Open Journal of Medicinal Chemistry
Vol.3 No.2(2013), Article ID:33154,9 pages DOI:10.4236/ojmc.2013.32006
Molecular Recognition of Human Telomeric DNA by Phenanthroline-Based G-Quadruplex Ligands*
1Dipartimento di Scienze della Salute, Università degli Studi “Magna Græcia”, Campus “S. Venuta”, Catanzaro, Italy
2Department of Pharmaceutical and Pharmacological Sciences, University of Padova, Padova, Italy
Email: #alcaro@unicz.it
Copyright © 2013 Anna Artese et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received April 20, 2013; revised May 18, 2013; accepted June 2, 2013
Keywords: phenanthroline; docking; G-quadruplex
ABSTRACT
G-quadruplexes (G4) are non-canonical DNA structures assumed by guanine rich sequences. G4 are stabilized by the presence of cations and are characterized by a high degree of structural polymorphism with different patterns of groove, loop arrangement, strand orientations and stoichiometry. G-rich sequences are over-represented in the promoter regions of many oncogenes as well as at human telomeres, d(TTAGGG) repeats, ranging in size from 3 to 15 kb, involved in protecting chromosomal ends. A specialized enzyme, called telomerase, provides a telomere maintenance mechanism by elongating the end of the G-strand and it is activated in the majority of cancer cells. Therefore there are two general strategies of telomerase targeting in cancer treatment. One is a direct targeting of telomerase to cause its inhibition; the other one is the use of G4 stabilizers which block telomerase access to telomere, thus causing an indirect enzyme inhibition. Here, we evaluated the molecular recognition of some phenanthroline-based ligands against four different experimental models of the human telomeric sequence d[AG3(T2AG3)3] by means of docking simulations. Our theoretical analysis was able to reproduce the experimental affinity measurements, with a linear squared correlation factor r2 equal to 0.719 among all the studied models. These findings highlighted the importance to consider the polymorphism of the DNA G4. Interestingly, this correlation resulted always improved with respect to that of the single folds, with the exception of the parallel structure, thus suggesting a key role of this G4 conformation in the interaction network of the tested binders. Moreover, we identified the moieties of the phenanthroline scaffold directly involved in the complex formation. This allowed to rationalize the improved binding affinity always associated with a bis-phenanthroline system and to explain why a phenanthroline substituted with a pyridine ring is favored with respect to the pyrimidine one.
1. Introduction
Telomeres are specialized structures at the end of human chromosomes that cap their ends and protect them from end-to-end fusions, degradation, and genetic instability [1]. Human and vertebrate telomeres consist of highly conserved tandem repeats of the hexanucleotide d (TTAGGG)n 5 - 10 kb in length, ending in a singlestranded G rich 3’-overhang [2-5]. To maintain proper function of telomeres, the 150 - 250 nucleotide-long single stranded G-rich 3’ overhang forms higher order structures like, a T-loop, and binds to a nuclear-protein complex [6]. Under normal aging conditions, telomeres become shorter at each cell division [7]. When a critical amount of telomere shortening has occurred, the genetic program of cell senescence, or cell ageing, is triggered. This event can be overcome by activation of telomerase, a reverse transcriptase which elongates telomere terminals. Telomerase activity in normal human cells is generally undetectable leading to successive telomere shortening with each cell division, which ultimately limits their proliferative capacity in vitro and in vivo [8]. The enzyme is active only in immortal cells like germ line cells [9], embryonic stem cells [10], and 90% of all tumor cells in humans [11]. For this reason, telomerase can be a potentially highly selective and attractive drug target for anticancer strategies. In addition, the induction of a telomere structure not recognized by the telomerase can indirectly target the enzyme resulting in inhibition of its catalytic activity. This working model is based on the G-rich character of telomeres. It has been experimentally confirmed that they can fold into peculiar structures called G-quadruplexes (G4s) which are not processed by the enzyme. The G4 conformation, as the name suggests, has a core that is made up of guanine bases only, with four guanines arranged in a rotationally symmetric manner.
In addition to the human telomere, G4-forming sequences are over-represented in the promoter regions of many oncogenes, such as c-myc [12], c-kit [13], bcl-2 [14], VEGF [15], and HIF-1a [16], whereas the occurrence of G4-forming sequences in tumor suppressor genes tends to be much lower [14].
G-quadruplexes are characterized by a remarkable structural polymorphism and can be classified primarily based on strand stoichiometry. In particular tetramolecular G-quadruplexes, formed by four guanine-rich DNA or RNA sequences, adopt only a parallel conformation with all four strands in the same direction, appearing to be the least polymorphic [17,18]. Bimolecular G-quadruplexes, formed from the dimerization of two guanine-rich sequences, are quite diverse providing three possible conformations, one parallel and two antiparallel structures. Finally, unimolecular G-quadruplexes, formed from the folding of a single guanine rich sequence into a fourstranded quadruple helical structure with three connecting loops, are capable of adopting multiple topologies due to different glycosidic conformations of guanines, which in turn define specific patterns of groove, loop arrangement, strand orientations and stoichiometry [19]. The first human telomeric structure was solved using NMR spectroscopy in Na+ solution (PDB model 143D) [20]. It corresponds to an antiparallel G4 consisting of three stacked G-quartets with two lateral and a diagonal connecting loops. Subsequently, X-ray crystallography experiments showed that in K+ containing sample, the human telomeric sequence adopted a parallel topology (PDB model 1KF1) [21] consisting of three stacked G-quartets and three double chain-reversal loops. In addition to these conformations, two mixed topologies, called Form 1 and Form 2 which differ from each other only by the order of loop arrangements, were observed.
These folds are also defined as hybrid-1 (2GKU-5’- T2-core-A-3’, 2HY9-5’-A3-core-A2-3’, and 2JSM-5’-TAcore-3’) [22-24], and hybrid-2 (2JSL-5’-TAcore-T2-3’ 2JPZ-5’-T2A-core-T2-3’) [24,25].
A substantial effort has been made to identify synthetic and natural compounds able to lock telomeric DNA in a G4 conformation and thus impede telomere elongation in vivo. Most of known G4 stabilizers are characterized by an extended planar aromatic array, in which the π-delocalized system allows stacking interactions with the external guanine tetrads, and positively charged side chains, that enhance the ligand affinity by interacting with the negative phosphates of the DNA backbone [26,27]. Among them, the trisubstituted acridine BRACO-19, the polycyclic compound RHPS4 and the natural ligand telomestatin are three of the most commonly studied agents [28]. Nevertheless, there are several other classes of indirect telomerase inhibitors characterized by a remarkable chemical diversity, such as porphyrins, perylene diimides, fluoroquinolones, indoloquinolines, cryptolepines, quindolines, phenanthrolines, triazines, carbazole derivatives, ethidium derivatives, bisamido-anthraquinones, fluorenones, acridones and acridines [29]. Recently, a new class of naphthalene diimides (NDIs), capable of reversibly binding and subsequently alkylating telomeric DNA, has been identified [30].
Phenanthroline derivatives and pyridostatin have been used in several cell-culture studies, in which they have been demonstrated to bind within the cells and disrupt the expression of targeted DNA-quadruplex-forming sequences [31-33]. These compounds have also turned out to be excellent tools for assessing the biological relevance of quadruplexes in vivo [34,35].
An alternative approach to increase the planar stacking surface of a G4 binder comprises the coordination of planar ligands by transition metal ions. Indeed, several metal complexes have shown remarkable G4 recognition properties [36-38]. In previous works, we focused our attention on the G4 binding by phenanthroline-based derivatives. These studies confirmed that phenanthrolinemetal ion coordination can largely improve G4 recognition compared to the free ligands [39,40]. Interestingly, the nature of metal ion played a key role in driving such an effect. However, during this investigation, some ligands with interesting G4 affinity also in the absence of metal ion coordination have been identified [41]. To better understand the rationale behind these results, in this manuscript we analyzed a set of phenanthroline compounds in absence of coordinating metal ions by means of a docking approach and we related the experimental data acquired in solution to their theoretical molecular recognition with respect to four different receptor folds of the human telomeric sequence d[AG3(T2AG3)3]. Specifically, the aim of our study was to find a theoretical procedure able to reproduce the experimental affinity measurements in order to use it as predictive tool in drug design and lead optimization processes toward the G4 target. Our analysis allowed us to identify the key moieties of the phenanthroline scaffold, highlighting the optimal position for the positively charged basic side chain and possible structural modifications aimed to improve the affinity of these ligands versus G4 different folds.
2. Material and Methods
2.1. Analysis of Compounds 1-8
The phenanthroline-based G4 ligands 1-8 were built by means of Maestro GUI (Maestro Graphics User Interface) version 9.8 [42]. For all the studied analogues, the existence probability of diverse tautomeric and protomeric forms was evaluated at pH 7.4 and the most probable forms, reported in Table 1, were considered. All structures were submitted to 2000 iterations of full energy minimization adopting the Polake-Ribiere Conjugated Gradient (PRCG) algorithm and the “all atoms” notation of the OPLS_2005 force field [43].
Solvent effects were considered by adopting the implicit solvation model GB/SA water [44]. The optimization process was performed up to the derivative convergence criterion of 0.05 kcal Å-1·mol-1. All the calculations were computed by using the version 9.8 of the MacroModel software [45].
2.2. PDB Model Pre-Treatment
The PDB X-ray structure 1KF1 [21] and the NMR models 143D [20], 2HY9 [23] and 2JPZ [25], related to the telomeric sequence d[AG3(T2AG3)3], were downloaded from the Protein Data Bank [46]. Co-crystallized water molecules and counter ions were removed from the X-ray structure. In their sequences, the hybrid NMR structures 2HY9 and 2JPZ presented head and tail caps,

Table 1. 2D chemical structures of compounds 1-8 (Lig).
resulting both formed by 26-mer. Thus, to obtain a similar analysis with respect to the first two models, the hybrid PDB structures were modified by deleting these caps, that is, considering them as conformational templates for the canonical 22-mer d[AG3(T2AG3)3]. The 27 experimental conformations extracted from the four PDB models were energy-optimized exactly in the same conditions (force field, implicit solvation model, iterations and convergence criterion) adopted for the ligands. For each G4 fold, the lowest energy conformation was chosen as receptor for the next docking study (for details see Supporting Information Tables S1-S4).
2.3. Docking Simulations and Thermodynamic Evaluation
Docking simulations were performed using the ligand flexible algorithm of Glide [47] at Standard Precision (SP) level. The docking binding site was defined by means of a regular box, which included the whole receptor structure, with a volume of 125000 A3. Glide grid maps were computed using the standard precision algorithm. The optimized ligands were submitted to flexible Glide docking simulations, evaluating their recognition against all G4 conformations. The best poses complexed to all considered G4 structures were optimized by means of a full energy minimization procedure, carried out using the same force field and environment reported for the ligands optimization. Analysis of the results was carried out taking into account the thermodynamic estimate of the state equations (free energy, enthalpy, and entropy of complex formation) computed at 300 K (Table 2). The evaluation of hydrogen bonds (HBs) and VdW contacts was performed by means of Maestro graphical interface
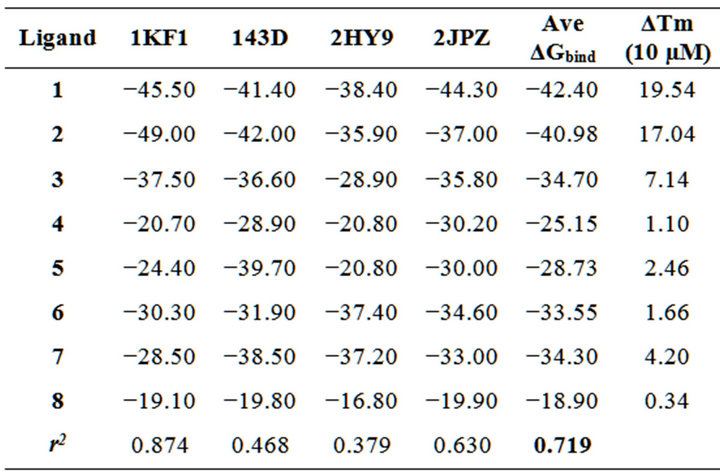
Table 2. Free energy of complexation, reported as ΔGbind and expressed in kcal mol-1, of compounds 1-8 against the PDB human telomeric sequence d[AG3(T2AG3)3] folds. Ave ΔGbind, ΔTm and r2 indicate, respectively, the average free energy of complexation considering all the G-quadruplex folds, the melting temperature shift measured at 10 µM and the linear squared regression coefficient.
(Maestro Graphics User Interface, version 9.8, Schrödinger, LLC).
2.4. Ligands
Stock solutions (4 mM) of 7 were prepared in deionized water; all other ligands were dissolved in DMSO. They were diluted to the appropriate concentration in the working buffer prior to use.
2.5. Fluorescence Melting Studies
Melting experiments were performed in a Roche Light Cycler, using an excitation source at 488 nm and recording the fluorescence emission at 520 nm. Target DNA was HTS (AGGGTTAGGGTTAGGGTTAGGGT, labelled with dabcyl and fluorescin at 5’ and 3’ end, respectively). It was synthesised and HPLC purified by Oswel Research Products Ltd. (Southampton, UK). Mixtures (20 mL) contained 0.25 mM of target DNA and variable concentrations of tested derivatives in 50 mM potassium buffer (10 mM LiOH; 50 mM KCl pH 7.4 with H3PO4). They were first denatured by heating to 95◦C for 5 min and then cooled to 30◦C at a rate of 0.5◦C min−1. Then, temperature was slowly increased (0.2◦C min−1) up to 90◦C and again lowered at the same rate to 30◦C. Recordings were taken during both these melting and annealing reactions to check for hysteresis. Tm values were determined from the first derivatives of the melting profiles using the Roche LightCycler software. Each curve was repeated at least three times and errors were ± 0.4◦C.
3. Results and Discussion
In our previous works we have investigated the G4 binding properties of two families of Phen derivatives. The first one comprises a Phen heterocycle with different substitution pattern on the aromatic system (compounds 6-8). For this group, an efficient G4 interaction was observed only when two ligands were accommodated around one metal center. We proposed that this complex most likely bound with an extended aromatic surface available for stacking interactions likely with the G-tetrads. To sustain this model, we subsequently examined a second series of derivatives in which two Phen moieties were covalently linked through an amine or thioether bond (compounds 1-3). In agreement with our prediction, these compounds were able to bind G4 structure even in the absence of coordinating-metal ions. Moreover, the simultaneous involvement of both Phen moieties in DNA recognition was suggested by the relevant efficiency in comparison to the mono-derivatives. These results are well described by monitoring the variation of the melting temperature of a DNA containing four repeats of the human telomeric sequence folded into a G4 conformation. Indeed, as reported in Figure 1, only derivatives 1-3 increased it to a remarkable extent (≈ 20◦C) in the low micromolar range.
Here, we applied the same experimental protocol to two novel compounds which are expected to fill the gap between the above described two series of derivatives. These are represented by compounds 4 and 5 which contain a pyridine or a pyrimidine linked to Phen by a thioether bond, the same linker which provided 1 with optimal G4 interaction properties. As a result, we assumed they can present to the DNA G-quartets an intermediated planar surface area. Interestingly, these compounds performed better than the leading Phen but worse than compounds 6 and 7. his is most likely due to the absence of protonable side chains in the novel compounds, thus suggesting that the partly increased stacking is not sufficient to overcome the loss of ionic interactions with the macromolecule.
Nevertheless, we cannot exclude that no favorable reciprocal orientation of the two aromatic systems is occurring.
To fully rationalize the binding energies involved in the recognition process, we have developed a computational protocol to theoretically generate and evaluate the molecular recognition of phenanthroline-based ligands against the G4 human telomeric repeated sequence. The computational approach assumes the target DNA in equilibrium among different known G4 folds. The configurational ensembles generated in the docking experiments were submitted to a full energy minimization procedure (for details see Material and methods), obtaining free energy estimations directly comparable to each other and to the experimental melting data.
Different conformations of the DNA human telomeric repeat sequence d[AG3(T2AG3)3] have been experiment tally determined. In order to consider its conformational
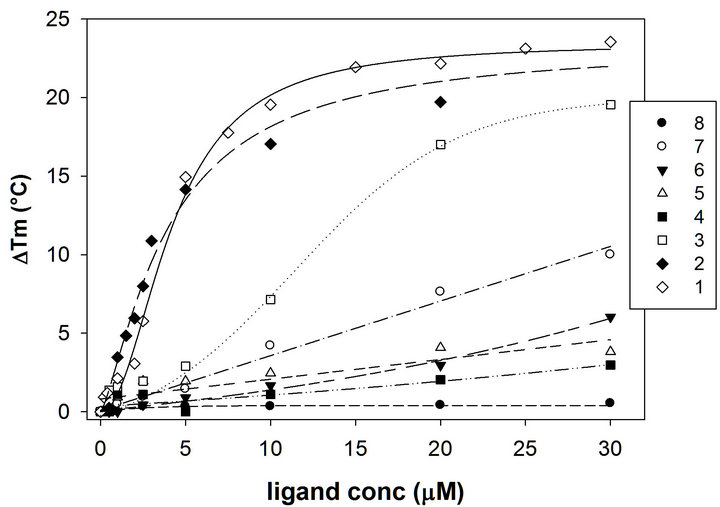
Figure 1. Variation of the melting temperature of the oligonucleotide HTS induced by increasing concentration of compounds 1-8 determined in 50 mM potassium buffer, pH 7.4.
polymorphism, we included in our study four PDB entries (codes 1KF1 [21], 143D [20], 2HY9 [23] and 2JPZ [25]) among Xray and NMR structures, using all the conformations stored in each experimental structure, as reported in our previous experiences [48-50]. After the geometric optimization procedure of the studied ligands, for each of them the energy minimized conformation was selected for the docking simulations. The obtained best pose of compounds 1-8 was fully optimized and thermodynamically estimated. Subsequently, in order to investigate the performance of our docking protocol with reference to the melting measurements, we correlated the theoretical binding affinities of the phenanthroline binders with their experimentally determined efficiencies, summarized as ΔTm (10 µM), which represent the variation of the melting temperature induced by a 10 mM concentration of tested ligands on the telomeric G4.
For the two data sets, we computed the linear squared correlation factor r2. In particular, as shown in Table 2, we obtained a significant correlation (r2 equal to 0.874) using as receptor the parallel fold and a moderate value was also associated to the hybrid-2 model 2JPZ (r2 equal to 0.630). The observation that the parallel and the mixed folds can give better correlation results is in agreement with the remark that the structure of telomeric G-quadruplexes in K+ solution is most important, since K+ is much more abundant than Na+ in cellular environments. Moreover recent studies highlighted the hybrid-type intramolecular G4 structures as the major conformations formed in human telomeric sequences in K+ solution, with a dynamic equilibrium between hybrid-1 and hybrid-2 conformations [22-25,51]. As indicated in Table 2, the average correlation between theoretical and experimental data yielded an r2 value equal to 0.719 among all the studied models, confirming the importance of considering the polymorphism of the DNA G4 when docking experiments are performed on this target. Interestingly, this correlation was always improved with respect to those of the single folds, with the exception of the 1KF1 model (r2 equal to 0.874), thus suggesting a key role of the parallel structure in the interaction network of 1-8 G-binders. As shown in Table 2, the presence of the bis-phenanthroline system was associated with an improved binding affinity, 1 and 2 being the most efficient in stabilizing G4. Interestingly, even if in 1 the amine side chain is substituted by a thioether linker, its recognition against all G4 fold was not disadvantaged. By analyzing their best poses toward the parallel telomeric structure, we observed they are both involved in end stacking interactions with the G4 core (Figure 2). Distinctly, the thermodynamic evaluation of 1 versus its simplified analogue 5 underlined a remarkably unfavorable profile for the latter. Such an observation could be justified by its reduced stacking interaction network, as
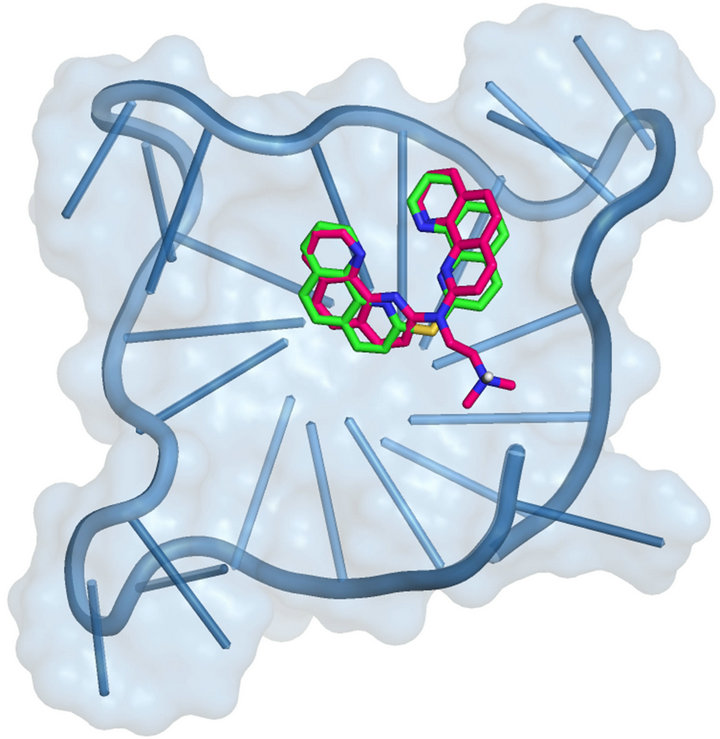
Figure 2. 1 and 2 best pose against 1KF1 PDB model obtained by Glide docking method ensemble optimization. 1 and 2 are indicated, respectively, as green and warm pink carbon stick representation. The DNA G4 is represented as sky blue transparent cartoon. Nonpolar hydrogen atoms are omitted for sake of clarity.
reported in Figure 3, where the smaller 5 was shown to fit into a lateral loop of the G4 parallel fold. The recognition of phenanthroline binders towards the hybrid-2 2JPZ fold indicated 1 as the best one in stabilizing such a structure. In particular, by comparing the poses of 1 and 2, we observed 1 involved in one hydrogen bond with adenine at position 7 and two with guanine at position 16.
By contrast 2 was able to establish only one hydrogen bond with G16, thus suggesting a somehow reduced binding affinity (Figure 4).
Interestingly, in order to rationalize the better binding affinity of 7 if compared to that of 6, we analyzed their optimized poses obtained in the recognition of the G4 antiparallel structure 143D, since docking results suggested this one as the most discriminated target. As reported in Figures 5, 7 resulted better embedded in the DNA structure, since it is accommodated in a kind of internal pocket and is involved in a wide stacking interactions network with the guanine core (at positions 2, 3, 14 and 15).
By contrast 6 is able to recognize only the bottom site of 143D model, making stacking interactions with the nucleobases T5, T6, A7, G8 and A19 and thus losing most of the stabilizing contacts with the guanine tetrad.
Such an observation could be justified by a reduced steric hindrance of 7 and by the different position of the charged basic side chain, that is favored when inserted at

Figure 3. 1 and 5 best pose against 1KF1 PDB model obtained by Glide docking method ensemble optimization. 1 and 5 are indicated, respectively, as green and magenta carbon stick representation. The DNA G4 is represented as sky blue transparent cartoon. Nonpolar hydrogen atoms are omitted for sake of clarity.
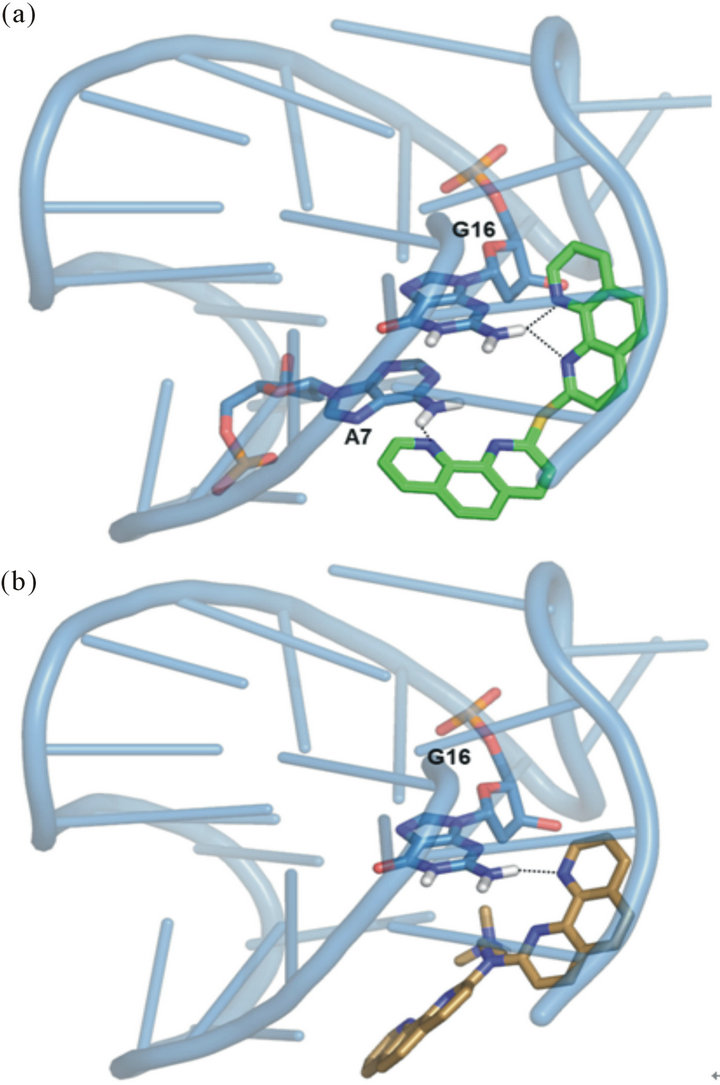
Figure 4. (a) 1 and (b) 2 best pose against 2JPZ PDB model obtained by Glide docking method ensemble optimization. 1 and 2 are indicated, respectively, as green and light orange carbon stick representation. The DNA G4 is represented as sky blue transparent cartoon. The nucleobases involved in hydrogen bonds, indicated as black dashed lines, are reported as slate carbon sticks. Nonpolar hydrogen atoms are omitted for sake of clarity.
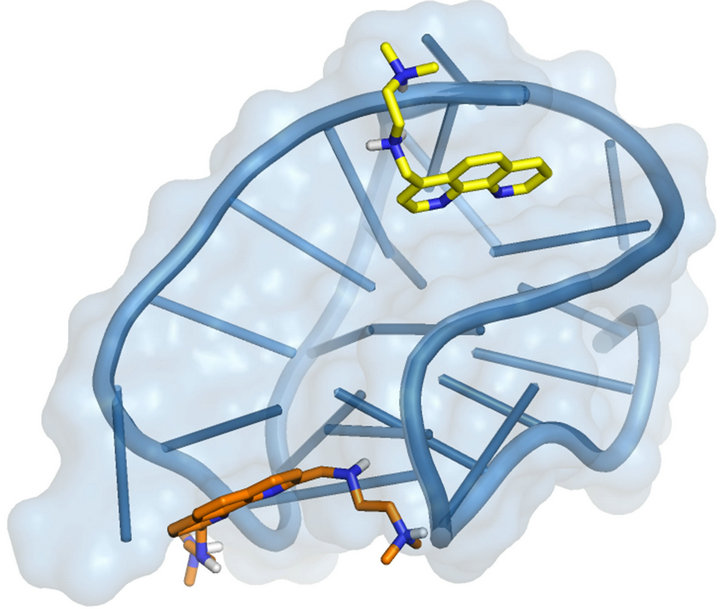
Figure 5. 6 and 7 best pose against 143D PDB model obtained by Glide docking method ensemble optimization. 6 and 7 are indicated, respectively, as orange and yellow carbon stick representation. The DNA G4 is represented as sky blue transparent cartoon. Nonpolar hydrogen atoms are omitted for sake of clarity.
position 4 of the phenanthroline scaffold if compared to positions 2 and 9. Analyzing the complexes of 4, 5 and 8, associated to the most unproductive thermodynamic profiles, we observed a remarkably decreased number of Van der Waals interactions of these ligands with all the G4 considered conformations (data not shown), according to their low ΔTm measurements.
Specifically 4 and 5 best poses against 2HY9 hybrid model, reported in Figure 1 of the Supplementary Information, were associated with the poorest recognition of such DNA conformation.
However 5 resulted related to a better thermodynamic profile in the recognition of the parallel and, in particular, of the antiparallel G4 structures if compared to 4, suggesting the favorable role of the pyridine ring with respect to the pyrimidine one.
4. Conclusions
In conclusion, in this manuscript the conformational polymorphism of the G4 DNA human telomeric repeat sequence has been considered analyzing X-ray and NMR PDB models. The set of G4 experimental models included four structures for a total of 54 target conformations.
Our docking analysis was able to reproduce the experimental affinity measurements, with a linear squared correlation factor r2 equal to 0.719 among all the studied models. This correlation was always improved with respect to that of the single folds, with the exception of the parallel structure, thus suggesting a key role of this G4 conformation in the interaction network of the studied tested binders.
The second step of our analysis will be the application of the same computational procedure to the studied phenanthroline compounds in presence of coordinating metal ions, with the aim of evaluating their role in the target recognition. Moreover, the obtained results suggested us to analyze some structural modifications onto the phenanthroline scaffold, such as the introduction of a second basic side chain at position 7 in ligand 7, in order to improve its G4 affinity, already favored with a single substitution, with respect to that of 6.
Finally, with the aim of enriching the theoreticalexperimental correlation factors, mo lecular dynamics simulations are currently under consideration in our laboratory and will be useful for the drug design of novel ligands toward specific G4 folds.
5. Acknowledgements
This work was funded by the Italian Ministry of Education projects PRIN 2009 (code 2009MFRKZ8), FIRB IDEAS (code RBID082ATK) and by University of Padova (grant # CPDA114388).
REFERENCES
- E. H. Blackburn, “Switching and Signaling at the Telomere,” Cell, Vol. 106, No. 6, 2001, pp. 661-673. doi:10.1016/S0092-8674(01)00492-5
- E. H. Blackburn and J. G. Gall, “A Tandemly Repeated Sequence at the Termini of Extrachromosomal Ribosomal RNA Genes in Tetrahymena,” Journal of Molecular Biology, Vol. 120, No. 1, 1978, pp. 33-53. doi:10.1016/0022-2836(78)90294-2
- R. C. Allshire, M. Dempster and N. D. Hastie, “Human Telomeres Contain at Least 3 Types of G-Rich Repeat Distributed Non-Randomly,” Nucleic Acids Research, Vol. 17, No. 12, 1989, pp. 4611-4627. doi:10.1093/nar/17.12.4611
- T. de Lange, L. Shiue, R. M. Myers, D. R. Cox, S. L. Naylor, et al., “Structure and Variability of Human Chromosome Ends,” Molecular and Cellular Biology, Vol. 10, No. 2, 1990, pp. 518-527. doi:10.1128/MCB.10.2.518
- R. K. Moyzis, J. M. Buckingham, L. S. Cram, M. Dani, L. L. Deaven, et al., “A Highly Conserved Repetitive DNA Sequence, (TTAGGG)n, Present at the Telomeres of Human Chromosomes,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 85, No. 18, 1988, pp. 6622-6626. doi:10.1073/pnas.85.18.6622
- Y. Xu, “Chemistry in Human Telomere Biology: Structure, Function and Targeting of Telomere DNA/RNA,” Chemical Society Reviews, Vol. 40, No. 5, 2011, pp. 2719- 2740. doi:10.1039/C0CS00134A
- H. Riethman, “Human Telomere Structure and Biology,” Annual Review of Genomics and Human Genetics, Vol. 9, 2008, pp. 1-19. doi:10.1146/annurev.genom.8.021506.172017
- C. B. Harley, A.B. Futcher and C. W. Greider, “Telomeres Shorten during Ageing of Human Fibroblasts,” Nature, Vol. 345, No. 6274, 1990, pp. 458-460. doi:10.1038/345458a0
- R. C. Allsopp, H. Vaziri, C. Patterson, S. Goldstein, E. V. Younglai, et al., “Telomere Length Predicts Replicative Capacity of Human Fibroblasts,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 89, No. 21, 1992, pp. 10114-10118. doi:10.1073/pnas.89.21.10114
- J. A. Thomson, J. Itskovitz-Eldor, S. S. Shapiro, M. A. Waknitz, J. J. Swiergiel, et al., “Embryonic Stem Cell Lines Derived from Human Blastocysts,” Science, Vol. 282, No. 5391, 1998, pp. 1145-1147. doi:10.1126/science.282.5391.1145
- N. W. Kim, M. A. Piatyszek, K. R. Prowse, C. B. Harley, M. D. West, et al., “Specific Association of Human Telomerase Activity with Immortal Cells and Cancer,” Science, Vol. 266, No. 5193, 1994, pp. 2011-2015. doi:10.1126/science.7605428
- A. Ambrus, D. Chen, J. X. Dai, R. A. Jones and D. Z. Yang, “Solution Structure of the Biologically Relevant g-Quadruplex Element in the Human c-MYC Promoter. Implications for G-Quadruplex Stabilization,” Biochemistry, Vol. 44, No. 6, 2005, pp. 2048-2058. doi:10.1021/bi048242p
- S. T. Hsu, P. Varnai, A. Bugaut, A. P. Reszka, S. Neidle, et al., ”A G-Rich Sequence within the c-Kit Oncogene Promoter Forms a Parallel G-Quadruplex Having Asymmetric G-Tetrad Dynamics,” Journal of the American Chemical Society, Vol. 131, No. 37, 2009, pp. 13399-13409. doi:10.1021/ja904007p
- J. Dai, D. Chen, R. A. Jones, L. H. Hurley and D. Yang, “NMR Solution Structure of the Major G-Quadruplex Structure Formed in the Human BCL2 Promoter Region,” Nucleic Acids Research, Vol. 34, No. 18, 2006, pp. 5133- 5144. doi:10.1093/nar/gkl610
- D. Y. Sun, K. X. Guo, J. J. Rusche and L. H. Hurley, “Facilitation of a Structural Transition in the Polypurine/ Polypyrimidine Tract within the Proximal Promoter Region of the Human VEGF Gene by the Presence of Potassium and G-Quadruplex-Interactive Agents,” Nucleic Acids Research, Vol. 33, No. 18, 2005, pp. 6070-6080. doi:10.1093/nar/gki917
- R. De Armond, S. Wood, D. Y. Sun, L. H. Hurley and S. W. Ebbinghaus, “Evidence for the Presence of a Guanine Quadruplex Forming Region within a Polypurine Tract of the Hypoxia Inducible Factor 1 Alpha Promoter,” Biochemistry, Vol. 44, No. 49, 2005, pp. 16341-16350. doi:10.1021/bi051618u
- G. Laughlan, A. I. Murchie, D. G. Norman, M. H. Moore, P. C. Moody, et al., “The High Resolution Crystal Structure of a Parallel-Stranded Guanine Tetraplex,” Science, Vol. 265, No. 5171, 1994, pp. 520-524. doi:10.1126/science.8036494
- K. Phillips, Z. Dauter, A. I. Murchie, D. M. Lilley and B. Luisi, “The Crystal Structure of a Parallel Stranded Guanine Tetraplex at 0.95Å Resolution,” Journal of molecular biology, Vol. 273, No. 1, 1997, pp. 171-182. doi:10.1006/jmbi.1997.1292
- M. Adrian, B. Heddi and A. T. Phan, “NMR Spectroscopy of G-Quadruplexes,” Methods, Vol. 57, No. 1, 2012, pp. 11-24. doi:10.1016/j.ymeth.2012.05.003
- Y. Wang and D. J. Patel, “Solution Structure of the Human Telomeric Repeat d[AG3(T2AG3)3] G-tetraplex,” Structure, Vol. 1, No. 4, 1993, pp. 263-282. doi:10.1016/0969-2126(93)90015-9
- G. N. Parkinson, M. P. H. Lee and S. Neidle, “Crystal Structure of Parallel Quadruplexes from Human Telomeric DNA,” Nature, Vol. 417, No. 6891, 2002, pp. 876- 880. doi:10.1038/nature755
- K. N. Luu, A. T. Phan, V. Kuryavyi, L. Lacroix and D. J. Patel, “Structure of the Human Telomere in K+ Solution: an Intramolecular (3+1) G-Quadruplex Scaffold,” Journal of the American Chemical Society, Vol. 128, No. 30, 2006, pp. 9963-9970. doi:10.1021/ja062791w
- J. Dai, C. Punchihewa, A. Ambrus, D. Chen, R. A. Jones, et al., “Structure of the Intramolecular Human Telomeric G-Quadruplex in Potassium Solution: A Novel Adenine Triple Formation,” Nucleic Acids Research, Vol. 35, No. 7, 2007, pp. 2440-2450. doi:10.1093/nar/gkm009
- A. T. Phan, V. Kuryavyi, K. N. Luu and D. J. Patel “Structure of Two Intramolecular G-Quadruplexes Formed by Natural Human Telomere Sequences in K+ Solution,” Nucleic Acids Research, Vol. 35, No. 19, 2007, 6517- 6525. doi:10.1093/nar/gkm706
- J. Dai, M. Carver, C. Punchihewa, R. A. Jones and D. Yang, “Structure of the Hybrid-2 Type Intramolecular Human Telomeric G-Quadruplex in K+ Solution: Insights into Structure Polymorphism of the Human Telomeric Sequence,” Nucleic Acids Research, Vol. 35, No. 15, 2007, pp. 4927-4940. doi:10.1093/nar/gkm522
- D. Monchaud and M. P. Teulade-Fichou, “A Hitchhiker’s Guide to G-Quadruplex Ligands,” Organic & Biomolecular Chemistry, Vol. 6, No. 4, 2008, pp. 627-636. doi:10.1039/B714772B
- S. Neidle, “Human Telomeric G-Quadruplex: The Current Status of Telomeric G-Quadruplexes as Therapeutic Targets in Human Cancer,” The FEBS Journal, Vol. 277, No. 5, 2010, pp. 1118-1125. doi:10.1111/j.1742-4658.2009.07463.x
- M. Ruden and N. Puri, “Novel Anticancer Therapeutics Targeting Telomerase,” Cancer Treatment Reviews, Vol. 39, No. 5, 2013, pp. 444-456. doi:10.1016/j.ctrv.2012.06.007
- A. De Cian, L. Lacroix, C. Dourre, N. Temime-Smaali, C. Trentesaux, et al., “Targeting Telomeres and Telomerase,” Biochimie, Vol. 90, No. 1, 2008, pp. 131-155. doi:10.1016/j.biochi.2007.07.011
- F. Doria, M. Nadai, M. Folini, M. Di Antonio, L. Germani, et al., “Hybrid Ligand-Alkylating Agents Targeting Telomeric G-Quadruplex Structures,” Organic & Biomolecular Chemistry, Vol. 10, No. 14, 2012, pp. 2798- 2806. doi:10.1039/C2OB06816H
- R. Rodriguez, S. Müller, J. A. Yeoman, C. Trentesaux, J. F. Riou, et al., “A Novel Small Molecule That Alters Shelterin Integrity and Triggers a DNA-Damage Response at Telomeres,” Journal of the American Chemical Society, Vol. 130, No. 47, 2008, pp. 15758-15759. doi:10.1021/ja805615w
- A. De Cian, E. DeLemos, J. L. Mergny, M. P. TeuladeFichou and D. Monchaud “Highly Efficient G-Quadruplex Recognition by Bisquinolinium Compounds,” Journal of the American Chemical Society, Vol. 129, No. 7, 2007, pp. 1856-1857. doi:10.1021/ja067352b
- R. Halder, J. F. Riou, M. P. Teulade-Fichou, T. Frickey and J. Hartig “Bisquinolinium Compounds Induce Quadruplex-Specific Transcriptome Changes in HeLa S3 Cell Lines,” BMC Research Notes, Vol. 5, 2012, 138. doi:10.1186/1756-0500-5-138
- S. Müller, S. Kumari, R. Rodriguez and S. Balasubramanian, “Small-Molecule-Mediated G-Quadruplex Isolation from Human Cells,” Nature Chemistry, Vol. 2, No. 12, 2010, pp. 1095-1098. doi:10.1038/nchem.842
- R. Rodriguez, K. M. Miller, J. V. Forment, C. R. Bradshaw, M. Nikan, et al., ”Small-Molecule-Induced DNA Damage Identifies Alternative DNA Structures in Human Genes,” Nature chemical biology, Vol. 8, No. 3, pp. 301- 310. doi:10.1038/nchembio.780
- J. E. Reed, A. J. White, S. Neidle and R. Vilar, “Effect of Metal Coordination on the Interaction of Substituted Phenanthroline and Pyridine Ligands with Quadruplex DNA,” Dalton Transactions, No. 14, 2009, pp. 2558-2568. doi:10.1039/B820086F
- J. E. Reed, A. A. Arnal, S. Neidle and R. Vilar, “Stabilization of G-Quadruplex DNA and Inhibition of Telomerase Activity by Square-Planar Nickel(II) Complexes,” Journal of the American Chemical Society, Vol. 128, No. 18, 2006, pp. 5992-5993. doi:10.1021/ja058509n
- D. Monchaud, P. Yang, L. Lacroix, M. P. Teulade-Fichou and J. L. Mergny, “A Metal-Mediated Conformational Switch Controls G-Quadruplex Binding Affinity,” Angewandte Chemie, Vol. 120, No. 26, 2008, pp. 4858-4861. doi:10.1002/ange.200800468
- C. Musetti, L. Lucatello, S. Bianco, A. P. Krapcho, S. A. Cadamuro, et al., “Metal Ion-Mediated Assembly of Effective Phenanthroline-Based G-Quadruplex Ligands,” Dalton Transactions, No. 19, 2009, pp. 3657-3660. doi:10.1039/B904630P
- C. Musetti, A. P. Krapcho, M. Palumbo and C. Sissi, “Effect of G-Quadruplex Polymorphism on the Recognition of Telomeric DNA by a Metal Complex,” Plos One, Vol. 8, No. 3, 2013. doi:10.1371/journal.pone.0058529
- S. Bianco, C. Musetti, A. Waldeck, S. Sparapani, J. D. Seitz, et al., “Bis-phenanthroline Derivatives as Suitable Scaffolds for Effective G-Quadruplex Recognition,” Dalton Transactions, Vol. 39, No. 25, 2010, pp. 5833-5841. doi:10.1039/C0DT00038H
- Maestro Graphics User Interface, version 9.8, Schrö- dinger, LLC.
- J. L. Banks, H. S. Beard, Y. Cao, A. E. Cho, W. Damm, et al., “Integrated Modeling Program, Applied Chemical Theory (IMPACT),” Journal of Computational Chemistry, Vol. 26, No. 16, 2005, pp. 1752-1780. doi:10.1002/jcc.20292
- W. C. Still, A. Tempczyk, R. C. Hawley and T. Hendrickson, “Semianalytical Treatment of Solvation for Molecular Mechanics and Dynamics,” Journal of the America Chemical Society, Vol. 112, No. 16, 1990, pp. 6127-6129. doi:10.1021/ja00172a038
- F. Mohamadi, N. G. J. Richards, W. C. Guida, R. Liskamp, M. Lipton, et al., “Macromodel—An Integrated Software System for Modeling Organic and Bioorganic Molecules Using Molecular Mechanics,” Journal of Computational Chemistry, Vol. 11, No. 4, 1990, pp. 440-467. doi:10.1002/jcc.540110405
- The Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB). http://www.rcsb.org/pdb.
- R. A. Friesner, J. L. Banks, R. B. Murphy, T. A. Halgren, J. J. Klicic, et al., “Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy,” Journal of Medicinal Chemistry, Vol. 47, No. 7, 2004, pp. 1739-1749. doi:10.1021/jm0306430
- S. Alcaro, C. Musetti, S. Distinto, M. Casatti, G. Zagotto, et al., “Identification and Characterization of New DNA G-Quadruplex Binders Selected by a Combination of Ligand and Structure-Based Virtual Screening Approaches,” Journal of Medicinal Chemistry, Vol. 56, No. 3, 2013, pp. 843-855. doi:10.1021/jm3013486
- S. Alcaro, G. Costa, S. Distinto, F. Moraca, F. Ortuso, et al., “The Polymorphisms of DNA G-Quadruplex Investigated by Docking Experiments with Telomestatin Enantiomers,” Current Pharmaceutical Design, Vol. 18, No. 14, 2012, pp. 1873-1879. doi:10.2174/138161212799958495
- S. Alcaro, A. Artese, J. N. Iley, S. Missailidis, F. Ortuso, et al., “Rational Design, Synthesis, Biophysical and Antiproliferative Evaluation of Fluorenone Derivatives with DNA G-Quadruplex Binding Properties,” ChemMedChem, Vol. 5, No. 4, 2010, pp. 575-583. doi:10.1002/cmdc.200900541
- A. Ambrus, D. Chen, J. X. Dai, T. Bialis, R. A. Jones, et al., “Human Telomeric Sequence Forms a Hybrid-Type Intramolecular G-Quadruplex Structure with Mixed Parallel/Antiparallel Strands in Potassium Solution, ” Nucleic Acids Research, Vol. 34, No. 9, 2006, pp. 2723-2735. doi:10.1093/nar/gkl348
NOTES
*Supplementary information is available online.
#Corresponding author.

