World Journal of Neuroscience
Vol.3 No.1(2013), Article ID:27900,9 pages DOI:10.4236/wjns.2013.31001
ERβ interacting protein ERAP 140 shows age dependent alteration in its interaction and expression in male mouse brain
![]()
1Biochemistry and Molecular Biology Laboratory, Centre of Advanced Study, Department of Zoology, Banaras Hindu University, Varanasi, India
2Department of Zoology, Indira Gandhi National Tribal University, Amarkantak, India
Email: *mkt_bhu@yahoo.com
Received 8 July 2012; revised 9 August 2012; accepted 8 October 2012
Keywords: Estrogen Receptor β; Alpha Helices and Coils; Interacting Proteins; SOPMA; PSIPRED; Aging Brain
ABSTRACT
Estrogen mediates multiple functions in the brain through the interaction of estrogen receptor (ER)α and ERβ with a host of nuclear proteins that regulate specific gene transcription. We have identified ERAP 140, AIB 1, Trk A, Src, pCREB and CREB as ERβ interacting proteins in the mouse brain. Earlier we showed that the interaction of ERβ with ERAP 140 decreased whereas its expression increased with aging in the brain of female mice. Here we report that the pattern of interaction and expression is different in male mice as compared to females. The interaction of ERAP 140 with ERβ decreased in adult male mouse brain as compared to young and remained almost similar in old whereas its expression was higher in adult than young and old, which were almost similar. Further in silico secondary structure analysis by selfoptimized prediction method alignment (SOPMA) and PSIPRED revealed that ERβ interacting proteins were rich in alpha helices and coils. Such findings might help to design ER modulators which can regulate specific functions of estrogen in the brain during aging and degenerative diseases.
1. INTRODUCTION
Estrogen mediates its multiple functions in almost all reproductive and non-reproductive organs including the brain through its two well characterized estrogen receptor (ER)α and ERβ and their isoforms [1-3]. Both ERs are expressed in ovary, prostate, testis, lung, thymus, spleen and various regions of the brain [4]. ERα is involved in mainly the reproductive function whereas ERβ in various brain functions [5].
The ER mediated gene regulation is complex, particularly the involvement of interacting proteins in genomic and non-genomic pathways [6]. These proteins control the estrogen dependent transcriptional responses and are named coregulators. The study of coregulators is important because of the existence of receptor subtypes with differential tissue distribution, ligand responses and celldependent functions in brain. Many coregulators have been identified to affect the ER-mediated transcriptional response and this number is increasing [7,8]. Like other members of the nuclear receptor superfamily, ERβ contains three functional domains, namely, transactivation domain (TAD), DNA binding domain (DBD) and ligand binding domain (LBD) [9]. Coregulators bind to activation function (AF)-1 of TAD and AF-2 of LBD of ER, resulting in either stimulation (coactivators) or suppression (corepressors) of expression of specific genes [3, 10].
During aging, the brain undergoes a number of changes including a sharp decline in the level of estrogen, which in turn, affects the multitude of functions [11]. Also, the secondary structure of proteins is reported to change during aging [12]. The ERβ interacting proteins and their secondary structure in the brain are poorly studied. We have recently identified estrogen receptor associated protein (ERAP) 140 [13], amplified in breast cancer (AIB)1 [14], Trk A and Src [15], pCREB and CREB as interacting partners of ERβ in mouse brain [16]. ERAP140 is characterized as a conserved tissue specific nuclear receptor coactivator, abundantly expressed in brain, exclusively in neurons [17]. It interacts with ERα, thyroid receptor β and retinoic acid receptor α, and shows no sequence similarity with other coactivators, though its homologues are present in both invertebrates as well as vertebrates including human [18].
Another interacting protein is AIB1, a member of the nuclear p 160 steroid receptor co-activator (SRC) family [19], which was discovered independently by several groups and given various names; AIB1, ACTR (activator of thyroid hormone and retinoid receptor), RAC-3 (receptor associated co-activator-3), and TRAM-1 (thyroid hormone receptor activating molecule) [20]. It promotes the transcriptional activity of multiple nuclear receptors such as ER [21], and a number of other transcription factors [22]. Recently, we have reported the interaction of AIB1 with ERβTAD in mouse brain and variation in the level of its interaction and expression as a function of age in both sexes [14].
Other proteins interacting with ERβLBD in mouse brain are Trk A, Src, CREB and pCREB [23-25]. Trk A and Src are involved in various functions in brain and cancer [26]. CREB is expressed in the brain and regulates a wide range of biological functions including cellular growth, memory and neuronal proliferation in response to a variety of intracellular signaling events [24].
Here, we show alteration in the level of interaction of ERAP 140 with ERβ and its expression during aging in male mouse brain. Further, we predict the possible secondary structure of ERβ interacting proteins including ERAP 140 using the software SOPMA and PSIPRED, which showed richness in alpha helices and coils. Such study might help to design ER modulators which can regulate specific functions of estrogen in the brain during aging and degenerative diseases.
2. MATERIALS AND METHODS
2.1. Animals
Young (6 weeks), adult (25 weeks) and old (70 weeks) male mice of AKR strain maintained under controlled laboratory conditions were used for the experiments. They were used according to the strict guidelines of the Institutional Animal Ethical Committee, Banaras Hindu University, Varanasi, India. All animals were killed by cervical dislocation and the brain was pooled from three mice of each age group. The tissue was washed to remove the adhering blood and used to prepare the nuclear extract.
2.2. Preparation of Nuclei and Nuclear Extract
To identify the interacting proteins, the nuclei were prepared from the brain of young, adult and old male mice according to Burgoyne and Hewish [27]. Briefly, the brain was homogenized in solution A containing 0.34 M sucrose, 0.5 mM EGTA, 2 mM EDTA and 0.5% Triton X-100 in buffer A (60 mM KCl, 15 mM NaCl, 0.15 mM spermine, 0.5 mM spermidine, 14 mM β-mercaptoethanol, 15 mM HEPES, pH 7.5). The homogenate was kept on ice for 5 min, filtered and centrifuged at 5000 × g for 15 min at 4˚C. The resulting pellet was suspended in solution B containing 1.8 M sucrose, 0.5 mM EGTA and 0.2 mM EDTA in buffer A and layered over cushion of solution B (half the volume of solution A used in the first step). The tube containing sample was centrifuged at 50,000 × g for 1 h at 4˚C. The pellet containing clean nuclei was suspended in solution C containing 0.34 M sucrose in buffer A and observed under the microscope. For the preparation of nuclear extract, the method of Dignam et al. [28] with some modifications was followed. Briefly, the nuclei were suspended in nuclear protein extraction buffer containing 20 mM HEPES (pH 7.6), 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM PMSF, 0.5 mM dithiothreitol, 0.4 mg/ml complete EDTA-free protease inhibitor (Roche Diagnostics, Germany) and stirred for 30 min on ice. Then the suspension was centrifuged at 50,000 × g for 40 min at 4˚C. The supernatant containing nuclear extract was checked quantitatively by Bradford method [29] and qualitatively by silver staining.
2.3. Immunoprecipitation
To know the age dependent level of interaction of ERβ with ERAP 140, the nuclear extract (50 µg) of brain from young, adult and old male mice were incubated with protein A beads at 4˚C for 4h and then centrifuged at 10,000 × g for 10 min at 4˚C. The pre-cleared supernatant was incubated overnight with beads and 5 µl of anti-ERβ in 500 µl immunoprecipitation (IP) buffer (50 mM HEPES pH 7.6, 50 mM NaCl, 0.1% Triton-100, 10% glycerol, 1 mM PMSF) in the presence of 10 nM 17β-estradiol. ERβ antibody (H-150: sc8974, Santacruz Biotechnology, USA). It is a polyclonal antibody raised against epitope corresponding to amino acid of ERβ 1 - 150 N-terminus residue. The unbound proteins were removed from the beads by washing with IP buffer. Finally, the bound proteins were eluted and denatured by boiling with SDS sample buffer. The interacting proteins were resolved by 7.5% SDS-PAGE, blotted using wet transfer apparatus (Bangalore Genei, India) onto PVDF membrane and probed with anti-ERAP 140 (The anti-ERAP- 140 polyclonal antibody was raised in rabbit against a GST-λ7 fusion protein. This was received as a kind gift from Prof Brown Myle, Dana Farber/Harvard Center, USA). IgG was used as a control in IP experiments. Same procedure was followed to detect β tubulin III.
2.4. Immunoblotting
Further, age dependent expression of ERAP 140 in the brain of mouse was checked by immunoblotting. Briefly, nuclear extract (50 µg) of the brain from young, adult and old male mice was resolved by 7.5% SDS-PAGE and transferred onto PVDF membrane using wet transfer apparatus (Bangalore Genei, India). Then the blot was incubated in blocking buffer (5% fat free milk in 1X PBS), followed by fresh blocking buffer containing anti-ERAP antibody (1:1000) overnight at 4˚C. After washing in PBS (3 × 5 min), the blot was again incubated in blocking buffer containing anti-rabbit antibody HRPO conjugate (1:2000) for 2 h at room temperature. The signal was finally detected by enhanced chemilumiscence method. After stripping the blot, same procedure was followed to detect β tubulin III.
2.5. Secondary Structure Prediction of Proteins by SOPMA and PSIPRED
We have identified ERAP 140, Trk A, Src, pCREB and CREB as ERβ interacting proteins and hypothesized that these proteins should have consensus secondary structures. We predicted the probable secondary structure of these proteins using sequence based secondary structure prediction for the protein with SOPMA, an established software tool. It is self-optimized prediction method (SOPM) to improve the success rate in the prediction of the secondary structure of proteins. This improved SOPM method (SOPMA) correctly predicts 69.5% of amino acids for a three-state description of the secondary structure, namely alpha-helix, beta-sheet and coil. The joint prediction with SOPMA and a neural network method correctly predict 82.2% of residues for 74% of co-predicted amino acids. In addition, we have analyzed the secondary structure of ERβ interacting proteins using PSIPRED as mentioned [30].
2.6. Densitometric Scanning and Statistical Analysis
Now the signals detected by immunoprecipitation and immunoblotting on the autoradiogram were scanned densitometrically and analyzed using Alpha imager software V3.1.2 2200. Nine animals were used for each experiment and repeated three times. The relative density value (RDV) was calculated after normalizing with β tubulin III. The histogram was plotted from mean RDV and standard errors of three independent experiments. The statistical analysis of the data was done using Sigma Stat 2.0 followed by all pairwise multiple comparison procedure and post hoc student-newmann-keuls (SNK) method. The data with p < 0.05 was considered as significant. Young was used as a control for comparison to adult and old.
3. RESULTS AND DISCUSSION
Estrogen signaling is complex in the brain and the involvement of coregulators increases the complexity of ER mediated gene transcription in brain [3,8]. Now it is well established that coregulators play a vital role in the differential expression of ERs leading to specific brain functions. Previously, we have studied the expression of ERα and ERβ and the binding of ERα promoter with transcription factors in aging mouse brain [31,32]. In continuation, we reported that mouse ERaLBD interacts with PELP1, RIP140, PGC1α, BAF60 and ADA3 [33, 34], and ERaTAD with metastasis associated protein (MTA) 1 and p68 RNA helicase [35], and these coregulators vary in their levels of interaction and expression during aging of mouse brain. For the identification ERβ interacting proteins in mouse brain, purified [36] and functionally active [37] ERβ was used. Recently, we reported age and sex dependent interaction of AIB1 with ERβTAD and its expression in mouse brain [14].
Here we have examined the effect of age on the interaction of ERβ with ERAP 140 in male mouse brain, and compared the level of interaction among young, adult and old mice by immunoprecipitation and immunoblotting. The level of interaction of ERAP 140 with ERβ decreased in adult as compared to young and remained almost similar in old (p < 0.05) (Figure 1(a)). In contrast, ERAP 140 expression was higher in adult than young and old male mouse brain (p < 0.05) (Figure 1(b)). The pattern of interaction of ERAP 140 with ERβ and its expression in male was different from the same in female brain. Such discrepancy may be assigned to differences in levels of estradiol, neurogenesis and differentiation in male as compared to female [38]. The mecha-

Figure 1. (a) Immunoprecipitation (IP) of nuclear extract from the brain of young (6 weeks), adult (25 weeks) and old (70 weeks) mice using ERβ antibody and immunoblotted with ERAP 140; (b) Immunoblotting (IB) of nuclear extract from the brain of young (6 weeks), adult (25 weeks) and old (70 weeks) of mice showing expression of ERAP 140. For both IP and IB beta tubulin III was used as a loading control. Histogram represents RDV with SEM obtained from the data of three independent experiments. Data were statistically analyzed using a one-way ANOVA comparing age, followed by all pairwise multiple comparison procedure (Student-Newmann-Keuls method). Young was taken as control for comparison with adult and old. The p values < 0.05 were considered as significant. * represents significant difference.
nism involved in the interaction of ERβ wih ERAP 140 is not fully understood. It is likely that the interaction is through alpha helix as known for other coregulators. Immunoprecipitation showed the level of recruited ERAP 140 to ERβ, which was influenced by the level of estradiol and change in the secondary structure of proteins [39]. Thus the level of ERβ and ERAP 140 did not correspond. The ERβ interacting ERAP 140 is abundantly expressed in neurons of thalamus, hypothalamus, hippocampus, cerebellum, striatum and choroid plexus in the brain [18]. ERAP 140 is believed to be similar to other nuclear receptor coactivators and directly regulate estrogen responsive genes involved in neurodevelopment and neuroprotection [40]. As these brain regions also show expression of ERβ, it is likely that ERAP 140 regulates estrogen functions through ERβ in these regions in age dependent manner.
Furthermore, the secondary structure prediction tool SOPMA and PSIPRED suggest that ERAP 140 and other ERβ interacting proteins were rich in alpha helices and coils; ERAP 140 (55.33% alpha helices, 44.33% coils), AIB 1 (37% alpha helices, 48% coils), TrkA (33% alpha helices, 43% coils), Src (10% alpha helices, 57% coils), CREB (39% alpha helices, 41% coils) (Figure 2(a), Table 1). The secondary structure prediction of ERβ interacting proteins by PSIPRED also showed richness in alpha helices and coils (Figure 2(b)). The secondary structure prediction SOPMA is an improvement of SOPM method [41,42], which is based on amino acid homology. It uses the information from an alignment of amino acid sequences of proteins belonging to the same family. In the absence of homologous sequences, SOPMA prediction is similar to SOPM with more than 70% accuracy. The secondary structure prediction is one of the most important goals pursued by bioinformatics and theoretical chemistry, and is highly useful to design drugs and novel enzymes. Thus the secondary structure prediction of ERβ interacting proteins, which are rich in alpha helices and coils, may be helpful in understanding the regulation of ERβ dependent signaling in the context
 (a)
(a) (b)
(b)
Figure 2. ERβ and its domains interacting proteins ERAP140, AIB1, Trk A, Src and CREB showing predominantly alpha helices and coils predicted by secondary structure prediction self-optimized prediction method alignment (SOPMA) (a) and PSIPRED (b).
Table 1. List of ERβ interacting proteins, ERAP 140, AIB1, TrkA, Src and CREB showing richness in alpha helix and random coil.
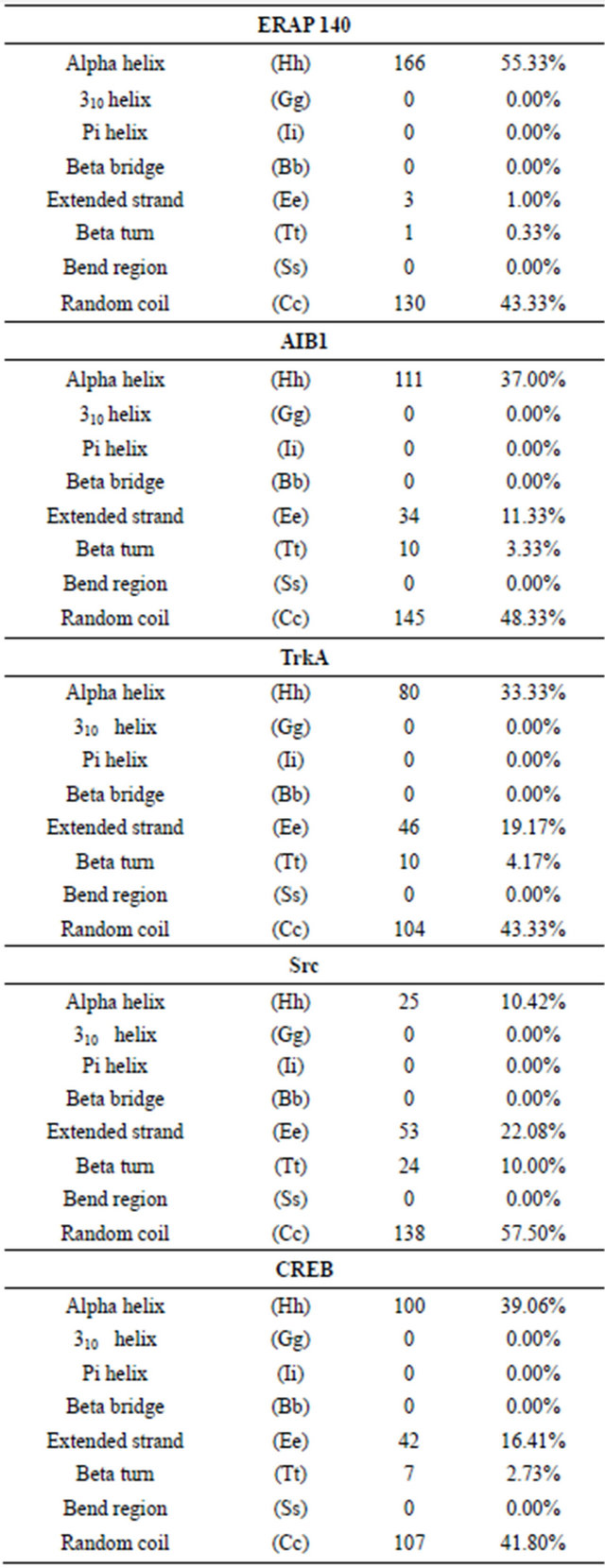
Footnote: The parameters for predicting the secondary structure are as follows: Window width: 17; Similarity threshold: 8; Number of states: 4.
of coregulators in brain, and designing ER modulators which can regulate specific functions of estrogen in the brain during aging and degenerative diseases.
5. ACKNOWLEDGEMENTS
The authors thank Prof Brown Myles, USA for the kind gift of ERAP 140 antibody. Financial support from the Department of Biotechnology (DBT) and Indian Council of Medical Research (ICMR), Government of India (BT/PR3593/Med/14/468/2003) to MKT and fellowship from ICMR to VP are highly acknowledged.
REFERENCES
- Koehler, K.F., Helguero, L.A., Haldosén, L.A., Warner, M. and Gustafsson, J.A. (2005) Reflections on the discovery and significance of estrogen receptor β. Endocrine Reviews, 26, 465-478. doi:10.1210/er.2004-0027
- Nagler, J.J., Cavileer, T., Sullivan, J., Cyr, D.G. and Rexroad III, C. (2007) The complete nuclear estrogen recaptor family in the rainbow trout: Discovery of the novel ERα2 and both ERβ isoforms. Gene, 392, 164-173. doi:10.1016/j.gene.2006.12.030
- Sugiyama, N., Barros, R.P.A., Warner, M., Gustafsson, J.A. (2010) ERβ: Recent understanding of estrogen signaling. Trends in Endocrinology and Metabolism, 21, 545-552. doi:10.1016/j.tem.2010.05.001
- Beato, M. and Klug, J. (2000) Steroid hormone receptors: An update. Human Reproduction Update, 6, 225-236. doi:10.1093/humupd/6.3.225
- Mckenna, N.J. and O’Malley, B.W. (2002) Combinatorial control of gene expression by nuclear receptors and coregulators. Cell, 108, 465-474. doi:10.1016/S0092-8674(02)00641-4
- Edwards, D.P. (2005) Regulation of signal transduction pathways by estrogen and progesterone. Annual Review of Physiology, 67, 335-376.
- Lonard, D.M. and O’Malley, B.W. (2008) SRC-3 transcription-coupled activation, degradation and the ubiquitin clock. Science Signaling, 1, e16.
- Thakur, M.K. and Paramanik, V. (2009) Role of steroid hormone coregulators in health and disease. Hormone Research, 71, 194-200. doi:10.1159/000201107
- Kumar, V., Green, S., Stack, G., Berry, M., Jin, J.R. and Chambon, P. (1987) Functional domains of the human estrogen receptor. Cell, 51, 941-951. doi:10.1016/0092-8674(87)90581-2
- Smirnov, A.N. (2002) Review: Nuclear receptors: Nomenclatute, ligands, mechanisms of their effects on gene expression. Biochemistry (Moscow), 67, 957-977. doi:10.1023/A:1020545200302
- Thakur, M.K. and Sharma, P.K. (2006) Aging of brain: Role of estrogen. Neurochemical Research, 31, 1389- 1398. doi:10.1007/s11064-006-9191-y
- Harding, J.J. (1972) Conformational changes in human lens proteins in cataract. Biochemistry Journal, 129, 97- 100.
- Paramanik, V. and Thakur, M.K. (2010) Interaction of estrogen receptor associated protein (ERAP) 140 with ERβ decreases but its expression increases in aging mouse cerebral cortex. Cellular and Molecular Neurobiology, 30, 961-966. doi:10.1007/s10571-010-9526-8
- Paramanik, V. and Thakur, M.K. (2011) AIB1 shows variation in interaction with ERβTAD and expression as a function of age in mouse brain. Biogerontology, 12, 321- 328. doi:10.1007/s10522-011-9330-y
- Thakur, M.K. and Paramanik, V. (2012) Expression of Trk A and Src and their interaction with ERβ ligand binding domain show age and sex dependent alteration in mouse brain. Neurochemical Research, 37, 448-453. doi:10.1007/s11064-011-0631-y
- Paramanik, V. and Thakur, M.K. (2012) Estrogen recaptor β and its domains interact with casein kinase 2, phosphokinase C and N-myristoylation sites of mitochondrial and nuclear proteins in mouse brain. The Journal of Biological Chemistry, 287, 22305-22316. doi:10.1074/jbc.M112.351262
- Greiner, E.F., Kirfel, J., Greschik, H., Huang, D., Becker, P., Kapfhammer, J.P. and Schüle, R. (2000) Differential ligand-dependent protein-protein interactions between nuclear receptors and a neuronal-specific cofactor. Proceeding of the National Academy of Sciences, 97, 7160-7165. doi:10.1073/pnas.97.13.7160
- Shao, W., Halachmi, S. and Brown, M. (2002) ERAP 140, a conserved tissue-specific nuclear receptor coactivator. Molecular and Cellular Biology, 22, 3358-3372. doi:10.1128/MCB.22.10.3358-3372.2002
- Voegel, J.J., Heine, M.J., Zechel, C., Chambon, P. and Gronemeyer, H. (1996) TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO Journal, 15, 3667- 3675.
- Anzick, S.L., Kononen, J., Walker, R.L., Azorsa, D.O., Tanner, M.M., Guan, X.Y., Sauter, G., Kallioniemi, O.P., Trent, J.M. and Meltzer, P.S. (1997) AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science, 277, 965-968. doi:10.1126/science.277.5328.965
- Suen C.S., Berrodin, T.J., Mastroeni, R., Cheskis B.J., Lyttle, C.R. and Frail, D.E. (1998) A transcriptional coactivator, steroid receptor coactivator-3, selectively augments steroid receptor transcriptional activity. The Journal of Biological Chemistry, 273, 27645-27653. doi:10.1074/jbc.273.42.27645
- Yan, J., Yu, C.T., Ozen, M., Ittmann, M., Tsai, S.Y. and Tsai M.J. (2006) Steroid receptor coactivator-3 and activator protein-1 coordinately regulate the transcription of components of the insulin-like growth factor/AKT signaling pathway. Cancer Research, 66, 11039-11046. doi:10.1158/0008-5472.CAN-06-2442
- Matocha, M.F., Fitzpatrick, S.W., Atack, J.R. and Rapoport, S.I. Matocha, M.F., Fitzpatrick, S.W., Atack, J.R., Rapoport, S.I. (1999) pp60c-src kinase expression in brain of adult rats in relation to age. Experimental Gerontology, 25, 47-54. doi:10.1016/0531-5565(90)90008-P
- Johannessen, M., Delghandi, M.P. and Moens, U. (2004) What turns CREB on? Cell Signal. Cell Signaling, 16, 1211-1227. doi:10.1016/j.cellsig.2004.05.001
- Costantini, C., Scrable, H. and Puglielli, L. (2006) An aging pathway controls the TrkA to p75NTR receptor switch and amyloid beta-peptide generation. EMBO Journal, 25, 1997-2006. doi:10.1038/sj.emboj.7601062
- Benitah, S.A. (2001) Intracellular signalling pathways and carcinogenesis. Clinical and. Translational Oncology, 3, 274-278.
- Burgoyne, L.A. and Hewish, D.R. (1972) The cell nucleus IV. Academic Press, New York.
- Dignam, J.D., Lebovitz, R.M. and Roeder, R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Research, 11, 1475-1489. doi:10.1093/nar/11.5.1475
- Bradford, M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Annals of Biochemistry, 72, 248-254.
- McGuffin, L.J., Bryson, K. and Jones, D.T. (2000) The PSIPRED protein structure prediction server. Bioinformatics, 16, e404-e405.
- Sharma, P.K. and Thakur, M.K. (2006) Expression of estrogen receptor (ER) α and β in mouse cerebral cortex: Effect of age, sex and gonadal steroids. Neurobiology of Aging, 27, 880-887. doi:10.1016/j.neurobiolaging.2005.04.003
- Sharma, P.K. and Thakur, M.K. (2008) Binding of estrogen receptor α promoter to nuclear proteins of mouse cerebral cortex: Effect of age, sex and gonadal steroids. Biogerontology, 9, 467-478. doi:10.1007/s10522-008-9166-2
- Ghosh, S. and Thakur, M.K. (2009) Interaction of ERα ligand binding domain with nuclear proteins of aging mouse brain. Journal of Neuroscience Research, 87, 2591- 2600. doi:10.1002/jnr.22068
- Ghosh, S. and Thakur, M.K. (2009b) Interaction of ERα transactivation domain with nuclear proteins of mouse brain: P68 RNA helicase shows ageand sex-specific change. Journal of Neuroscience Research, 6, 1323-1328. doi:10.1002/jnr.21948
- Thakur, M.K. and Ghosh, S. (2009) Interaction of ERα transactivation domain with MTA1 decreases in old mouse brain. Journal of Molecular Neurosciences, 37, 269-273. doi:10.1007/s12031-008-9131-1
- Paramanik, V. and Thakur, M.K. (2010b) Overexpression of mouse estrogen receptor β decreases but its transactivation and ligand binding domains increase the growth characteristics of E. coli. Molecular Biotechnology, 47, 26-33. doi:10.1007/s12033-010-9308-z
- Paramanik, V. and Thakur, M.K. (2011b) NMR analysis reveals 17β-estradiol induced conformational change in ERβ ligand binding domain expressed in E. Coli. Molecular Biology Reports, 38, 4657-4661. doi:10.1007/s11033-010-0600-6
- Arai, H., Ozaki, T., Niizuma, H., Nakamura, Y., Ohira, M., Takano, K., Matsumoto, M., Nakagawara. A. (2008) ERAP 140/Nbla10993 is a novel favorable prognostic indicator for neuroblastoma induced in response to retinoic acid. Oncology Reports, 19, 1381-1388.
- Levine, R.L. and Stadtman, E.R. (2001) Oxidative modifications of proteins during aging. Experimental Gerontology, 36, 1495-1502. doi:10.1016/S0531-5565(01)00135-8
- Middleton, F.A., Pato, M.T., Gentile, K.L., Morley, C.P., Zhao, X., Eisener, A.F., Brown, A., Petryshen, T.L., Kirby, A.N., Medeiros, H., Carvalho, C., Macedo, A., Dourado, A., Coelho, I., Valente, J., Soares, M.J., Ferreira, C.P., Lei, M., Azevedo, M.H., Kennedy, J.L., Daly, M.J., Sklar, P. and Pato, C.N. (2004) Genome wide linkage analysis of bipolar disorder by use of a high-density single-nucleotide-polymorphism (SNP) genotyping Assay: A comparison with microsatellite marker assays and finding of significant linkage to chromosome 6q22. The American Journal of Human Genetics, 74, 886-897. doi:10.1086/420775
- Geourjon, C. and Deleage, G. (1994) SOPM: A self-optimised method for protein secondary structure prediction. Protein Engineering, 7, 157-164. doi:10.1093/protein/7.2.157
- Geourjon, C. and Deleage, G. (1995) ANTHEPROT 2.0: A three-dimensional module fully coupled with protein sequence analysis methods. Journal of Molecular Graphics, 13, 209-212. doi:10.1016/0263-7855(95)00035-5
NOTES
*Corresponding author.

