World Journal of Neuroscience
Vol.2 No.2(2012), Article ID:19404,4 pages DOI:10.4236/wjns.2012.22009
Adult and neonatal astrocytes exhibit diverse gene expression profiles in response to beta amyloid ex vivo
![]()
1Department of Neurobiology, A.I. Virtanen Institute for Molecular Sciences, University of Eastern Finland, Kuopio, Finland
2Biocenter Kuopio, University of Eastern Finland, Kuopio, Finland
3School of Pharmacy, Faculty of Health Sciences, University of Eastern Finland, Kuopio, Finland
4Department of Biosciences, University of Eastern Finland, Kuopio, Finland
5Medeia Therapeutics Ltd., Kuopio, Finland
6Department of Oncology, Kuopio University Hospital, Kuopio, Finland
Email: jari.koistinaho@uef.fi
Received 7 December 2011; revised 29 December 2011; accepted 16 February 2012
Keywords: Alzheimer’s Disease; Astrocytes; Mice; Amyloid Beta; Gene Expression; Microarray Analysis; Endocytosis; Proteolysis
ABSTRACT
Astrocytes are implicated in the neuropathology of Alzheimer’s disease (AD) by clustering with other activated inflammatory cells at the sites of amyloid beta (Aβ) deposits formed in the cortex and hippocampus. Astrocytes are known to contribute to the clearance of Aβ in the AD brain. Also, adult but not neonatal mouse astrocytes are able to clear Aβ deposits from the tissue sections of transgenic AD mice and human brain ex vivo. Because these findings suggest that cultured neonatal astrocytes may not represent a relevant cell for modeling the function of astrocytes in neurodegenerative diseases, we studied whether neonatal and adult astrocytes show different responses in gene expression when exposed to brain sections burdened by deposits of human Aβ. Whole genome microbarrays demonstrated greater alteration of gene expression in adult astrocytes than in neonatal astrocytes. When exposed to Aβ burdened brain sections adult but not neonatal astrocytes up-regulated genes related to peptidase (such as MMP13, MMP12, Phex, Htra1), scavenger receptor (Scara5, Enpp2) and glutathioine transferase (Gsta1, Gsta2, Gclm) activity, suggesting increased ability to degrade and endocytose Aβ peptides and protect against oxidative bursts. Quantitative RT-PCR analysis confirmed the significant alteration in gene expression of key peptidases, scavenger receptors and cholesterol synthesis. Our data suggest that adult astrocytes in culture are more sensitive to disease-relevant stress showing more extensive genetic response compared to neonatal astrocytes. In addition, the identified peptidases and scavenger receptors which increase expression selectively in adult astrocytes suggest their major role in astrocyte-mediated clearance of Aβ deposits in AD.
1. INTRODUCTION
Alzheimer’s disease (AD) is the most common age-related form of dementia and a devastating neurodegenerative disease characterized by neurofibrillary tangles and senile plaques in the neocortex and hippocampus. While AD usually occurs sporadically, about 2% of the AD cases are familial being associated with mutations in genes encoding proteins involved in the production of Aβ. Aβ is generated when amyloid precursor protein (APP) is subsequently cleaved by β and γ-secretases [1]. Even though there is no correlation between overall Aβ deposition and disease severity [2-5], soluble Aβ oligomers correlate well with behavioural deficits in transgenic (tg) mouse models of AD [6] and levels of Aβ oligomers in the cerebrospinal fluid correlate with the severity of dementia in AD patients [7]. The hypothesis that Aβ is a major contributory factor in the neuropathology of AD is also supported by the observation that in tg mice expressing mutated APP, presenilin and tau, Aβ deposition precedes the formation of neurofibrillary tangles and cognitive impairment [8]. Moreover, Aβ immunotherapy in the same triple tg mice reduces not only Aβ burden, but also early tau pathology [9], indicating that targeting Aβ pathology could also reduce the effects of abnormally phosphorylated tau.
In addition to numerous general tasks of supporting the brain homeostasis and neuronal integrity, such as synaptic formation and function, brain astrocytes have been suggested to play an important role in AD. Astrocytes are the most abundant glial cell type in the CNS and activated astrocytes are found surrounding Aβ deposits in the AD brain. During the development of AD astrocytes adopt inflammatory functions but also represent a source of neurotrophic factors. Recently, astrocytes have been found to be potential Aβ clearing cells. Astrocytes contain fragments of Aβ peptides in aged human brain [10] and Aβ42-positive material originating from local degenerated dendrites and synapses has been found in astrocytes in the entorhinal cortex of AD patients [11]. Contrary to neonatal astrocytes, adult astrocytes are able to clear human Aβ ex vivo from AD mouse brain sections [12-14] by an apolipoprotein E dependent mechanism [12]. Adult astrocytes clear Aβ also from postmortem human AD brain sections and transplanted astrocytes are capable of internalizing Aβ in vivo in a tg AD mouse model [15]. The difference in Aβ clearing capacity between neonatal and adult astrocytes is not surprising, as astrocytes are known to substantially maturate and adopt adult phenotype after the first postnatal weeks [16]. The major waves of rodent brain synaptogenesis occur during the first 2 to 3 weeks of postnatal life [17-19] corresponding to the time when astrocytes are dynamically growing and reach their mature morphology. By the 4th week of postnatal life astrocytes exhibit refined processes that occupy only unique spatial domains in the brain space. While the mechanisms of this postnatal maturation process of astrocytes are poorly known, the morphology, functions and mobility are substantially altered by the adulthood [16].
Because cultured astrocytes represent an important model to investigate the functions and role of these cells in brain diseases, such as AD, it is important to clarify whether the response of these cells to pathological environment, such as Aβ in AD, differ between the astrocytes derived from adult and neonatal rodents. A vast majority of studies in general are performed on neonatal astrocytes due to ease of culture and yield of cells. In addition, defining genetic responses underlying the functional differences between the adult and neonatal astrocytes in clearing Aβ in vitro may provide important clues for understanding the disease mechanisms and even for identifying potential targets for novel therapeutic approaches. As there are no comprehensive data regarding the genes activated in astrocytes during Aβ clearance, we decided to determine the gene expression patterns in adult and neonatal astrocytes cultured ex vivo on top of the tg APdE9 AD or wild-type (wt) mouse brain sections. For this purpose the cultured astrocytes were isolated from the ex vivo assay before mRNA isolation and accomplishment of the whole genome microarray analysis.
2. MATERIALS AND METHODS
2.1. Animals
The animal experiments were conducted according to the national regulations of the usage and welfare of laboratory animals and approved by the Animal Experiment Committee in State Provincial Office of Southern Finland. The mice were group-housed under a 12-h light-dark cycle and allowed free access to standard rodent chow and water. Astrocyte cultures were prepared from C57Bl/ 6j tg mice expressing enhanced green fluorescence protein, eGFP [20], purchased from Jackson Laboratories (Maine, USA) and maintained in C57Bl/6j strain in the facilities of the Lab Animal Center in Kuopio, University of Eastern Finland.
The mouse brain sections used for ex vivo assays were prepared from tg APdE9 mouse line (created by co-injection of chimeric mouse/human APPSwe and PS1-dE9 vectors, both controlled by their own mouse prion protein promoter element (APdE9 mice [21] provided by Prof. Heikki Tanila) and their wt C57Bl/6j littermates. In the tg APdE9 mouse line, the first Aβ deposits appear at 4 - 5 months of age.
2.2. Astrocyte Cultures
Hippocampi and cortices were isolated either from 2 to 3-day-old or 6 to 8-week-old tg eGFP mice and the tissue was suspended in DMEM/F12 (3:1, Gibco BRL, NY, USA) containing 10% heat-inactivated fetal bovine serum (Gibco BRL) and 100 U/ml penicillin-streptomycin (Gibco BRL). The suspension was triturated 10 × and centrifuged at 1500 rpm for 5 min at RT. After adding 0.25% trypsin-EDTA (Gibco BRL) the suspension was incubated at 37˚C for 30 min. Fresh culture medium was added and the suspension was centrifuged at 1500 rpm for 5 min. The cells were treated with Percoll (SigmaAldrich, St. Louis, USA), centrifuged at 1500 rpm for 10 min at RT or until the phases separated clearly and washed once with fresh culture media. The cells were plated onto poly-L-lysine coated flasks in DMEM/F12 (3:1) containing 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin-streptomycin and G5 supplement (Gibco BRL) at the density of 4 × 104 cells/ml. Before the experiments these mixed glial cell cultures were shaken to remove microglia at 200 rpm for 2 h. For the neonatal astrocyte cultures the culture medium was DMEM (+4500 mg/l glucose, + L-glutamine, -pyruvate, Gibco BRL) without G5 supplement and the isolation method included no Percoll treatment.
The cell density for purity analyses was 2 × 104/well in a 48-well plate. Standard anti-GFAP (anti-glial fibrillary acidic protein, 1:500; Dako Cytomation, Glostrup, Denmark) immunocytochemistry was used to confirm the purity of astrocyte cultures. The possible microglial contamination of adult and neonatal eGFP expressing cells used for ex vivo experiments was evaluated using anti-Iba (2 µg/ml; Wako, Japan) and anti-CD11b (1:500 dilution; Serotec, UK). The cells were incubated with biotinylated goat anti-rabbit IgG (1:200 dilution; Vector Laboratories, CA) secondary antibody and Vectastain ABC Elite kit was used (Vector Laboratories) according to manufacturer’s protocol. Hydrogen peroxide (H2O2) and nickel-enhanced diaminobenzidine (Ni-DAB) were used to visualize the immunoreactivity. Three fields per well and at least three wells per cell type were analyzed for the percentage of Ni-DAB stained cells of all cellular profiles in the field. Both adult and neonatal astrocyte cultures contained on average 99.6% ± 0.4% GFAP-immunoreactive astrocytes. The cultures contained less than 0.4% of microglial cells in all experiments.
2.3. Separating the Cultured Astrocytes from Top of the Mouse Brain Section
We examined the possible differences in gene expression profiles in eGFP expressing neonatal and adult astrocytes as depicted in Figure 1. Isolated neonatal or adult astrocytes (4 × 105 cells/well) were cultured on top of APdE9 or wt mouse brain sections. Six sections per cell and section type were processed and before the mRNA isolation process, the astrocyte samples from two sections were pooled to yield n = 3. After 22 h culture co-incubation of plated astrocytes and brain sections the medium was
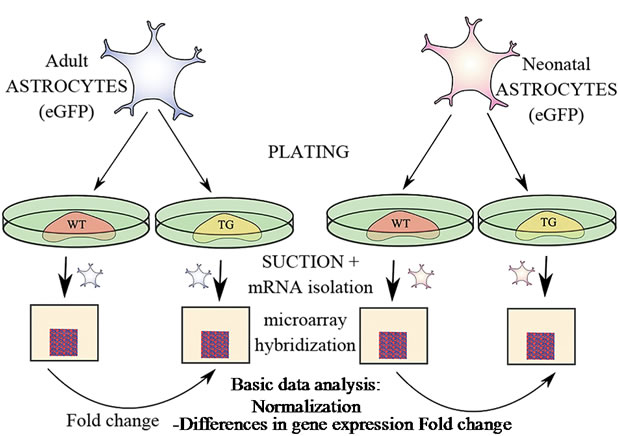
Figure 1. Configuration of the microarray experiment. Adult and neonatal eGFP expressing astrocytes were plated and cultured on brain sections either from WT mouse or APdE9 TG mouse for 22 h. Subsequently, eGFP expressing astrocytes were removed, mRNA isolated and hybridized to microarrays. For each sample group three biological replicate arrays were scanned and the data were normalized with RMA normalizetion. Probe-specific fold changes between results from astrocytes plated to WT and TG APdE9 brain sections were calculated.
removed from the wells and the brain sections with cultured eGFP astrocytes were incubated with 1 × trypsin-EDTA (500 µl/well) for up to 15 min at 37˚C. The detachment (at this point, the astrocytes do not lose the grip completely) of the astrocytes from the underlying brain section was carefully monitored with a standard light microscope. To inactivate trypsin, the brain sections with cultured astrocytes were carefully removed into a small Petri dish filled with fetal bovine serum containing culture medium. Subsequently, the brain sections were immediately examined with a fluorescence microscope (Olympus IX-71, Japan). The eGFP expressing astrocytes were separated from the underlying brain section using a special suction hose modified for this purpose from suction hoses typically used for the microinjection techniques. One end of the tube contained a pipet tip as a mouthpiece, the other end a stretched glass capillary (prepared with a Bunsen burner) and in the middle of the tube there was a 0.22 µm filter. The eGFP expressing astrocytes were gently picked up from the underlying brain section using wavelength 468 nm of the fluorescence microscope and simultaneous light microscopy. Two parallel samples were collected into one 15 ml tube and the cells were centrifuged at 1500 rpm for 5 min. The supernatant was removed and the remaining pellet was resuspended in 1 ml of culture medium. The cell suspension was removed into an Eppendorf tube and centrifuged again at 12,000 rpm for 3 min.
2.4. mRNA Isolation
RibopureTM Kit (Ambion, Applied Biosystems, USA) was used for the total mRNA isolation according to the manufacturer’s instructions. The cell pellet was lysed by triturating with 100 µl TRI reagent.
2.5. Microarray Procedure and Analysis
The microarray procedure was performed at Biomedicum Genomics (BMGen), University of Helsinki, Finland. The qualities of mRNA samples were evaluated using Bioanalyzer/Nano RNA assessments. After the samples passed the quality control criteria, BMGen performed gene expression analyses using 12 × Affymetrix GeneChip Mouse Genome 430 2.0 hybridizations (2-Cycle method). Three replicate arrays for each sample group were used and scanned with an Affymetrix GeneChip scanner. All data from arrays passed the quality control (Chipster v1.4.7; CSC, Finland; [22]) performed according the manual of the software (http://chipster.csc.fi/manual/tools. html#quality). The rest of data analysis was made with Genespring GX 11.0 (Agilent, Santa Clara, Calif., USA) by preprocessing the raw data with the robust multi-chip algorithm [23], normalizing per chip to the median and filtering the differentially expressed data of all 12 microarrays containing 45,101 probes by fold change >2.0 or <0.5. The change in the astrocyte gene expression was identified according to the sections the astrocytes were incubated with (wt or tg; Figure 1). The differenttially expressed probe sets were further processed using statistical analysis with Welch unpaired t-test assuming unequal variances (p < 0.05; fold change >2.0 or <0.5). Benjamini-Hochberg false-discovery rate (FDR) correction for multiple testing was used to control the number of false-positives results. By using MS Excel, replicate probes for individual genes were combined by taking averages of values and probes without gene annotations were excluded. Differentially expressed genes were grouped with their molecular function and biological process terms of Gene Ontology (GO) annotations by using visualization program Cytoscape [24] and its plugin BiNGO 2.4.2 (p < 0.05, Benjamini-Hochberg FDR, whole GO annotation as reference set) [25]. Molecular function groups were categorized with CateGOrizer 3.2.18 [26] by making the classification with modified Generic GO_slim-list (http://www.geneontology.org/GO.slims.shtml).
2.6. Comparison of the Dataset to Former Studies on Astrocyte Microarray
The expression profiles of the astrocytes were compared to publicly available datasets [27], created by using the same Affymetrix GeneChip Mouse Genome 430 2.0 Array (GEO accession: GSE9566). The additional data consisted of expression data of highly purified brain cells included astrocytes from early postnatal age at postnatal day 1, and later postnatal age at day 30 when astrocytes are morphologically fully differentiated. In addition, the datasets included expression profiles from neurons and oligodendrocytes. One dataset was a mixed sample of mouse forebrain. The additional datasets were loaded as original CEL files and normalized with the same protocol as the present data. The combined datasets were clustered using standard correlation coefficient.
2.7. Quantitative Real-Time PCR
Synthesis of cDNA was made by using 400 ng of total RNA, Maxima reverse transcriptase and random hexamer primers (Fermentas life sciences). Six samples were prepared for each sample group. The relative expression levels of mRNA encoding several genes were measured according to manufacturer’s protocol by quantitative RT-PCR (StepOnePlus™ Real-Time PCR System; Applied Biosystems, USA) by using specific assays-on-demand (Applied Biosystems, USA) target mixes. The mean expression levels were normalized to Beta-2 microglobulin (B2m) and the four relative values representing the four sample groups (neonatal or adult astrocytes on wt or tg APdE9 brain sections) for each gene were scaled to range 0 - 100 ± SEM.
2.8. Statistical Analysis
A statistical probability of p < 0.05 using MS Excel (two-sample t-test) and Genespring GX 11.0 (Welch unpaired t-test) was considered significant. The results of the qRT-PCR are represented as the mean ± SEM and other results are shown as basic mean values.
3. RESULTS
3.1. Verifying Microarray Data Homogeneity by Clustering and Reviewing Marker Genes
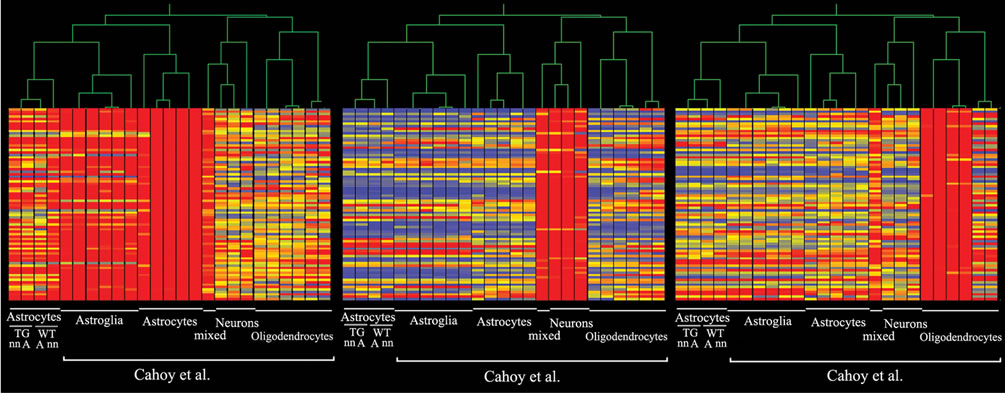
To verify that the GFAP-immunoreactive cells analyzed in microarray experiment were indeed astrocytes, we compared our data (GEO accession number: GSE29317) to published microarray data sets of highly purified astrocytes, neurons and oligodendrocytes [27]. Our data sets clustered well with control data of in vivo purified adult astrocytes and in vitro cultured neonatal astroglia (Figure 2). Minor deviations observed possibly indicated difference in culture conditions and ages of tissues used for cell isolation.
3.2. Differentially Expressed Genes and Gene Ontology Classification
By comparing expression profiles between the astrocytes cultured on top of wt and tg APdE9 brain sections, 877 genes in adult and 132 genes in neonatal astrocytes were differentially expressed two fold or greater (Figure 3(a)). Up-regulation here means that gene expression in astrocytes cultured on tg APdE9 brain section was higher than in astrocytes cultured on wt section, down-regulation conversely. While one third of the genes showing altered expression when cultured on tg APdE9 brain sections were up-regulations both in adult and neonatal astrocytes, only 13 genes of the up-regulated and 30 genes of the down-regulated genes were the identical. To illustrate the differences in gene expression related to certain cellular functions, the genes with altered expression were classified with Gene Ontology (GO) annotations (Figures 3(b), (c); 4(a), (b)). Adult astrocytes upand down-regulated grossly equal number of genes belonging to categories of catalytic activity and hydrolase activity when cultured on top of APdE9 brain sections. However, there was a clear shift towards up-regulation of peptidase activity, scavenger receptor activity and glutathione transferase activity (Figures 3(b), 5(a)) which are cellular functions that could enhance clearance of Aβ or reduce its toxic effects. Correspondingly, several peptidase inhibitors were specifically down-regulated by adult as-
 (a) (b) (c)
(a) (b) (c)
Figure 2. Microarray results clustered with control data. The data set from microarray was compared to data (Cahoy et al. 2008) of a study on the same platform that defines genes specific for highly purified astrocytes (a), neurons (b) and oligodendrocytes (c). The profile of the 4 groups: neonatal (nn) or adult (a) astrocytes and WT or TG ApdE9 brain section are at the leftmost columns of each cluster group (a, b, c).
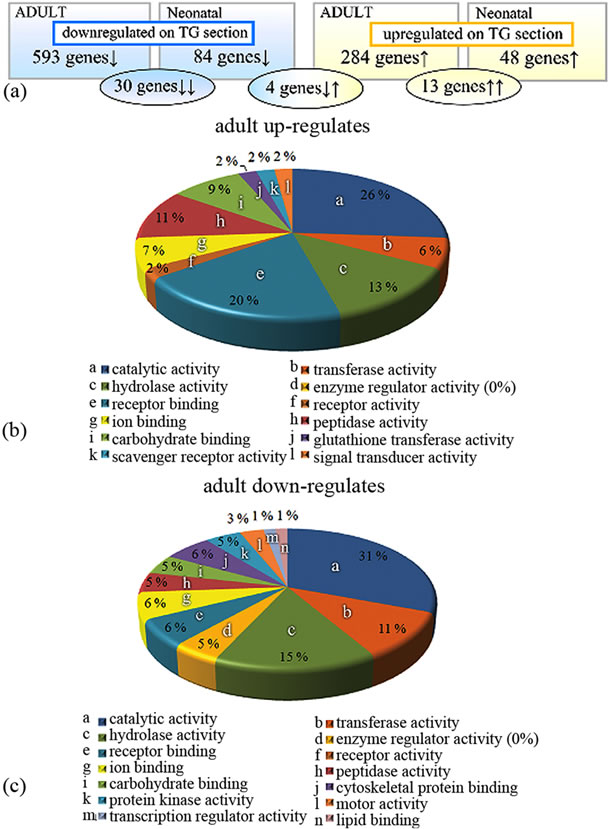
Figure 3. Gene Ontology analysis of microarray data. (a) Differentially expressed genes of astrocytes cultured on top APdE9 tg brain section compared to astrocytes cultured on wt brain sections. Adult and neonatal astrocytes altered 877 and 132 genes, respectively. Altogether, there are 30 genes down-regulated (blue ellipse) and 13 genes up-regulated (yellow ellipse) by both adult and neonatal astrocytes. Four genes were upregulated by adult but down-regulated by neonatal astrocytes (the middle ellipse). (b), (c) Expression profiles of adult astrocytes represented by categorized molecular functions. For more information, see the text.
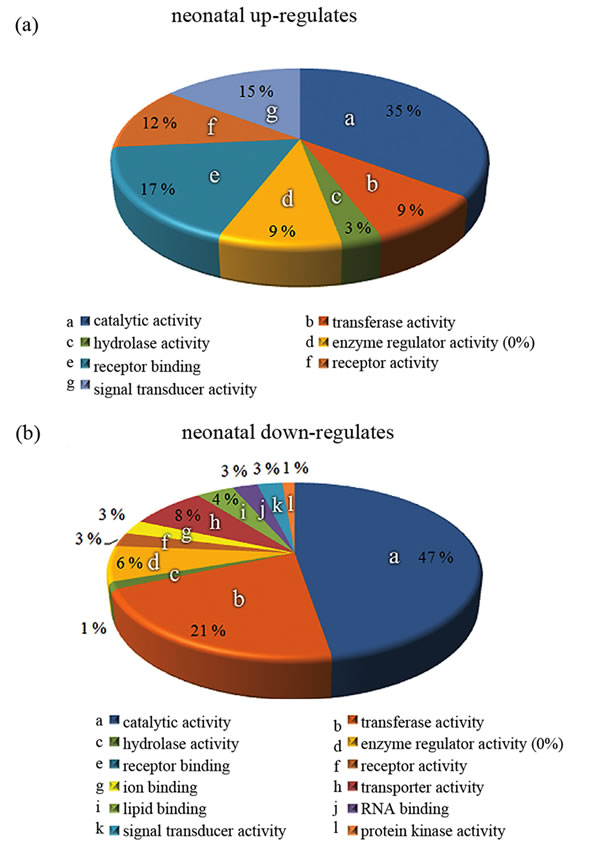
Figure 4. Gene Ontology analysis of microarray data. (a), (b) Expression profiles of neonatal astrocytes represented by categorized molecular functions. For more information, see the text.
trocytes grown on tg APdE9 brain section (Figure 5(a)). Even though gene expressions of peptidase inhibitors were down-regulated also in neonatal astrocytes, GO analysis did not generate any enriched groups of peptidases or proteases in these cells when cultured on top of APdE9 brain sections (Figures 4(b), 5(b)). GO class transferase activity was highly enriched among the genes down-regulated in neonatal astrocytes (Figure 4(b)) and most of those genes were related to cholesterol synthesis (Figure 5(b)). Based on microarray results, neonatal astrocytes down-regulated but also up-regulated several genes associated with endocytosis when cultured on top of APdE9 brain sections (Figure 5(b)). For example, the array revealed an increased expression level of macrophage scavenger receptor 1 (Msr1), which has been shown to mediate endocytosis of Aβ in microglia [28].
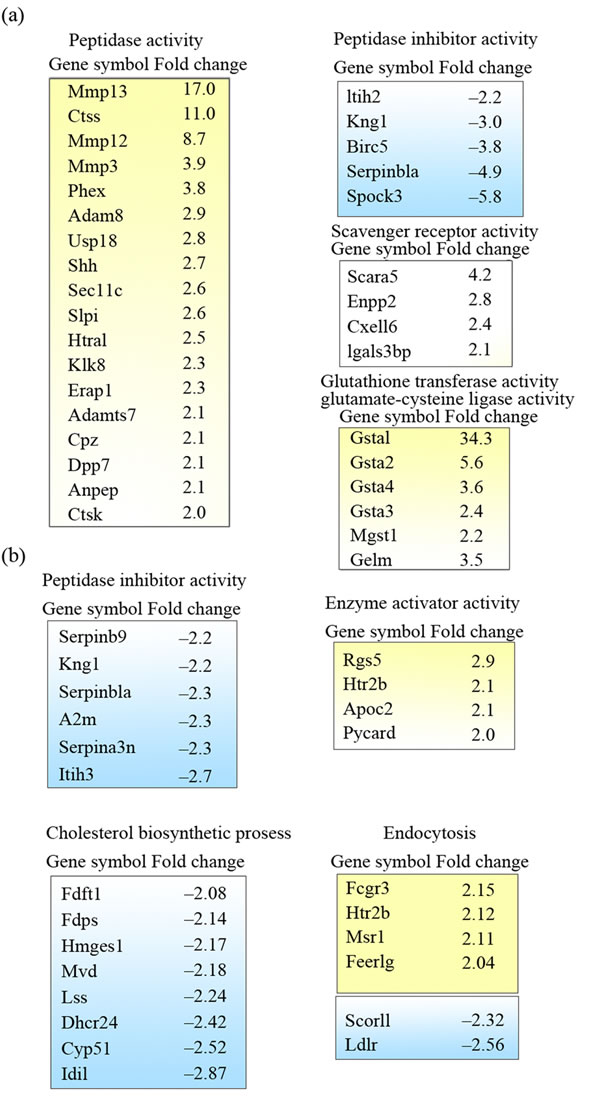
Figure 5. Gene Ontology clusters of differently expressed genes in the microarray study. (a) Enriched GO classes of the genes with altered expression in adult astrocytes; (b) GO classes of genes with altered expression in neonatal astrocytes. Up-regulated and down-regulated genes are colored yellow and blue, respectively. Positive or negative fold changes are indicated on the right. The genes selected for qRT-PCR are bolded.
3.3. Quantification of mRNA Levels by qRT-PCR
To confirm the results obtained from the microarray analysis, we selected 13 genes and analyzed their expression by qRT-PCR. Selection was made by evaluating the differences and similarities between adult and neonatal expression profiles as well as selecting genes possibly related to clearance of Aβ based on GO analysis and literature. The genes with very low expression levels in microarray data were excluded from further study, for example Cathepsin S (Ctss) had very high fold change value but expression levels were extremely low.
Adult astrocytes grown on tg APdE9 brain sections had very high, over 200 fold up-regulation of Scavenger receptor class A, member 5 (Scara5), which has been found to mediate phagocytosis of bacterial material and ferritin bound iron [29,30]. Scara5 was expressed at extremely low levels in neonatal astrocytes and adult astrocytes grown on wt brain section (Figure 6). Apolipoprotein E (apoE) has been shown to be the major susceptibility gene for AD [reviewed in 31-34]. Our microarray results demonstrated that adult up-regulated apoE over 3 fold, but only down-regulation in neonatal astrocytes was confirmed by qRT-PCR. However, expression of apoE was 10 times higher in neonatal than in adult astrocytes grown on tg APdE9 brain sections (Figure 6). Also, neonatal astrocytes seemed to down-regulate two genes encoding enzymes of cholesterol synthesis: 3-hydroxy-3- methylglutaryl-CoA synthase 1 (Hmgcs1) and 24-dehydrocholesterol reductase (Dhcr24) in same extent as apoE (Figure 6).
Phosphate regulating gene with homologies to endopeptidases on the X chromosome (Phex) has been proved to degrade synthetic Aβ40, even though only to minor extent [35]. When grown on tg APdE9 brain sections, adult astrocytes up-regulated Phex over 10 fold whereas expression by neonatal astrocytes was very low (Figure 6). A member of high temperature requirement family of serine proteases, Htra1, has also been shown to decrease Aβ levels in vitro and cell cultures (Figure 6) [36]. Both adult and neonatal astrocytes expressed moderate levels of HtrA serine peptidase 1 (Htra1), but only adult astrocytes up-regulated this protease on tg ApdE9 brain sections compared to astrocytes grown on wt mouse brain sections. Based on microarray results, among all matrix metallopeptidases and well-known Aβ degrading enzymes, neonatal astrocytes expressed very high level of matrix metallopeptidase 9 (Mmp9; data not shown). Quantitative RT-PCR confirmed significant up-regulation of Mmp9 in adult astrocytes cultured on tg APdE9 mouse brain section compared to astrocytes cultured on wt mouse brain section (Figure 6).
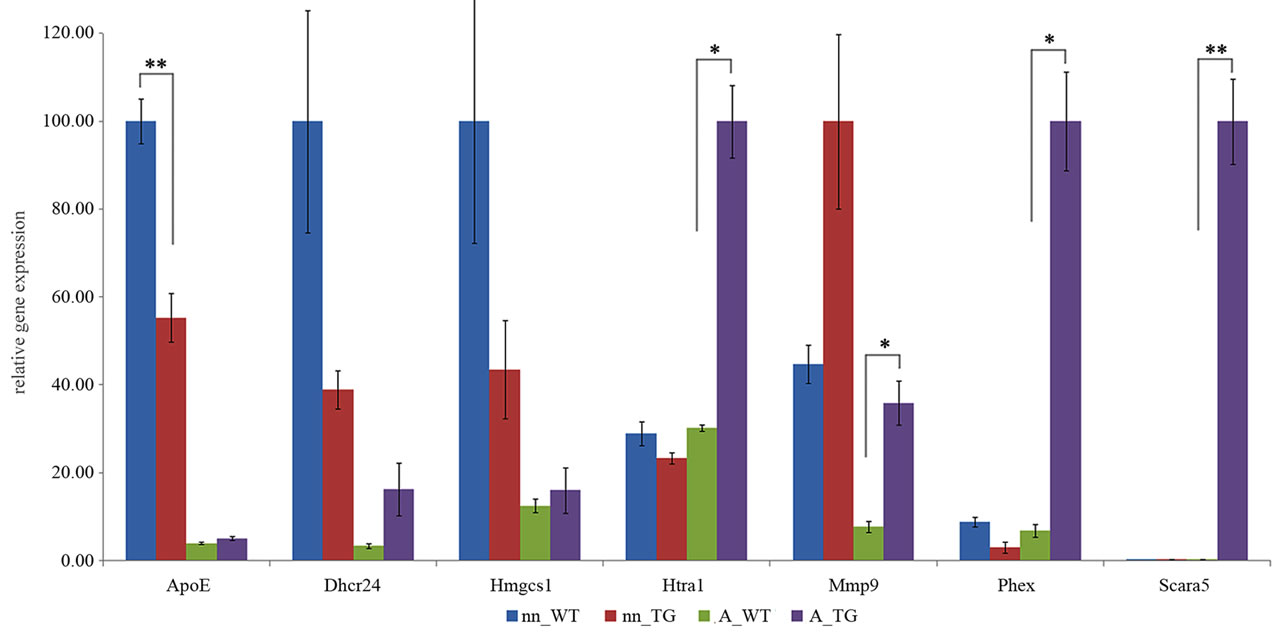
Figure 6. Quantification of mRNA levels for selected genes, comparison by real time RT-PCR. Sample groups (each n = 6): neonatal (nn) or adult (A) astrocytes on wt or tg APdE9 brain section. The mRNA levels are presented as mean relative expression levels normalized to beta-2 microglobulin (B2m) ± SEM by two sample t-test (*p < 0.05; **p < 0.01).
4. DISCUSSION
Astrocytes are the most abundant cell type in the mammalian brain having multiple functions both in healthy and diseased brain. Because astrocyte cultures can be easily prepared from neonatal rodent brain, astrocytes of newborn mice and rats are almost exclusively used as cellular models for research of astrocyte functions. Considering that astrocytes in rodents take their mature morphology by weeks 3 to 4 after extensive and dynamic interaction with the synapses in developing neurons [16], it is evident that adult astrocytes may be substantially different from the neonatal astrocytes, especially in functions related to neuronal survival and neurodegenerative diseases taking place in late adulthood and old age. Supporting this hypothesis we have previously reported that adult but not neonatal rodent astrocytes clear Aβ deposits present in mouse and human brain tissues ex vivo and similar findings have also been reported by other laboratories [12-15]. Importantly, the phenotype difference between neonatal and adult astrocyte appear to remain through passaging the cells several times, indicating that maturation of astrocytes to adult phenotype requires in vivo interaction with cells including neurons in the brain. The major finding of our study is that also genetic response of adult astrocytes to external Aβ deposits, a toxic stimulus implicated in AD, is substantially greater in adult compared to neonatal astrocytes. In addition, the gene expression analyses demonstrate differences in genes related to proteases and scavenging receptors as well as genes involved in synthesis of glutathione, the major intracellular antioxidant in astrocytes [37], which help understand the ability of adult astrocytes to clear Aβ and tolerate that process ex vivo and possibly in vivo.
Our microarray data implicates several changes in gene expression that may be related to uptake and degradation of Aβ. When cultured on top of APdE9 brain sections, adult astrocytes showed an up-regulation of four scavenger receptors and 18 peptidases/proteases. In addition, both adult and neonatal astrocytes showed down-regulation of several peptidase/protease inhibitors. Expressions of well known Aβ degrading proteases such as neprilysin, endothelin-converting enzyme (ECE-1, ECE-2) and insulin degrading enzyme (IDE) [38] were mostly unaltered in our microarray results. It is possible, but unlikely that expression levels of the main Aβ degrading enzymes would be unchanged and only activation of those enzymes would be increased in astrocytes exposed to APdE9 brain sections. Even though it is not completely clear to which extent the function of up-regulated genes may be involved in Aβ clearance and only some of these expression changes were confirmed by RT-PCR, the results suggest a distinct phenotype shift of adult astrocytes towards Aβ clearing function whereas no such alterations were detected in neonatal astrocytes.
Scara5, a member of the scavenger receptor type A family, was selectively and strongly up-regulated in adult astrocytes grown on top of APdE9 brain sections. While members of this scavenger receptor family are known to mediate endocytosis of synthetic Aβ [28,39-41] even by neonatal mouse astrocytes [39,40], class A scavenger receptors mediated uptake of Aβ in adult astrocytes has not determined ex vivo. Amounts of several metal ions, including ferritin bound ferrous iron are increased and adhered to Aβ plaques in AD [42-45]. Scara5 has been found to endocytose ferritin bound iron [30], implying importance of Scara5 in the clearance of free iron and possibly Aβ from the extracellular space.
The increased expression of peptidases/proteases Phex (phosphate regulating gene with homologies to endopeptidases on the X chromosome) and Htra1 (a member of high temperature requirement family of serine proteases) that are known to be able to degrade synthetic Aβ [35,36] and decrease Aβ levels in vitro and cell cultures were confirmed to be up-regulated in adult but not neonatal astrocytes grown on top of APdE9 sections. It is well possible that these two genes/peptidases and the peptidases seen to be up-regulated in microarray data but not confirmed by rt-PCR contribute to the ability of adult astrocytes to clear Aβ. However, the role of extracellular proteases in Aβ clearance by astrocytes is currently unclear. Our preliminary studies suggest a minor role for extracellular proteases in Aβ clearance by adult astrocytes (Pihlaja et al., unpublished data). Interestingly, Mmp9, a peptidase capable of degrading Aβ fibrils [46], was expressed at over two fold higher levels in neonatal astrocytes compared to adult astrocytes, although neonatal astrocytes do not possess Aβ degrading or clearing capacity ex vivo.
Based on our previous studies the uptake and clearance of Aβ is apoE-dependent in adult astrocytes [12]. Interestingly, we found that adult astrocytes expressed considerably lower levels of apoE than neonatal astrocytes. Possibly adult astrocytes compensate the low expression level of apoE by recycling it, which has been shown to be apoE-isoform dependent [47]. According to our data, it is evident that the overall expression level of apoE is not a limiting factor preventing neonatal astrocytes to clear Aβ.
Under physiological conditions, cholesterol uptake in the brain is efficiently prevented by the blood-brain barrier, and mature neurons are thought to rely on glial cells for their cholesterol supply. Accumulating data supports the concept that alterations of cholesterol metabolism might influence the development of AD. In addition, Aβ42 has been shown to exert an inhibitory effect on the expression of some cholesterol transporter in cultured neonatal astrocytes [48]. Brain apoE mediates also transport of cholesterol from astrocytes to neurons by forming HDL particles [49-51]. Our finding that neonatal astrocytes down-regulated genes of cholesterol synthesis (Dhcr24, Hmgsc1) and apoE when cultured on top of APdE9 brain sections is in line with the results of Canepa et al. (2011) [48] suggesting further that cholesterol synthesis is down-regulated in these cells. Our microarray results suggest that several genes involved in cholesterol biosynthesis were down-regulated also in adult astrocytes grown on APdE9 brain section (data not shown). ApoE expression and cholesterol synthesis are both regulated by oxysterols through LXR transcription factors [reviewed in 52] and their activity might be altered in neonatal astrocytes in the presence of Aβ or other substances of tg ApdE9 brain section. 24S-hydroxycholesterol (24OHC) and 27-hydroxycholesterol, are two most studied oxysterols involved in neurogenerative diseases and their levels are altered in AD brains [53, reviewed in 54]. In addition, 24OHC decreases the expression of genes related to cholesterol synthesis through transcription factors LXR and SREBP-2 in neuroblastoma cells [55].
In conclusion, the genetic response of adult astrocytes to external Aβ deposits is substantially greater in adult compared to neonatal astrocytes and involves genes encoding proteases and scavenging receptors as well as proteins with antioxidant functions. Our data also indicates a distinct phenotype shift of adult astrocytes towards Aβ clearing function upon exposure to natural human Aβ depostis whereas no such alterations were detected in neonatal astrocytes. Adult rather than neonatal astrocytes should be used as a cell culture model for AD and possibly other neurodegenerative diseases and may reveal relevant targets for novel therapeutic approaches.
5. ACKNOWLEDGEMENTS
This work was supported by the Academy of Finland, the Finnish Funding Agency for Technology and Innovation, Sigrid Juselius Foundation, Saastamoinen Foundation and Biocenter Finland.
REFERENCES
- Cole, S.L. and Vassar, R. (2007) The Alzheimer’s disease beta-secretase enzyme, BACE1. Molecular Neurodegeneration, 2, 22. doi:10.1186/1750-1326-2-22
- Giannakopoulos, P., Herrmann, F.R., Bussière, T., Bouras, C., Kövari, E., Perl, D.P., Morrison, J.H., Gold, G. and Hof, P.R. (2003) Tangle and neuron numbers, but not amyloid load, predict cognitive status in Alzheimer’s disease. Neurology, 60, 1495-500.
- Delaère, P., Duyckaerts, C., Masters, C., Beyreuther, K., Piette, F. and Hauw, J.J. (1990) Large amounts of neocortical beta A4 deposits without neuritic plaques nor tangles in a psychometrically assessed, non-demented person. Neuroscience Letters, 116, 87-93. doi:10.1016/0304-3940(90)90391-L
- Dickson, D.W., Crystal, H.A., Mattiace, L.A., Masur, D.M., Blau, A.D., Davies, P., Yen, S.H. and Aronson, M.K. (1992) Identification of normal and pathological aging in prospectively studied nondemented elderly humans. Neurobiology of Aging, 13, 179-189. doi:10.1016/0197-4580(92)90027-U
- Davis, D.G., Schmitt, F.A., Wekstein, D.R. and Markesbery, W.R. (1999) Alzheimer neuropathologic alterations in aged cognitively normal subjects. Journal of Neuropathology & Experimental Neurology, 58, 376-388.
- Lesné, S., Koh, M.T., Kotilinek, L., Kayed, R., Glabe, C.G., Yang, A., Gallagher, M. and Ashe, K.H. (2006) A specific amyloid-beta protein assembly in the brain impairs memory. Nature, 440, 352-357. doi:10.1038/nature04533
- Fukumoto, H., Tokuda, T., Kasai, T., Ishigami, N., Hidaka, H., Kondo, M., Allsop, D. and Nakagawa, M. (2010) High-molecular-weight beta-amyloid oligomers are elevated in cerebrospinal fluid of Alzheimer patients. FASEB Journal, 24, 2716-2726. doi:10.1096/fj.09-150359
- Oddo, S., Caccamo, A., Kitazawa, M., Tseng, B.P. and LaFerla, F.M. (2003) Amyloid deposition precedes tangle formation in a triple transgenic model of Alzheimer’s disease. Neurobiology of Aging, 24, 1063-1070. doi:10.1016/j.neurobiolaging.2003.08.012
- Oddo, S., Billings, L., Kesslak, J.P., Cribbs, D.H. and LaFerla, F.M. (2004) Abeta immunotherapy leads to clearance of early, but not late, hyperphosphorylated tau aggregates via the proteasome. Neuron, 43, 321-332.
- Funato, H., Yoshimura, M., Yamazaki, T., Saido, T.C., Ito, Y., Yokofujita, J., Okeda, R. and Ihara, Y. (1998) Astrocytes containing amyloid beta-protein (Abeta)- positive granules are associated with Abeta40-positive diffuse plaques in the aged human brain. American Journal of Pathology, 152, 983-992.
- Nagele, R.G., D’Andrea, M.R., Lee, H., Venkataraman, V. and Wang, H.Y. (2003) Astrocytes accumulate A beta 42 and give rise to astrocytic amyloid plaques in Alzheimer’s disease brains. Brain Research, 971, 197-209. doi:10.1016/S0006-8993(03)02361-8
- Koistinaho, M., Lin, S., Wu, X., Esterman, M., Koger, D., Hanson, J., Higgs, R., Liu, F., Malkani, S., Bales, K.R. and Paul, S.M. (2004) Apolipoprotein E promotes astrocyte colocalization and degradation of deposited amyloid-beta peptides. Nature Medicine, 10, 719-726. doi:10.1038/nm1058
- Matsunaga, W., Shirokawa, T. and Isobe, K. (2003) Specific uptake of Abeta1-40 in rat brain occurs in astrocyte, but not in microglia. Neuroscience Letters, 342, 129-131. doi:10.1016/S0304-3940(03)00240-4
- Wyss-Coray, T., Loike, J.D., Brionne, T.C., Lu, E., Anankov, R., Yan, F., Silverstein, S.C. and Husemann, J. (2003) Adult mouse astrocytes degrade amyloid-b in vitro and in situ. Nature Medicine, 9, 453-457. doi:10.1038/nm838
- Pihlaja, R., Koistinaho, J., Malm, T., Sikkilä, H., Vainio, S. and Koistinaho, M. (2008) Transplanted astrocytes internalize deposited beta-amyloid peptides in a transgenic mouse model of Alzheimer’s disease. Glia, 56, 154-163. doi:10.1002/glia.20599
- Freeman, M.R. (2010) Specification and morphogenesis of astrocytes. Science, 330, 774-778. doi:10.1126/science.1190928
- Lund, R.D. and Lund, J.S. (1972) Development of synaptic patterns in the superior colliculus of the rat. Brain Research, 42, 1-20. doi:10.1016/0006-8993(72)90038-8
- Warton, S.S. and McCart, R. (1989) Synaptogenesis in the stratum griseum superficiale of the rat superior colliculus. Synapse, 3, 136-148.
- Ullian, E.M., Christopherson, K.S. and Barres, B.A. (2004) Role for glia in synaptogenesis. Glia, 47, 209-216. doi:10.1002/glia.20082
- Okabe, M., Ikawa, M., Kominami, K., Nakanishi, T. and Nishimune, Y. (1997) “Green mice” as a source of ubiquitous green cells. FEBS Letters, 407, 313-319. doi:10.1016/S0014-5793(97)00313-X
- Jankowsky, J.L., Fadale, D.J., Anderson, J., Xu, G.M., Gonzales, V., Jenkins, N.A., Copeland, N.G., Lee, M.K., Younkin, L.H., Wagner, S.L., Younkin, S.G. and Borchelt, D.R. (2004) Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Human Molecular Genetics, 13, 159-170. doi:10.1093/hmg/ddh019
- Kallio, M.A., Tuimala, J.T., Hupponen, T., Klemelä, P., Gentile, M., Scheinin, I., Koski, M., Käki, J. and Korpelainen, E.I. (2011) Chipster: user-friendly analysis software for microarray and other high-throughput data. BMC Genomics, 12, 507. doi:10.1186/1471-2164-12-507
- Irizarry, R.A., Hobbs, B., Collin, F., Beazer-Barclay, Y.D., Antonellis, K.J., Scherf, U. and Speed, T.P. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics, 4, 249-264. doi:10.1093/biostatistics/4.2.249
- Shannon, P., Markiel, A., Ozier, O., Baliga, N.S., Wang J.T., Ramage, D., Amin, N., Schwikowski, B. and Ideker, T. (2003) Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research, 13, 2498-2504. doi:10.1101/gr.1239303
- Maere, S., Heymans, K. and Kuiper, M. (2005) BiNGO: A cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics, 21, 3448-3449. doi:10.1093/bioinformatics/bti551
- Hu, Z.L., Bao, J. and Reecy, J.M. (2008) CateGOrizer: A web-based program to batch analyzegene ontology classification categories. Online Journal of Bioinformatics, 9, 108-112.
- Cahoy, J.D., Emery, B., Kaushal, A., Foo, L.C., Zamanian, J.L., Christopherson, K.S., Xing, Y., Lubischer, J.L., Krieg, P.A., Krupenko, S.A., Thompson, W.L. and Barres, B.A. (2008) A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. Journal of Neuroscience, 28, 264-278. doi:10.1523/JNEUROSCI.4178-07.2008
- Paresce, D.M., Ghosh, R.N. and Maxfield, F.R. (1996) Microglial cells internalize aggregates of the Alzheimer’s disease amyloid beta-protein via a scavenger receptor. Neuron, 17, 553-565. doi:10.1016/S0896-6273(00)80187-7
- Jiang, Y., Oliver, P., Davies, K.E. and Platt, N. (2006) Identification and characterization of murine SCARA5, a novel class A scavenger receptor that is expressed by populations of epithelial cells. Journal of Biological Chemistry, 281, 11834-11845. doi:10.1074/jbc.M507599200
- Li, J.Y., Paragas, N., Ned, R.M., Qiu, A., Viltard, M., Leete, T., Drexler, I.R., Chen, X., Sanna-Cherchi, S., Mohammed, F., Williams, D., Lin, C.S., Schmidt-Ott, K.M., Andrews, N.C. and Barasch, J. (2009) Scara5 is a ferritin receptor mediating non-transferrin iron delivery. Developmental Cell, 16, 35-46. doi:10.1016/j.devcel.2008.12.002
- Bertram, L., Lill, C.M. and Tanzi, R.E. (2010) The genetics of Alzheimer disease: Back to the future. Neuron, 68, 270-281. doi:10.1016/j.neuron.2010.10.013
- Rebeck, G.W., Reiter, J.S., Strickland, D.K., Hyman, B.T. (1993) Apolipoprotein E in sporadic Alzheimer’s disease: Allelic variation and receptor interactions. Neuron, 11, 575-580. doi:10.1016/0896-6273(93)90070-8
- Saunders, A.M., Strittmatter, W.J., Schmechel, D., George-Hyslop, P.H., Pericak-Vance, M.A., Joo, S.H., Rosi, B.L. and Gusella, (1993) Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology, 43, 1467-1472.
- Strittmatter, W.J., Saunders, A.M., Schmechel, D., Pericak-Vance, M., Enghild, J., Salvesen, G.S. and Roses, A.D. (1993) Apolipoprotein E: High-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proceedings of the National Academy of Sciences of the United States of America, 90, 1977-1981.
- Shirotani, K., Tsubuki, S., Iwata, N., Takaki, Y., Harigaya, W., Maruyama, K., Kiryu-Seo, S., Kiyama, H., Iwata, H., Tomita, T., Iwatsubo, T. and Saido, T.C. (2001) Neprilysin degrades both amyloid beta peptides 1-40 and 1-42 most rapidly and efficiently among thiorphanand phosphoramidon-sensitive endopeptidases. Journal of Biological Chemistry, 276, 21895-218901. doi:10.1074/jbc.M008511200
- Grau, S., Baldi, A., Bussani, R., Tian, X., Stefanescu, R., Przybylski, M., Richards, P., Jones, S.A., Shridhar, V., Clausen, T. and Ehrmann, M. (2005) Implications of the serine protease HtrA1 in amyloid precursor protein processing. Proceedings of the National Academy of Sciences of the United States of America, 102, 6021-6026. doi:10.1073/pnas.0501823102
- Chen, Y., Vartiainen, N.E., Ying, W., Chan, P.H., Koistinaho, J. and Swanson, R.A. (2001) Astrocytes protect neurons from nitric oxide toxicity by a glutathione-dependent mechanism. Journal of Neurochemistry, 77, 1601-1610. doi:10.1046/j.1471-4159.2001.00374.x
- De Strooper, B. (2010) Proteases and proteolysis in Alzheimer disease: A multi-factorial view on the disease process. Physiological Reviews, 90, 465-494. doi:10.1152/physrev.00023.2009
- Alarcón, R., Fuenzalida, C., Santibáñez, M. and von Bernhardi, R. (2005) Expression of scavenger receptors in glial cells. Comparing the adhesion of astrocytes and microglia from neonatal rats to surface-bound beta-amyloid. Journal of Biological Chemistry, 280, 30406-30415. doi:10.1074/jbc.M414686200
- Allaman, I., Gavillet, M., Bélanger, M., Laroche, T., Viertl, D., Lashuel, H.A. and Magistretti, P.J. (2010) Amyloid-beta aggregates cause alterations of astrocytic metabolic phenotype: Impact on neuronal viability. Journal of Neuroscience, 30, 3326-3338.
- Nakamura, K., Ohya, W., Funakoshi, H., Sakaguchi, G., Kato, A., Takeda, M., Kudo, T. and Nakamura, T. (2006) Possible role of scavenger receptor SRCL in the clearance of amyloid-beta in Alzheimer’s disease. Journal of Neuroscience Research, 84, 874-890. doi:10.1002/jnr.20992
- Baruch-Suchodolsky, R. and Fischer, B. (2008) Soluble amyloid-β(1-28) copper(I)/copper(II)/iron(II) complexes are potent antioxidants in cell-free systems. Biochemistry, 47, 7796-7806. doi:10.1021/bi800114g
- Faller, P., Hureau, C. (2009) Bioinorganic chemistry of copper and zinc ions coordinated to amyloid-beta peptide. Dalton Transactions, 21, 1080-1094. doi:10.1039/B813398K
- Grundke-Iqbal, I., Fleming, J., Tung, Y.C., Lassmann, H., Iqbal, K. and Joshi, J.G. (1990) Ferritin is a component of the neuritic (senile) plaque in Alzheimer dementia. Acta Neuropathologica, 81, 105-110. doi:10.1007/BF00334497
- Lovell, M.A., Robertson, J.D., Teesdale, W.J., Campbell, J.L., Markesbery, W.R. (1998) Copper, iron and zinc in Alzheimer’s disease senile plaques. Journal of the Neurological Sciences, 158, 47-52. doi:10.1016/S0022-510X(98)00092-6
- Yan, P., Hu, X., Song, H., Yin, K., Bateman, R.J., Cirrito, J.R., Xiao, Q., Hsu, F.F., Turk, J.W., Xu, J., Hsu, C.Y., Holtzman, D.M. and Lee, J.M. (2006) Matrix metalloproteinase-9 degrades amyloid-beta fibrils in vitro and compact plaques in situ. Journal of Biological Chemistry, 281, 24566-22574. doi:10.1074/jbc.M602440200
- Heeren, J., Grewal, T., Laatsch, A., Becker, N., Rinninger, F., Rye, K.A. and Beisiegel, U. (2004) Impaired recycling of apolipoprotein E4 is associated with intracellular cholesterol accumulation. Journal of Biological Chemistry, 279, 55483-55492. doi:10.1074/jbc.M409324200
- Canepa, E., Borghi, R., Viña, J., Traverso, N., Gambini, J., Domenicotti, C., Marinari, U.M., Poli, G., Pronzato, M.A. and Ricciarelli, R. (2011) Cholesterol and amyloid-β: Evidence for a cross-talk between astrocytes and neuronal cells. Journal of Alzheimer’s Disease, 25, 645- 653. doi:10.3233/JAD-2011-110053
- Gong, J.S., Morita, S.Y., Kobayashi, M., Handa, T., Fujita, S.C., Yanagisawa, K. and Michikawa, M. (2007) Novel action of apolipoprotein E (ApoE): ApoE isoform specifically inhibits lipid-particle-mediated cholesterol release from neurons. Molecular Neurodegeneration, 2, 9. doi:10.1186/1750-1326-2-9
- Ito, J., Zhang, L.Y., Asai, M. and Yokoyama, S. (1999) Differential generation of high-density lipoprotein by endogenous and exogenous apolipoproteins in cultured fetal rat astrocytes. Journal of Neurochemistry, 72, 2362- 2369.
- Krimbou, L., Denis, M., Haidar, B., Carrier, M., Marcil, M. and Genest, J. Jr. (2004) Molecular interactions between apoE and ABCA1: Impact on apoE lipidation. Journal of Lipid Research, 45, 839-848. doi:10.1194/jlr.M300418-JLR200
- Vaya, J. and Schipper, H.M. (2007) Oxysterols, cholesterol homeostasis, and Alzheimer disease. Journal of Neurochemistry, 102, 1727-1737. doi:10.1111/j.1471-4159.2007.04689.x
- Heverin, M., Bogdanovic, N., Lütjohann, D., Bayer, T., Pikuleva, I., Bretillon, L., Diczfalusy, U., Winblad, B. and Björkhem, I. (2004) Changes in the levels of cerebral and extracerebral sterols in the brain of patients with Alzheimer’s disease. Journal of Lipid Research, 45, 186-193. doi:10.1194/jlr.M300320-JLR200
- Leoni, V. and Caccia, C. (2011) Oxysterols as biomarkers in neurodegenerative diseases. Chemistry and Physics of Lipids, 164, 515-524. doi:10.1016/j.chemphyslip.2011.04.002
- Cartagena, C.M., Burns, M.P. and Rebeck, G.W. (2010) 24S-hydroxycholesterol effects on lipid metabolism genes are modeled in traumatic brain injury. Brain Research, 1319, 1-12. doi:10.1016/j.brainres.2009.12.080
NOTES
*Equal contribution.

