of Clostridium difficile-Associated Diarrhea among Outpatients at a Cancer Hospital. Infection Control and Hospital Epidemiology, 26, 680-684. http://dx.doi.org/10.1086/502602
- Pepin, J., Valiquette, L., Alary, M.E., Villemure, P., Pelletier, A. and Forget, K. (2004) Clostridium difficile-Associated Diarrhea in a Region of Quebec from 1991 to 2003: A Changing Pattern of Disease Severity. Canadian Medical Association Journal, 171, 466-472. http://dx.doi.org/10.1503/cmaj.1041104
- Loo, V.G., Poirier, L., Miller, M.A., Oughton, M., Libman, M.D., Michaud, S., Bourgault, A.M., Nguyen, T., Frenette, C., Kelly, M., Vibien, A., Brassard, P., Fenn, S., Dewar, K., Hudson, T.J., Horn, R., Rene, P., Monczak, Y. and Dascal, A. (2005) A Predominantly Clonal Multi-Institutional Outbreak of Clostridium difficile-Associated Diarrhea with High Morbidity and Mortality. New England Journal of Medicine, 353, 2442-2449. http://dx.doi.org/10.1056/NEJMoa051639
- McDonald, L.C., Killgore, G.E., Thompson, A., Owens Jr., R.C., Kazakova, S.V., Sambol, S.P., Johnson, S. and Gerding, D.N. (2005) An Epidemic, Toxin Gene-Variant Strain of Clostridium difficile. New England Journal of Medicine, 353, 2433-2441. http://dx.doi.org/10.1056/NEJMoa051590
- McDonald, L.C. and Jernigan, D.B. (2004) Increasing Incidence of Clostridium difficile in U.S. Acute Care Hospitals 1993-2001. Proceedings of 14th Annual Scientific Meeting of the Society for Healthcare Epidemiology of America, Philadelphia.
- McDonald, L.C. (2005) Clostridium difficile: Responding to a New Threat from an Old Enemy. Infection Control and Hospital Epidemiology, 26, 672-675. http://dx.doi.org/10.1086/502600
- Bartlett, J.G. (2006) Narrative Review: The New Epidemic of Clostridium difficile-Associated Enteric Disease. Annals of Internal Medicine, 145, 758-764. http://dx.doi.org/10.7326/0003-4819-145-10-200611210-00008
- Hubert, B., Loo, V.G., Bourgault, A.M., Poirier, L., Dascal, A., Fortin, E., Dionne, M. and Lorange, M. (2007) A Portrait of the Geographic Dissemination of the Clostridium difficile North American Pulsed-Field Type 1 Strain and the Epidemiology of C. difficile-Associated Disease in Quebec. Clinical Infectious Diseases, 44, 238-244. http://dx.doi.org/10.1086/510391
- Cloud, J. and Kelly, C.P. (2007) Update on Clostridium difficile Associated Disease. Current Opinions in Gastroenterology, 23, 4-9.
- McEllistrem, M.C., Carman, R.J., Gerding, D.N., Genheimer, C.W. and Zheng, L. (2005) A Hospital Outbreak of Clostridium difficile Disease Associated with Isolates Carrying Binary Toxin Genes. Clinical Infectious Diseases, 40, 265-272. http://dx.doi.org/10.1086/427113
- Warny, M., Pepin, J., Fang, A., Killgore, G., Thompson, A., Brazier, J., Frost, E. and McDonald, L.C. (2005) Toxin Production by an Emerging Strain of Clostridium difficile Associated with Outbreaks of Severe Disease in North America and Europe. Lancet, 366, 1079-1084. http://dx.doi.org/10.1016/S0140-6736(05)67420-X
- Rubin, M.S., Bodenstein, L.E. and Kent, K.C. (1995) Severe Clostridium difficile Colitis. Diseases of the Colon and Rectum, 38, 350-354. http://dx.doi.org/10.1007/BF02054220
- Clinical and Laboratory Standards Institute (2004) Methods for Antimicrobial Susceptibility Testing of Anaerobic Bacteria. 6th Edition, Clinical and Laboratory Standards Institute, Wayne.
- Fawley, W.N. and Wilcox, M.H. (2002) Pulsed-Field Gel Electrophoresis Can Yield DNA Fingerprints of Degradation-Susceptible Clostridium difficile Strains. Journal of Clinical Microbiology, 40, 3546-3547. http://dx.doi.org/10.1128/JCM.40.9.3546-3547.2002
- Tenover, F.C., Arbeit, R.D., Goering, R.V., Mickelsen, P.A., Murray, B.E., Persing, D.H. and Swaminathan, B. (1995) Interpreting Chromosomal DNA Restriction Patterns Produced by Pulsed-Field Gel Electrophoresis: Criteria for Bacterial Strain Typing. Journal of Clinical Microbiology, 33, 2233-2239.
- Tan, E.T., Robertson, C.A., Brynildsen, S., Bresnitz, E., Tan, C. and McDonald, C. (2007) Clostridium difficile-Associated Disease in New Jersey Hospitals, 2000-2004. Emerging Infectious Diseases, 13, 498-500. http://dx.doi.org/10.3201/eid1303.060294
- Harbarth, S., Samore, M.H. and Carmeli, Y. (2001) Antibiotic Prophylaxis and the Risk of Clostridium difficile-Associated Diarrhoea. Journal of Hospital Infections, 48, 93-97. http://dx.doi.org/10.1053/jhin.2001.0951
- Kuntz, J.L., Cavanaugh, J.E., Becker, L.K., Ward, M.A., Appelgate, D.M., Herwaldt, L.A. and Polgreen, P.M. (2007) Clostridium difficile-Associated Disease in Patients in a Small Rural Hospital. Infection Control and Hospital Epidemiology, 28, 1236-1239. http://dx.doi.org/10.1086/521662
- Muto, C.A., Pokrywka, M., Shutt, K., Mendelsohn, A.B., Nouri, K., Posey, K., Roberts, T., Croyle, K., Krystofiak, S., Patel-Brown, S., Pasculle, A.W., PAterson, D.L., Saul, M. and HArrison, L.H. (2005) A Large Outbreak of Clostridium difficile-Associated Disease with an Unexpected Proportion of Deaths and Colectomies at a Teaching Hospital Following Increased Fluoroquinolone Use. Infection Control and Hospital Epidemiology, 26, 273-280. http://dx.doi.org/10.1086/502539
- Mody, L.R., Smith, S.M. and Dever, L.L. (2001) Clostridium difficile-Associated Diarrhea in a VA Medical Center: Clustering of Cases, Association with Antibiotic Usage, and Impact on HIV-Infected Patients. Infection Control and Hospital Epidemiology, 22, 42-45. http://dx.doi.org/10.1086/501825
- McCusker, M.E., Harris, A.D., Perencevich, E. and Roghmann, M.C. (2003) Fluoroquinolone Use and Clostridium difficile-Associated Diarrhea. Emerging Infectious Diseases, 9, 730-733. http://dx.doi.org/10.3201/eid0906.020385
- Carroll, D.N. (2003) Moxifloxacin-Induced Clostridium difficile-Associated Diarrhea. Pharmacotherapy, 23, 1517- 1519. http://dx.doi.org/10.1592/phco.23.14.1517.31936
- Gaynes, R., Rimland, D., Killum, E., Lowery, H.K., Johnson, T.M., Killgore, G. andTenover, F.C. (2004) Outbreak of Clostridium difficile Infection in a Long-Term Care Facility: Association with Gatifloxacin Use. Clinical Infectious Diseases, 38, 640-645. http://dx.doi.org/10.1086/381551
- Changela, U., Cannon, J.P., Aneziokoro, C., Shah, P.S., Thottapurathu, L. and Lentino, J. (2004) Risk Factors and Mortality Associated with Clostridium difficile-Associated Diarrhoea at a VA Hospital. International Journal of Antimicrobial Agents, 24, 562-656. http://dx.doi.org/10.1016/j.ijantimicag.2004.07.011
- Chang, H.T., Krezolek, D., Johnson, S., Parada, J.P., Evans, C.T. and Gerding, D.N. (2007) Onset of Symptoms and Time to Diagnosis of Clostridium difficile-Associated Disease Following Discharge from an Acute Care Hospital. Infection Control and Hospital Epidemiology, 28, 926-931. http://dx.doi.org/10.1086/519178
- Curry, S.R., Marsh, J.W., Muto, C.A., O’Leary, M.M., Pasculle, A.W. and Harrison, L.H. (2007) tcdC Genotypes Associated with Severe TcdC Truncation in an Epidemic Clone and Other Strains of Clostridium difficile. Journal of Clinical Microbiology, 45, 215-221. http://dx.doi.org/10.1128/JCM.01599-06
- MacCannell, D.R., Louie, T.J., Gregson, D.B., Laverdiere, M., Labbe, A.C., Laing, R. and Henwick, S. (2006) Molecular Analysis of Clostridium difficile PCR Ribotype 027 Isolates from Eastern and Western Canada. Journal of Clinical Microbiology, 44, 2147-2152. http://dx.doi.org/10.1128/JCM.02563-05
- Spigaglia, P. and Mastrantonio, P. (2002) Molecular Analysis of the Pathogenicity Locus and Polymorphism in the Putative Negative Regulator of Toxin Production (TcdC) among Clostridium difficile Clinical Isolates. Journal of Clinical Microbiology, 40, 3470-3475. http://dx.doi.org/10.1128/JCM.40.9.3470-3475.2002
NOTES

*Corresponding Author.

