Open Journal of Genetics
Vol.3 No.2A1(2013), Article ID:33669,11 pages DOI:10.4236/ojgen.2013.32A1001
Vectors for gene therapy: A place for DNA transposon
![]()
Mer Molécules Santé EA2160, UFR Sciences et Techniques, Université du Maine, Le Mans, France
Email: bchenais@univ-lemans.fr
Copyright © 2013 Benoît Chénais. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 21 January 2013; revised 22 February 2013; accepted 10 March 2013
Keywords: Transposable Element; Gene Delivery; Non-Viral Vectors; Piggybac; Sleeping Beauty
ABSTRACT
Gene therapy offers important perspectives in current and future medicine but suffers from imperfect vectors for the delivery of the therapeutic gene. Most preclinical and clinical trials have been based on the use of viral vectors, which have evident advantages but also some serious disadvantages. In the past decade the use of DNA transposon-based systems for gene delivery has emerged as a non-viral alternative. DNA transposon vector engineering remains largely in a preclinical phase but some interesting results have been obtained. This mini-review aims to provide the current state of the art on DNA transposon vectors used in a gene therapy perspective.
1. INTRODUCTION
Gene therapy has emerged as a promising method to cure genetic diseases or at least improve the status of patients. If the idea of replacing an “abnormal” dysfunctional gene by its “normal” functional counterpart may look simple, technical difficulties are often encountered such as the stable and targeted insertion of the therapeutic gene into the host genome. Several gene therapy strategies can be distinguished that address different kinds of pathologies. Recessive monogenic hereditary diseases, such as cystic fibrosis and Duchenne’s muscular dystrophy, could be treated by the addition of the “normal” gene counterpart that will produce a sufficient amount of functional protein to reverse the disease. However, some other monogenic defects will require the replacement of the “abnormal” gene by its “normal” allele and not solely its addition. In some cases of acquired and/or polygenic diseases such as diabetes, the replacement of one of the deleterious alleles or the addition of one “normal” gene may be sufficient to fight the disease. Moreover, in complex pathologies that mix genetic predisposition with environmental factors (cancer diseases for example), different gene therapy strategies may be useful, such as the extinction of a targeted gene expression or the introduction of a suicide gene. The common challenge is to introduce the new DNA durably into the genome of pathologic cells without disturbing the regulation of “normal” gene expression. This is the main objective of vector development. Over 1340 human gene therapy clinical trials were completed in 28 countries between 1989 and 2007 [1]. However, very few phase-II and phase-III clinical trials have been conducted that mainly use viruses as vectors [2]. Following a brief description of virus-based vectors, the advantages offered by DNA transposons for gene therapy will be presented with some examples of successful applications.
2. VIRUS-BASED VECTORS: PROMISE AND DISAPPOINTMENT
Virus-based vectors are the most commonly used gene delivery system because of their ability to integrate the therapeutic gene into the host genome and ensure stable and long term gene expression. The second advantage of virus-based vectors is the combination of the DNA vector with a highly efficient DNA delivery system in the cell, i.e. the natural infection process. The main virusbased vectors used are derived from retroviruses (especially lentiviruses), adenoviruses and adeno-associated viruses [2,3]. For example, 68% of the gene therapy clinical trials completed in to date used virus-based vectors [1,2]. Unfortunately some fatal adverse events have clouded the sky. In 1999, a patient treated for ornithine transcarbamylase deficiency with an adenoviral gene transfer developed a fatal systemic inflammatory response syndrome [4]. In 2000, a French clinical trial using a retrovirus-based gene therapy on children suffering from X-linked severe combined immunodeficiency syndrome (X-SCID) seemed more promising, and 10 patients appeared successfully cured [5]. However, two years later two of the ten children treated developed T-cell lymphoma and one died [6]. This major adverse event was subsequently explained by the insertion of the retroviral vector in the vicinity of the LMO-2 gene that encodes a crucial regulator of haematopoeisis [7]. This insertional mutagenesis event resulted in the dysregulation of LMO- 2 gene expression that induced T-cell proliferation and then lymphoma [7,8]. So, virus-based vectors may have genotoxic effects, caused by deregulation of gene expression at the transcriptional or posttranscriptional level. In particular, integration of retroviral (lentiviral or gammaretroviral) vectors within transcribed genes has a significant potential to affect their expression by interfering with splicing and polyadenylation of primary transcripts [9].
Despite their recognized disadvantages, adenoviruses and retroviruses remained the two most widely-used vectors for gene therapy to date [1,2]. The adeno-associated virus (AAV) displays particularities making it a safer virus-vector [3]. AAV is a parvovirus, the proliferation of which depends on a helper virus, usually an adenovirus or a herpes virus [10]. Wild-type AAV is nonpathogenic with low immunogenicity and integrates into a specific point of the genome (19q 13qter) at high frequency and without genotoxicity to the host [11-14]. However this integration specificity is lost with engineered AAV, especially when increased cargo capacity is tried. Several AAV-based gene therapy trials have been launched for cancers, Parkinson’s disease, Alzheimer’s disease, and genetic diseases such as Leber’s congenital amaurosis (LCA). LCA is a common cause of blindness in infants and children and two AAV-based gene therapy trails reported improved vision without serious adverse effects [15,16]. Nevertheless the main drawback of AAV is its limited cargo capacity. Furthermore both AAV and adenoviruses remain largely episomal, requiring repeated delivery that may compromise efficacy [17] and might induce a severe immune response [18]. Furthermore the presence of AAV-neutralizing antibodies prevalent in humans can hamper the clinical success of AAV-based gene therapy. For a more comprehensive review of pros and cons of viral vectors the reader may refer to Patel and Misra, 2011 [19].
The fatal adverse effects associated with adenoviruses and retroviruses and the limited cargo capacity of AAV prompted the investigation of non-viral vectors, especially those based on DNA transposons.
3. DNA TRANSPOSON-BASED VECTORS
DNA transposons are a group of transposable elements present in most (if not all) organisms from bacteria to mammals [20], but poorly represented in the human genome [21]. The main families of DNA transposons (Tc1/ mariner, hAT and PiggyBac) have a simple structure with a unique gene flanked by inverted terminal repeats (Figure 1(a)). Their mobility is supported by the transposon-
 (a)
(a)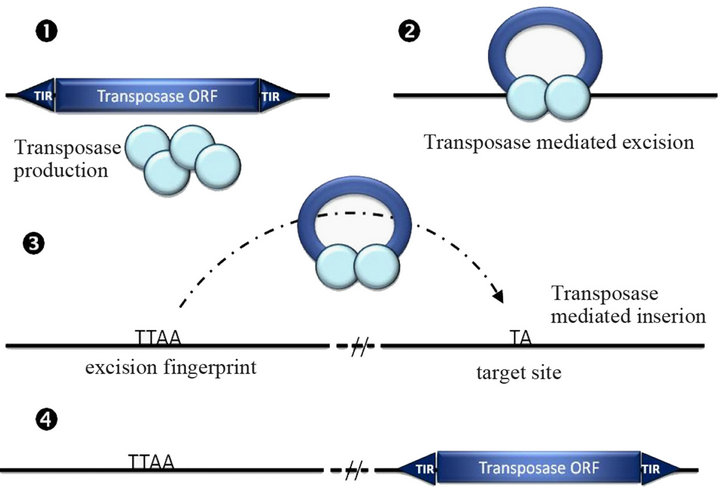 (b)
(b)
Figure 1. (a) Structure and (b) simplified “cut and paste” transposition mechanism of a DNA transposon. ORF, open reading frame; TIR, terminal inverted repeats; UTR, untranslated region. The light blue circles in B represent the transposase proteins encoded by the transposon ORF (b1). Transposase dimer (or more) binds to ITR (b2) and excises transposon DNA, then the transpososome is displaced (b3) to be integrated in another genomic site (b4).
encoded transposase enzyme in a “cut and paste” mechanism (Figure 1(b)). This simple genome integration mechanism makes DNA transposons very attractive as gene delivery tools. To achieve this goal a two-plasmid system should be developed consisting of a helper plasmid expressing the transposase and a donor plasmid with the terminal repeat sequence flanking the gene of interest (Figure 2) [22-24]. Using this system, several DNA transposons, such as P element, PiggyBac, Tol2, Hsmar1 and Sleeping Beauty, have been utilized for gene delivery and mutagenesis in invertebrate and vertebrate cells [24- 33]. Among the DNA transposons, PiggyBac, Tol2 and Sleeping Beauty have been evaluated for gene therapy in animal experiments, primary cell gene delivery and a few pre-clinical trials [34-37]. The advantages and disadvantages of DNA transposon-based gene delivery will be discussed below with respect to cargo capacity, efficiency, gene silencing and targeted insertion with special emphasis on PiggyBac, Tol2 and Sleeping Beauty.
3.1. Cargo Capacity
A large cargo capacity is desired for therapeutic application and DNA transposon-based vectors appear as good candidates with respect to this criterion. Indeed Tol2 and PiggyBac vectors are able to integrate up to 11 and 9.1 kb of foreign DNA respectively [28,38]. Furthermore the large cargo capacity of Tol2 was further evidenced with a ~70 kb BAC transfer in mouse and zebrafish genomes [39]. By contrast the transposition activity of wild type Sleeping Beauty is significantly decreased when foreign DNA exceeds the initial 1.7 kb size [40].
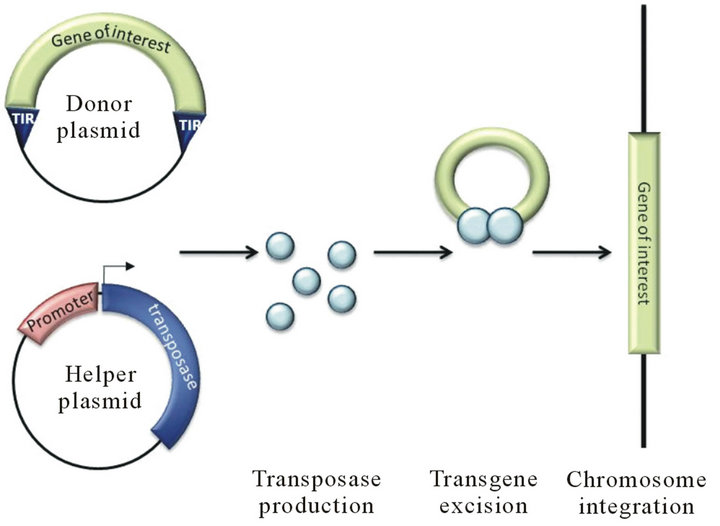
Figure 2. Schematic view of a DNA-transposon system for gene delivery. Both donor and helper plasmids are transfected into the host cells. The helper plasmid is an expression vector that allows transposase production and the donor plasmid bears the gene of interest that will be integrated into the host genome by the transposase. The light blue circles represent the transposase proteins produced from helper plasmid.
3.2. Transposition Efficiency
The transposition efficiency of PiggyBac, Tol2 and a mutant Sleeping Beauty transposase (SB11) was compared in several human cell lines. Tol2 and PiggyBac were highly active in all cell lines tested while SB11 was obviously less efficient and marginally or totally inactive in two cell lines, i.e. HeLa and H1299, respectively [30]. However, a new hyperactive Sleeping Beauty transposase (SB100X) displays transposition efficiency comparable to PiggyBac under non-restrictive conditions [41]. So, transposase mutagenesis may be an interesting way to increase DNA transposon-based vector efficiency as shown for Sleeping Beauty hyperactive mutants [41,42] and hyperactive or mouse-codon optimized PiggyBac [43,44].
3.3. Gene Silencing and Stability of Gene Insertion
Any transgene introduced into the host genome is a potential target for the position effect that causes transgene silencing. This phenomenon is observed particularly in viral vectors but also appears in plasmid and DNA-transposon vectors. However, in the study by Grabundzija et al. transgene silencing observed following DNA-based gene delivery using Tol2, PiggyBac or SB100X, was found to be very low (3.8% vs 26.5% in control) [45]. This result has to be moderated since earlier results reported progressive post-integrative gene silencing after Sleeping Beauty transposition [46]. Furthermore post-integration transgene silencing may be higher than previously observed as suggested by Meir and Wu [47] indicating 63% for Sleeping Beauty, 46% for Tol2 and 20% for PiggyBac. A comparative study between viral vectors and non viral vectors used under the same conditions may be of interest to decide whether DNA transposon-based vectors have advantages or not with respect to the gene silencing effect [47]. Interestingly, reducing the terminal repeats of PiggyBac to 40 bp and 67 bp from 245 bp and 311 bp of 3’ and 5’ terminal repeats respectively in the commonly used PiggyBac, enhances Piggy Bac’s transposition activity [48]. This suggests a possible interaction between epigenetic silencing factors and sequences present in the common PiggyBac but not in its shorter form. Furthermore, this shorter PiggyBac, called micro-PiggyBac, lacks many activator sequences initially present in the 3’ PiggyBac terminal repeats that influence neighboring gene expression [49].
In addition, transgene delivery without drug selection is advantageous for therapeutic purposes but may interfere with stable insertion. A recent study compared the stable insertion ability of the Tol2, PiggyBac and Sleeping Beauty DNA-transposon gene delivery systems in human peripheral blood and umbilical cord blood-derived T-cells [50]. This study showed that PiggyBac is the most efficient of the three DNA transposon systems in mediating stable gene insertion.
3.4. Insertion Site Targeting
Site targeting is a crucial point in gene transfer and especially in the human gene therapy perspective. Most of the integrating viral vectors have integration site preferences leading to an integration site bias that may explain some adverse effects such as gene silencing or activation of cancer-related genes [51-54]. By contrast, genome-wide targeting profiles of PiggyBac, Sleeping Beauty and Tol2 in various human cell lines and human primary cells clearly demonstrate that the three transposon systems display a lower frequency in targeting near cancer-related genes and less bias in targeting active genes compared to viral-based vectors [29,45,48,50,55, 56]. A detailed analysis of this aspect is available in Meir and Wu (2011) [47]. The ability of PiggyBac transposase to be molecularly engineered without a loss of activity allows the construction of PiggyBac transposase chimeras with a DNA binding domain, such as the zinc finger DNA binding domain that would recognize specific and safe integration sites.
4. PHENOTYPIC CORRECTION IN DISEASE MODELS AND PRECLINICAL TRIALS
Stable chromosomal insertion can be achieved from a transposon-based delivery system which results in robust, long-term expression of a therapeutic transgene in certain primary stem cells as well as in vivo (Table 1). This includes extended expression of human clotting factor IX in factor IX-deficient mice [57], in vivo correction of murine tyrosinemia type I [58], sustainable correction of junctional epidermolysis bullosa [59], long term expression of factor VIII and phenotypic correction in hemophilic mice [60-62], correction of lung allograft fibrosis
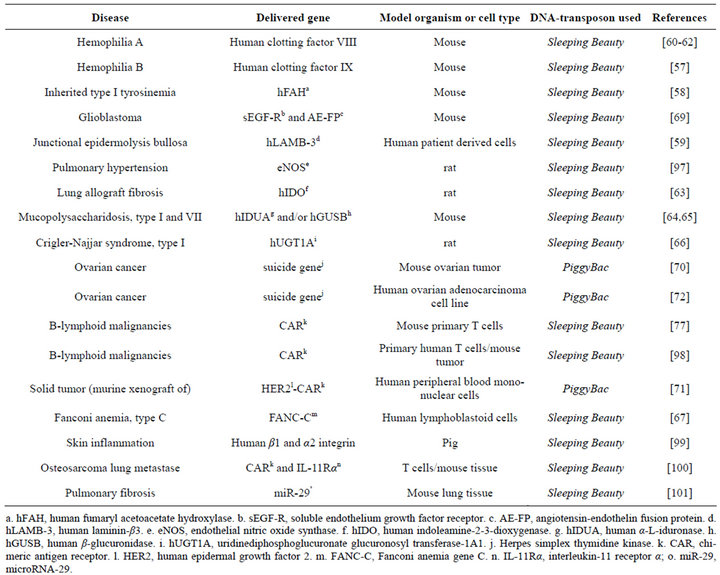
Table 1. Examples of successful DNA transposon-based gene delivery in disease models.
[63], prolonged expression of lysosomal enzymes in mouse liver and correction of mucopolysaccharidosis [64, 65]; long term reduction of jaundice in Gunn rats [66], correction of Fanconi anemia type C in human patientderived cells [67], and stable integration and continuous expression of pigment epithelium-derived factor gene in retinal and iris pigment epithelial cells [68]. Another example of successful Sleeping Beauty-based gene delivery is given by the antiangiogenic gene therapy of mice bearing intracranial human glioblastoma, which causes tumor regression and improves animal survival [69]. The PiggyBac system was also used in cancer cell gene therapy [70,71]. For example, Kang et al. applied a genedirected enzyme prodrug therapy using a PiggyBac-based vector to treat mouse ovarian cancer and human ovarian adenocarcinoma cells [70,72].
In the ex vivo gene delivery approach, the therapeutic gene construct is introduced into a selected cell population, removed from the patient and then the treated cells are infused back into the same patient. Efficient transfection and stable transgene expression using (Sleeping Beauty or PiggyBac) DNA-transposon based systems were achieved in several types of human stem cells, including cord blood-derived CD34+ hematopoietic progenitors [41, 73-76], primary T cells [50,71,77-81], and human embryonic stem cells [82,83]. So, transposon-based gene delivery systems such as Sleeping Beauty [78,81,83] or PiggyBac [71,79,80] allow stable gene transfer into T cells and an efficient expression of the therapeutic transgene.
The first clinical application of the Sleeping Beauty system was tested using autologous T cells genetically modified to redirect specificity to B-lineage malignancies [84]. The first generation Sleeping Beauty vector phase-I clinical trials carried a chimeric antigen receptor (CAR) to render the T cells cytotoxic specifically towards CD19+ B-lineages tumors (reviewed in [85]. In the next generation of clinical trials the chimeric antigen receptor carried by Sleeping Beauty vectors combined with a CD28-CD3-z fusion to provide T cells with an endodomain to achieve CD19-specific activation events of cytolysis, cytokine production, and proliferation [86]. In addition Torikai et al., 2012 recently reported an important step towards eliminating the need to generate patient-specific T cells by using genetically-edited T cells that express CAR without the endogenous αβ T cell receptor and are therefore suitable for administration in multiple recipients [87].
5. DRAWBACKS AND FUTURE CHALLENGES
The DNA transposon-based gene delivery systems encounter some drawbacks which must be overcome, some being shared with other viral and non-viral vectors. The behavior of DNA transposon in different cell types remains largely unknown, especially regarding PiggyBac and Tol2. One might worry some cytotoxicity of transposase overproduction, leading to overproduction inhibition. Studies with Sleeping Beauty have been conducted in numerous cell types showing a cell-type dependent activity and the interaction with human factors such as HMGB1 and Miz-1 [24,36,45]. Unlike Sleeping Beauty, PiggyBac transposition seems to be cell type independent. However cytotoxic interaction of PiggyBac with cellular factors cannot be ruled out since a little overproduction inhibition could be observed [30,44,47]. The existence of PiggyBac-like elements in the human genome [21] raises the question of safety concerns. Indeed a trans-mobilization of endogenous elements by the engineered transposase might be mutagenic. This does not apply to Tc-1 like elements such as Sleeping Beauty, which have no close relatives in the human genome. This is also one interest to develop new vectors based on distant sequences (e.g. from crabs∙∙∙) [22,88]. However the longer the foreign sequences introduced into the host genome, the greater the probability of evoking adverse consequences such as gene silencing and dysregulation of genes nearby. Therapeutic gene silencing and dysregulation of genes nearby are common drawback of DNA transposon-based vectors and viral vectors [3,47]. Epigenetic gene silencing was observed post-integration with Sleeping Beauty that was influenced by DNA methylation and histone deacetylation [46]. Epigenetic regulation of PiggyBac vector was also suggested by the results of Meir et al. [48].
Another concern shared by both non-viral and viral vectors, is the integration site specificity and its counterpart insertional mutagenesis. To overcome this point is the major challenge for gene therapy, and despite years of research, this has not been addressed to date. The addition of specific DNA binding domain, such as zinc-finger domain of known transcription factors, appears inefficient and reduces the transposition rate. Some outlook may come from artificial zinc-fingers which harbor modular structure and function making possible to recognize theoretically any sequence in the genome [37]. Finally, to my point of view, the major concern of DNA transposon-based delivery systems is to keep open the problem of DNA delivery into cells, with the drawbacks of all non-viral delivery systems (mainly toxicity and low efficiency). It is beyond the scope of this review to address the pros and cons of non-viral delivery systems (e.g. liposomes, polymers, electroporation∙∙∙) but it is a great challenge to develop efficient and non-cytotoxic reagents for DNA delivery across the cell membrane. Maybe the future resides in the hybrid technology combining the natural ability of viruses to cross cell membranes with efficient genomic insertion of DNA-transposon. Several Sleeping Beauty-virus combinations have been tried with herpes virus [89] or lentivirus [90,91] in human cells and adenovirus in mouse liver [92,93].
6. CONCLUDING REMARKS
Because of their lower immunogenicity and the fact that they are less prone to gene silencing than viral vectors, DNA transposon-based vectors appear to be a more desirable therapeutic gene delivery system. PiggyBac has some advantages over Sleeping Beauty or Tol2 because PiggyBac transposase can be molecularly modified without substantially losing its activity [47]. By contrast, any modification done on Tol2 and Sleeping Beauty transposases drastically reduces or completely eliminates their enzymatic activity [94]. According to Meir and Wu, 2011, PiggyBac seems currently the most promising DNA transposon to obtain a highly efficient gene delivery system capable of targeting the therapeutic gene at a predefined safe location in the host genome where the transgene can be stably and faithfully expressed without disturbing global gene expression. However, this ambitious goal has not yet been achieved and much progress is still required. Finally, improvement of cargo capacity, delivery efficiency and expression stability/persistence may be achieved with chimeric vectors combining viral (i.e. baculovirus or adenovirus) and non viral (i.e. DNA transposon) elements such as those recently developed [95,96].
7. ACKNOWLEDGEMENTS
The author is grateful to Marilyn Twell for revising the English text.
REFERENCES
- Edelstein, M.L., Abedi, M.R. and Wixon, J. (2007) Gene therapy clinical trials worldwide to 2007: An update. Journal of Gene Medicine, 9, 833-842. doi:10.1002/jgm.1100
- O’Reilly, M., Shipp, A., Rosenthal, E., Jambou, R., Shih, T., Montgomery, M., Gargiulo, L., Patterson, A. and Corrigan-Curay, J. (2012) NIH oversight of human gene transfer research involving retroviral, lentiviral, and adenoassociated virus vectors and the role of the NIH recombinant DNA advisory committee. Methods in Enzymology, 507, 313-335. doi:10.1016/B978-0-12-386509-0.00016-8
- Giacca, M. and Zacchigna, S. (2012) Virus-mediated gene delivery for human gene therapy. Journal of Controlled Release, 161, 377-388. doi:10.1016/j.jconrel.2012.04.008
- Raper, S.E., Chirmule, N., Lee, F.S., Wivel, N.A., Bagg, A., Gao, G.P., Wilson, J.M. and Batshaw, M.L. (2003) Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Molecular and Genetic Metabolism, 80, 148-158. doi:10.1016/j.ymgme.2003.08.016
- Cavazzana-Calvo, M., Hacein-Bey, S., de Saint Basile, G., Gross, F., Yvon, E., Nusbaum, P., Selz, F., Hue, C., Certain, S., Casanova, J.L., Bousso, P., Deist, F.L. and Fischer, A. (2000) Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science, 288, 669- 672. doi:10.1126/science.288.5466.669
- Hacein-Bey-Abina, S., von Kalle, C., Schmidt, M., Le Deist, F., Wulffraat, N., McIntyre, E., Radford, I., Villeval, J.L., Fraser, C.C., Cavazzana-Calvo, M. and Fischer, A. (2003) A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. New England Journal of Medicine, 348, 255-256. doi:10.1056/NEJM200301163480314
- Hacein-Bey-Abina, S., Von Kalle, C., Schmidt, M., McCormack, M.P., Wulffraat, N., Leboulch, P., Lim, A., Osborne, C.S., Pawliuk, R., Morillon, E., Sorensen, R., Forster, A., Fraser, P., Cohen, J.I., de Saint Basile, G., Alexander, I., Wintergerst, U., Frebourg, T., Aurias, A., Stoppa-Lyonnet, D., Romana, S., Radford-Weiss, I., Gross, F., Valensi, F., Delabesse, E., Macintyre, E., Sigaux, F., Soulier, J., Leiva, L.E., Wissler, M., Prinz, C., Rabbitts, T.H., Le Deist, F., Fischer, A. and Cavazzana-Calvo, M. (2003) LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science, 302, 415- 419. doi:10.1126/science.1088547
- Hacein-Bey-Abina, S., Garrigue, A., Wang, G.P., Soulier, J., Lim, A., Morillon, E., Clappier, E., Caccavelli, L., Delabesse, E., Beldjord, K., Asnafi, V., MacIntyre, E., Dal Cortivo, L., Radford, I., Brousse, N., Sigaux, F., Moshous, D., Hauer, J., Borkhardt, A., Belohradsky, B.H., Wintergerst, U., Velez, M.C., Leiva, L., Sorensen, R., Wulffraat, N., Blanche, S., Bushman, F.D., Fischer, A. and Cavazzana-Calvo, M. (2008) Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. Journal of Clinical Investigation, 118, 3132-3142. doi:10.1172/JCI35700
- Moiani, A. and Mavilio, F. (2012) Alternative splicing caused by lentiviral integration in the human genome. Methods in Enzymology, 507, 155-169. doi:10.1016/B978-0-12-386509-0.00008-9
- Goncalves, M.A. (2005) Adeno-associated virus: From defective virus to effective vector. Virology Journal, 2, 43. doi:10.1186/1743-422X-2-43
- Kotin, R.M., Menninger, J.C., Ward, D.C. and Berns, K.I. (1991) Mapping and direct visualization of a region-specific viral DNA integration site on chromosome 19q13- qter. Genomics, 10, 831-834. doi:10.1016/0888-7543(91)90470-Y
- Kotin, R.M., Siniscalco, M., Samulski, R.J., Zhu, X.D., Hunter, L., Laughlin, C.A., McLaughlin, S., Muzyczka, N., Rocchi, M. and Berns, K.I. (1990) Site-specific integration by adeno-associated virus. Proceedings of the National Academy of Sciences of the United States of America, 87, 2211-2215. doi:10.1073/pnas.87.6.2211
- Samulski, R.J., Zhu, X., Xiao, X., Brook, J.D., Housman, D.E., Epstein, N. and Hunter, L.A. (1991) Targeted integration of adeno-associated virus (AAV) into human chromosome 19. The EMBO Journal, 10, 3941-3950.
- Grieger, J.C. and Samulski, R.J. (2012) Adeno-associated virus vectorology, manufacturing, and clinical applications. Methods in Enzymology, 507, 229-254. doi:10.1016/B978-0-12-386509-0.00012-0
- Bainbridge, J.W., Smith, A.J., Barker, S.S., Robbie, S., Henderson, R., Balaggan, K., Viswanathan, A., Holder, G.E., Stockman, A., Tyler, N., Petersen-Jones, S., Bhattacharya, S.S., Thrasher, A.J., Fitzke, F.W., Carter, B.J., Rubin, G.S., Moore, A.T. and Ali, R.R. (2008) Effect of gene therapy on visual function in Leber’s congenital amaurosis. New England Journal of Medicine, 358, 2231-2239. doi:10.1056/NEJMoa0802268
- Maguire, A.M., Simonelli, F., Pierce, E.A., Pugh Jr., E.N., Mingozzi, F., Bennicelli, J., Banfi, S., Marshall, K.A., Testa, F., Surace, E.M., Rossi, S., Lyubarsky, A., Arruda, V.R., Konkle, B., Stone, E., Sun, J., Jacobs, J., Dell’Osso, L., Hertle, R., Ma, J.X., Redmond, T.M., Zhu, X., Hauck, B., Zelenaia, O., Shindler, K.S., Maguire, M.G., Wright, J.F., Volpe, N.J., McDonnell, J.W., Auricchio, A., High, K.A. and Bennett, J. (2008) Safety and efficacy of gene transfer for Leber’s congenital amaurosis. New England Journal of Medicine, 358, 2240-2248. doi:10.1056/NEJMoa0802315
- Riviere, C., Danos, O. and Douar, A.M. (2006) Longterm expression and repeated administration of AAV type 1, 2 and 5 vectors in skeletal muscle of immunocompetent adult mice. Gene Therapy, 13, 1300-1308. doi:10.1038/sj.gt.3302766
- Hartman, Z.C., Appledorn, D.M. and Amalfitano, A. (2008) Adenovirus vector induced innate immune responses: Impact upon efficacy and toxicity in gene therapy and vaccine applications. Virus Research, 132, 1-14. doi:10.1016/j.virusres.2007.10.005
- Patel, D.H. and Misra, A. (2011) Gene delivery using viral vectors. In: Misra, A., Ed., Challenges in Delivery of Therapeutic Genomics and Proteomics, Elsevier, Amsterdam, 207-270. doi:10.1016/B978-0-12-384964-9.00005-0
- Chénais, B., Caruso, A., Hiard, S. and Casse, N. (2012) The impact of transposable elements on eukaryotic genomes: From genome size increase to genetic adaptation to stressful environments. Gene, 509, 7-15. doi:10.1016/j.gene.2012.07.042
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., Funke, R., Gage, D., Harris, K., Heaford, A., Howland, J., Kann, L., Lehoczky, J., LeVine, R., McEwan, P., McKernan, K., Meldrim, J., Mesirov, J.P., Miranda, C., Morris, W., Naylor, J., Raymond, C., Rosetti, M., Santos, R., Sheridan, A., Sougnez, C., Stange-Thomann, N., Stojanovic, N., Subramanian, A., Wyman, D., Rogers, J., Sulston, J., Ainscough, R., Beck, S., Bentley, D., Burton, J., Clee, C., Carter, N., Coulson, A., Deadman, R., Deloukas, P., Dunham, A., Dunham, I., Durbin, R., French, L., Grafham, D., Gregory, S., Hubbard, T., Humphray, S., Hunt, A., Jones, M., Lloyd, C., McMurray, A., Matthews, L., Mercer, S., Milne, S., Mullikin, J.C., Mungall, A., Plumb, R., Ross, M., Shownkeen, R., Sims, S., Waterston, R.H., Wilson, R.K., Hillier, L.W., McPherson, J.D., Marra, M.A., Mardis, E.R., Fulton, L.A., Chinwalla, A.T., Pepin, K.H., Gish, W.R., Chissoe, S.L., Wendl, M.C., Delehaunty, K.D., Miner, T.L., Delehaunty, A., Kramer, J.B., Cook, L.L., Fulton, R.S., Johnson, D.L., Minx, P.J., Clifton, S.W., Hawkins, T., Branscomb, E., Predki, P., Richardson, P., Wenning, S., Slezak, T., Doggett, N., Cheng, J.F., Olsen, A., Lucas, S., Elkin, C., Uberbacher, E., Frazier, M., Gibbs, R.A., Muzny, D.M., Scherer, S.E., Bouck, J.B., Sodergren, E.J., Worley, K.C., Rives, C.M., Gorrell, J.H., Metzker, M.L., Naylor, S.L., Kucherlapati, R.S., Nelson, D.L., Weinstock, G.M., Sakaki, Y., Fujiyama, A., Hattori, M., Yada, T., Toyoda, A., Itoh, T., Kawagoe, C., Watanabe, H., Totoki, Y., Taylor, T., Weissenbach, J., Heilig, R., Saurin, W., Artiguenave, F., Brottier, P., Bruls, T., Pelletier, E., Robert, C., Wincker, P., Smith, D.R., Doucette-Stamm, L., Rubenfield, M., Weinstock, K., Lee, H.M., Dubois, J., Rosenthal, A., Platzer, M., Nyakatura, G., Taudien, S., Rump, A., Yang, H., Yu, J., Wang, J., Huang, G., Gu, J., Hood, L., Rowen, L., Madan, A., Qin, S., Davis, R.W., Federspiel, N.A., Abola, A.P., Proctor, M.J., Myers, R.M., Schmutz, J., Dickson, M., Grimwood, J., Cox, D.R., Olson, M.V., Kaul, R., Shimizu, N., Kawasaki, K., Minoshima, S., Evans, G.A., Athanasiou, M., Schultz, R., Roe, B.A., Chen, F., Pan, H., Ramser, J., Lehrach, H., Reinhardt, R., McCombie, W.R., de la Bastide, M., Dedhia, N., Blocker, H., Hornischer, K., Nordsiek, G., Agarwala, R., Aravind, L., Bailey, J.A., Bateman, A., Batzoglou, S., Birney, E., Bork, P., Brown, D.G., Burge, C.B., Cerutti, L., Chen, H.C., Church, D., Clamp, M., Copley, R.R., Doerks, T., Eddy, S.R., Eichler, E.E., Furey, T.S., Galagan, J., Gilbert, J.G., Harmon, C., Hayashizaki, Y., Haussler, D., Hermjakob, H., Hokamp, K., Jang, W., Johnson, L.S., Jones, T.A., Kasif, S., Kaspryzk, A., Kennedy, S., Kent, W.J., Kitts, P., Koonin, E.V., Korf, I., Kulp, D., Lancet, D., Lowe, T.M., McLysaght, A., Mikkelsen, T., Moran, J.V., Mulder, N., Pollara, V.J., Ponting, C.P., Schuler, G., Schultz, J., Slater, G., Smit, A.F., Stupka, E., Szustakowski, J., Thierry-Mieg, D., Thierry-Mieg, J., Wagner, L., Wallis, J., Wheeler, R., Williams, A., Wolf, Y.I., Wolfe, K.H., Yang, S.P., Yeh, R.F., Collins, F., Guyer, M.S., Peterson, J., Felsenfeld, A., Wetterstrand, K.A., Patrinos, A., Morgan, M.J., de Jong, P., Catanese, J.J., Osoegawa, K., Shizuya, H., Choi, S. and Chen, Y.J. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860-921. doi:10.1038/35057062
- Delauriere, L., Chénais, B., Hardivillier, Y., Gauvry, L. and Casse, N. (2009) Mariner transposons as genetic tools in vertebrate cells. Genetica, 137, 9-17. doi:10.1007/s10709-009-9370-2
- Ivics, Z., Li, M.A., Mates, L., Boeke, J.D., Nagy, A., Brad- ley, A. and Izsvak, Z., (2009) Transposon-mediated genome manipulation in vertebrates. Nature Methods, 6, 415-422. doi:10.1038/nmeth.1332
- Ivics, Z. and Izsvak, Z. (2010) The expanding universe of transposon technologies for gene and cell engineering. Mobile DNA, 1, 25. doi:10.1186/1759-8753-1-25
- Kawakami, K. (2007) Tol2: A versatile gene transfer vector in vertebrates. Genome Biology, 8, S1-S7. doi:10.1186/gb-2007-8-s1-s7
- O’Brochta, D.A. and Handler, A.M. (2008) Perspectives on the state of insect transgenics. Advances in Experimental Medicine and Biology, 627, 1-18. doi:10.1007/978-0-387-78225-6_1
- Thibault, S.T., Singer, M.A., Miyazaki, W.Y., Milash, B., Dompe, N.A., Singh, C.M., Buchholz, R., Demsky, M., Fawcett, R., Francis-Lang, H.L., Ryner, L., Cheung, L.M., Chong, A., Erickson, C., Fisher, W.W., Greer, K., Hartouni, S.R., Howie, E., Jakkula, L., Joo, D., Killpack, K., Laufer, A., Mazzotta, J., Smith, R.D., Stevens, L.M., Stuber, C., Tan, L.R., Ventura, R., Woo, A., Zakrajsek, I., Zhao, L., Chen, F., Swimmer, C., Kopczynski, C., Duyk, G., Winberg, M.L. and Margolis, J., (2004) A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nature Genetics, 36, 283-287. doi:10.1038/ng1314
- Ding, S., Wu, X., Li, G., Han, M., Zhuang, Y. and Xu, T., (2005) Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell, 122, 473-483. doi:10.1016/j.cell.2005.07.013
- Wilson, M.H., Coates, C.J. and George Jr., A.L. (2007) PiggyBac transposon-mediated gene transfer in human cells. Molecular Therapy, 15, 139-145. doi:10.1038/sj.mt.6300028
- Wu, S.C., Meir, Y.J., Coates, C.J., Handler, A.M., Pelczar, P., Moisyadi, S. and Kaminski, J.M. (2006) PiggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America, 103, 15008-15013. doi:10.1073/pnas.0606979103
- Miller, W.J. and Capy, P. (2004) Mobile genetic elements as natural tools for genome evolution. Methods in Molecular Biology, 260, 1-20.
- Schetelig, M.F., Gotschel, F., Viktorinova, I., Handler, A.M. and Wimmer, E.A. (2011) Recombination technologies for enhanced transgene stability in bioengineered insects. Genetica, 139, 71-78. doi:10.1007/s10709-010-9494-4
- Keravala, A., Liu, D., Lechman, E.R., Wolfe, D., Nash, J.A., Lampe, D.J. and Robbins, P.D. (2006) Hyperactive Himar1 transposase mediates transposition in cell culture and enhances gene expression in vivo. Human Gene Therapy, 17, 1006-1018. doi:10.1089/hum.2006.17.1006
- Hackett Jr., P.B., Aronovich, E.L., Hunter, D., Urness, M., Bell, J.B., Kass, S.J., Cooper, L.J. and McIvor, S. (2011) Efficacy and safety of Sleeping Beauty transposon-mediated gene transfer in preclinical animal studies. Current Gene Therapy, 11, 341-349. doi:10.2174/156652311797415827
- Hackett, P.B., Largaespada, D.A. and Cooper, L.J. (2010) A transposon and transposase system for human application. Molecular Therapy, 18, 674-683. doi:10.1038/mt.2010.2
- Ivics, Z. and Izsvak, Z. (2011) Nonviral gene delivery with the sleeping beauty transposon system. Human Gene Therapy, 22, 1043-1051. doi:10.1089/hum.2011.143
- Izsvak, Z., Hackett, P.B., Cooper, L.J. and Ivics, Z. (2010) Translating sleeping beauty transposition into cellular therapies: Victories and challenges. BioEssays, 32, 756- 767. doi:10.1002/bies.201000027
- Balciunas, D., Wangensteen, K.J., Wilber, A., Bell, J., Geurts, A., Sivasubbu, S., Wang, X., Hackett, P.B., Largaespada, D.A., McIvor, R.S. and Ekker, S.C. (2006) Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genetics, 2, e169. doi:10.1371/journal.pgen.0020169
- Suster, M.L., Sumiyama, K. and Kawakami, K. (2009) Transposon-mediated BAC transgenesis in zebrafish and mice. BMC Genomics, 10, 477. doi:10.1186/1471-2164-10-477
- Izsvak, Z., Ivics, Z. and Plasterk, R.H. (2000) Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. Journal of Molecular Biology, 302, 93-102. doi:10.1006/jmbi.2000.4047
- Mates, L., Chuah, M.K., Belay, E., Jerchow, B., Manoj, N., Acosta-Sanchez, A., Grzela, D.P., Schmitt, A., Becker, K., Matrai, J., Ma, L., Samara-Kuko, E., Gysemans, C., Pryputniewicz, D., Miskey, C., Fletcher, B., VandenDriessche, T., Ivics, Z. and Izsvak, Z. (2009) Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nature Genetics, 41, 753-761. doi:10.1038/ng.343
- Geurts, A.M., Yang, Y., Clark, K.J., Liu, G., Cui, Z., Dupuy, A.J., Bell, J.B., Largaespada, D.A. and Hackett, P.B. (2003) Gene transfer into genomes of human cells by the sleeping beauty transposon system. Molecular Therapy, 8, 108-117. doi:10.1016/S1525-0016(03)00099-6
- Cadinanos, J. and Bradley, A. (2007) Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Research, 35, e87. doi:10.1093/nar/gkm446
- Yusa, K., Zhou, L., Li, M.A., Bradley, A. and Craig, N.L. (2011) A hyperactive piggyBac transposase for mammalian applications. Proceedings of the National Academy of Sciences of the United States of America, 108, 1531-1536. doi:10.1073/pnas.1008322108
- Grabundzija, I., Irgang, M., Mates, L., Belay, E., Matrai, J., Gogol-Doring, A., Kawakami, K., Chen, W., Ruiz, P., Chuah, M.K., VandenDriessche, T., Izsvak, Z. and Ivics, Z. (2010) Comparative analysis of transposable element vector systems in human cells. Molecular Therapy, 18, 1200-1209. doi:10.1038/mt.2010.47
- Garrison, B.S., Yant, S.R., Mikkelsen, J.G. and Kay, M.A. (2007) Postintegrative gene silencing within the Sleeping Beauty transposition system. Molecular and Cellular Biology, 27, 8824-8833. doi:10.1128/MCB.00498-07
- Meir, Y.J. and Wu, S.C. (2011) Transposon-based vector systems for gene therapy clinical trials: Challenges and considerations. Chang Gung Medicine Journal, 34, 565- 579.
- Meir, Y.J., Weirauch, M.T., Yang, H.S., Chung, P.C., Yu, R.K. and Wu, S.C. (2011) Genome-wide target profiling of piggyBac and Tol2 in HEK 293: Pros and cons for gene discovery and gene therapy. BMC Biotechnology, 11, 28. doi:10.1186/1472-6750-11-28
- Shi, X., Harrison, R.L., Hollister, J.R., Mohammed, A., Fraser Jr., M.J. and Jarvis, D.L. (2007) Construction and characterization of new piggyBac vectors for constitutive or inducible expression of heterologous gene pairs and the identification of a previously unrecognized activator sequence in piggyBac. BMC Biotechnology, 7, 5. doi:10.1186/1472-6750-7-5
- Huang, X., Guo, H., Tammana, S., Jung, Y.C., Mellgren, E., Bassi, P., Cao, Q., Tu, Z.J., Kim, Y.C., Ekker, S.C., Wu, X., Wang, S.M. and Zhou, X. (2010) Gene transfer efficiency and genome-wide integration profiling of Sleeping Beauty, Tol2, and piggyBac transposons in human primary T cells. Molecular Therapy, 18, 1803-1813. doi:10.1038/mt.2010.141
- Mitchell, R.S., Beitzel, B.F., Schroder, A.R., Shinn, P., Chen, H., Berry, C.C., Ecker, J.R. and Bushman, F.D. (2004) Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biology, 2, e234. doi:10.1371/journal.pbio.0020234
- Nakai, H., Montini, E., Fuess, S., Storm, T.A., Grompe, M. and Kay, M.A. (2003) AAV serotype 2 vectors preferentially integrate into active genes in mice. Nature Genetics, 34, 297-302. doi:10.1038/ng1179
- Nienhuis, A.W., Dunbar, C.E. and Sorrentino, B.P. (2006) Genotoxicity of retroviral integration in hematopoietic cells. Molecular Therapy, 13, 1031-1049. doi:10.1016/j.ymthe.2006.03.001
- Wu, X., Li, Y., Crise, B. and Burgess, S.M. (2003) Transcription start regions in the human genome are favored targets for MLV integration. Science, 300, 1749-1751. doi:10.1126/science.1083413
- Galvan, D.L., Nakazawa, Y., Kaja, A., Kettlun, C., Cooper, L.J., Rooney, C.M. and Wilson, M.H. (2009) Genome-wide mapping of PiggyBac transposon integrations in primary human T cells. Journal of Immunotherapy, 32, 837-844. doi:10.1097/CJI.0b013e3181b2914c
- Yant, S.R., Wu, X., Huang, Y., Garrison, B., Burgess, S.M. and Kay, M.A. (2005) High-resolution genomewide mapping of transposon integration in mammals. Molecular and Cellular Biology, 25, 2085-2094. doi:10.1128/MCB.25.6.2085-2094.2005
- Yant, S.R., Meuse, L., Chiu, W., Ivics, Z., Izsvak, Z. and Kay, M.A. (2000) Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nature Genetics, 25, 35- 41. doi:10.1038/75568
- Montini, E., Held, P.K., Noll, M., Morcinek, N., AlDhalimy, M., Finegold, M., Yant, S.R., Kay, M.A. and Grompe, M. (2002) In vivo correction of murine tyrosinemia type I by DNA-mediated transposition. Molecular Therapy, 6, 759-769. doi:10.1006/mthe.2002.0812
- Ortiz-Urda, S., Lin, Q., Yant, S.R., Keene, D., Kay, M.A. and Khavari, P.A. (2003) Sustainable correction of junctional epidermolysis bullosa via transposon-mediated nonviral gene transfer. Gene Therapy, 10, 1099-1104. doi:10.1038/sj.gt.3301978
- Liu, L., Mah, C. and Fletcher, B.S. (2006) Sustained FVIII expression and phenotypic correction of hemophilia A in neonatal mice using an endothelial-targeted sleeping beauty transposon. Molecular Therapy, 13, 1006- 1015. doi:10.1016/j.ymthe.2005.11.021
- Ohlfest, J.R., Frandsen, J.L., Fritz, S., Lobitz, P.D., Perkinson, S.G., Clark, K.J., Nelsestuen, G., Key, N.S., McIvor, R.S., Hackett, P.B. and Largaespada, D.A. (2005) Phenotypic correction and long-term expression of factor VIII in hemophilic mice by immunotolerization and nonviral gene transfer using the Sleeping Beauty transposon system. Blood, 105, 2691-2698. doi:10.1182/blood-2004-09-3496
- Kren, B.T., Unger, G.M., Sjeklocha, L., Trossen, A.A., Korman, V., Diethelm-Okita, B.M., Reding, M.T. and Steer, C.J. (2009) Nanocapsule-delivered Sleeping Beauty mediates therapeutic Factor VIII expression in liver sinusoidal endothelial cells of hemophilia A mice. Journal of Clinical Investigation, 119, 2086-2099.
- Liu, H., Liu, L., Fletcher, B.S. and Visner, G.A. (2006) Sleeping Beauty-based gene therapy with indoleamine 2,3-dioxygenase inhibits lung allograft fibrosis. FASEB Journal, 20, 2384-2386. doi:10.1096/fj.06-6228fje
- Aronovich, E.L., Bell, J.B., Khan, S.A., Belur, L.R., Gunther, R., Koniar, B., Schachern, P.A., Parker, J.B., Carlson, C.S., Whitley, C.B., McIvor, R.S., Gupta, P. and Hackett, P.B. (2009) Systemic correction of storage disease in MPS I NOD/SCID mice using the sleeping beauty transposon system. Molecular Therapy, 17, 1136-1144. doi:10.1038/mt.2009.87
- Aronovich, E.L., Bell, J.B., Belur, L.R., Gunther, R., Koniar, B., Erickson, D.C., Schachern, P.A., Matise, I., McIvor, R.S., Whitley, C.B. and Hackett, P.B. (2007) Prolonged expression of a lysosomal enzyme in mouse liver after Sleeping Beauty transposon-mediated gene delivery: Implications for non-viral gene therapy of mucopolysaccharidoses. Journal of Gene Medicine, 9, 403-415. doi:10.1002/jgm.1028
- Wang, X., Sarkar, D.P., Mani, P., Steer, C.J., Chen, Y., Guha, C., Chandrasekhar, V., Chaudhuri, A., Roy-Chowdhury, N., Kren, B.T. and Roy-Chowdhury, J. (2009) Long-term reduction of jaundice in Gunn rats by nonviral liver-targeted delivery of Sleeping Beauty transposon. Hepatology, 50, 815-824. doi:10.1002/hep.23060
- Hyland, K.A., Olson, E.R., Clark, K.J., Aronovich, E.L., Hackett, P.B., Blazar, B.R., Tolar, J. and Scott McIvor, R. (2011) Sleeping Beauty-mediated correction of Fanconi anemia type C. Journal of Gene Medicine, 13, 462-469. doi:10.1002/jgm.1589
- Johnen, S., Izsvak, Z., Stocker, M., Harmening, N., Salz, A.K., Walter, P. and Thumann, G. (2012) Sleeping Beauty transposon-mediated transfection of retinal and iris pigment epithelial cells. Investigative Ophthalmology and Visual Science, 53, 4787-4796. doi:10.1167/iovs.12-9951
- Ohlfest, J.R., Demorest, Z.L., Motooka, Y., Vengco, I., Oh, S., Chen, E., Scappaticci, F.A., Saplis, R.J., Ekker, S.C., Low, W.C., Freese, A.B. and Largaespada, D.A. (2005) Combinatorial antiangiogenic gene therapy by nonviral gene transfer using the sleeping beauty transposon causes tumor regression and improves survival in mice bearing intracranial human glioblastoma. Molecular Therapy, 12, 778-788. doi:10.1016/j.ymthe.2005.07.689
- Kang, Y., Zhang, X., Jiang, W., Wu, C., Chen, C., Zheng, Y., Gu, J. and Xu, C. (2009) Tumor-directed gene therapy in mice using a composite nonviral gene delivery system consisting of the piggyBac transposon and polyethyleneimine. BMC Cancer, 9, 126. doi:10.1186/1471-2407-9-126
- Nakazawa, Y., Huye, L.E., Salsman, V.S., Leen, A.M., Ahmed, N., Rollins, L., Dotti, G., Gottschalk, S.M., Wilson, M.H. and Rooney, C.M. (2011) PiggyBac-mediated cancer immunotherapy using EBV-specific cytotoxic Tcells expressing HER2-specific chimeric antigen receptor. Molecular Therapy, 19, 2133-2143. doi:10.1038/mt.2011.131
- Kang, Y., Zhang, X.Y., Jiang, W., Wu, C.Q., Chen, C.M., Gu, J.R., Zheng, Y.F. and Xu, C.J. (2009) The piggyBac transposon is an integrating non-viral gene transfer vector that enhances the efficiency of GDEPT. Cell Biology International, 33, 509-515. doi:10.1016/j.cellbi.2009.01.017
- Hollis, R.P., Nightingale, S.J., Wang, X., Pepper, K.A., Yu, X.J., Barsky, L., Crooks, G.M. and Kohn, D.B. (2006) Stable gene transfer to human CD34+ hematopoietic cells using the Sleeping Beauty transposon. Experimental Hematology, 34, 1333-1343. doi:10.1016/j.exphem.2006.05.023
- Izsvak, Z., Chuah, M.K., Vandendriessche, T. and Ivics, Z. (2009) Efficient stable gene transfer into human cells by the Sleeping Beauty transposon vectors. Methods, 49, 287-297. doi:10.1016/j.ymeth.2009.07.001
- Sumiyoshi, T., Holt, N.G., Hollis, R.P., Ge, S., Cannon, P.M., Crooks, G.M. and Kohn, D.B. (2009) Stable transgene expression in primitive human CD34+ hematopoietic stem/progenitor cells, using the Sleeping Beauty transposon system. Human Gene Therapy, 20, 1607-1626. doi:10.1089/hum.2009.109
- Xue, X., Huang, X., Nodland, S.E., Mates, L., Ma, L., Izsvak, Z., Ivics, Z., LeBien, T.W., McIvor, R.S., Wagner, J.E. and Zhou, X. (2009) Stable gene transfer and expression in cord blood-derived CD34+ hematopoietic stem and progenitor cells by a hyperactive Sleeping Beauty transposon system. Blood, 114, 1319-1330. doi:10.1182/blood-2009-03-210005
- Huang, X., Guo, H., Kang, J., Choi, S., Zhou, T.C., Tammana, S., Lees, C.J., Li, Z.Z., Milone, M., Levine, B.L., Tolar, J., June, C.H., Scott McIvor, R., Wagner, J.E., Blazar, B.R. and Zhou, X. (2008) Sleeping Beauty transposon-mediated engineering of human primary T cells for therapy of CD19+ lymphoid malignancies. Molecular Therapy, 16, 580-589. doi:10.1038/sj.mt.6300404
- Huang, X., Wilber, A., McIvor, R.S. and Zhou, X. (2009) DNA transposons for modification of human primary T lymphocytes. Methods in Molecular Biology, 506, 115- 126. doi:10.1007/978-1-59745-409-4_9
- Manuri, P.V., Wilson, M.H., Maiti, S.N., Mi, T., Singh, H., Olivares, S., Dawson, M.J., Huls, H., Lee, D.A., Rao, P.H., Kaminski, J.M., Nakazawa, Y., Gottschalk, S., Kebriaei, P., Shpall, E.J., Champlin, R.E. and Cooper, L.J. (2010) Piggybac transposon/transposase system to generate CD19-specific T cells for the treatment of B-lineage malignancies. Human Gene Therapy, 21, 427-437. doi:10.1089/hum.2009.114
- Nakazawa, Y., Huye, L.E., Dotti, G., Foster, A.E., Vera, J.F., Manuri, P.R., June, C.H., Rooney, C.M. and Wilson, M.H. (2009) Optimization of the PiggyBac transposon system for the sustained genetic modification of human T lymphocytes. Journal of Immunotherapy, 32, 826-836. doi:10.1097/CJI.0b013e3181ad762b
- Singh, H., Manuri, P.R., Olivares, S., Dara, N., Dawson, M.J., Huls, H., Hackett, P.B., Kohn, D.B., Shpall, E.J., Champlin, R.E. and Cooper, L.J. (2008) Redirecting specificity of T-cell populations for CD19 using the Sleeping Beauty system. Cancer Research, 68, 2961-2971. doi:10.1158/0008-5472.CAN-07-5600
- Orban, T.I., Apati, A., Nemeth, A., Varga, N., Krizsik, V., Schamberger, A., Szebenyi, K., Erdei, Z., Varady, G., Karaszi, E., Homolya, L., Nemet, K., Gocza, E., Miskey, C., Mates, L., Ivics, Z., Izsvak, Z. and Sarkadi, B. (2009) Applying a “double-feature” promoter to identify cardiomyocytes differentiated from human embryonic stem cells following transposon-based gene delivery. Stem Cells, 27, 1077-1087. doi:10.1002/stem.45
- Wilber, A., Linehan, J.L., Tian, X., Woll, P.S., Morris, J.K., Belur, L.R., McIvor, R.S. and Kaufman, D.S. (2007) Efficient and stable transgene expression in human embryonic stem cells using transposon-mediated gene transfer. Stem Cells, 25, 2919-2927. doi:10.1634/stemcells.2007-0026
- Williams, D.A. (2008) Sleeping beauty vector system moves toward human trials in the United States. Molecular Therapy, 16, 1515-1516. doi:10.1038/mt.2008.169
- Jena, B., Dotti, G. and Cooper, L.J. (2010) Redirecting T-cell specificity by introducing a tumor-specific chimeric antigen receptor. Blood, 116, 1035-1044. doi:10.1182/blood-2010-01-043737
- Kebriaei, P., Huls, H., Jena, B., Munsell, M., Jackson, R., Lee, D.A., Hackett, P.B., Rondon, G., Shpall, E., Champlin, R.E. and Cooper, L.J. (2012) Infusing CD19-directed T cells to augment disease control in patients undergoing autologous hematopoietic stem-cell transplantation for advanced B-lymphoid malignancies. Human Gene Therapy, 23, 444-450. doi:10.1089/hum.2011.167
- Torikai, H., Reik, A., Liu, P.Q., Zhou, Y., Zhang, L., Maiti, S., Huls, H., Miller, J.C., Kebriaei, P., Rabinovitch, B., Lee, D.A., Champlin, R.E., Bonini, C., Naldini, L., Rebar, E.J., Gregory, P.D., Holmes, M.C. and Cooper, L.J. (2012) A foundation for universal T-cell based immunotherapy: T cells engineered to express a CD19-specific chimeric-antigen-receptor and eliminate expression of endogenous TCR. Blood, 119, 5697-5705. doi:10.1182/blood-2012-01-405365
- Delauriere, L., Chénais, B., Pradier, E., Hardivillier, Y., Renault, S. and Casse, N. (2009) DNA binding specificity and cleavage activity of Pacmmar transposase. Biochemistry, 48, 7279-7286. doi:10-.1021/bi900609v
- de Silva, S., Lotta Jr., L.T., Burris, C.A. and Bowers, W.J. (2010) Virion-associated cofactor high-mobility group DNA-binding protein-1 facilitates transposition from the herpes simplex virus/Sleeping Beauty amplicon vector platform. Human Gene Therapy, 21, 1615-1622. doi:10.1089/hum.2010.022
- Moldt, B., Miskey, C., Staunstrup, N.H., Gogol-Doring, A., Bak, R.O., Sharma, N., Mates, L., Izsvak, Z., Chen, W., Ivics, Z. and Mikkelsen, J.G. (2011) Comparative genomic integration profiling of Sleeping Beauty transposons mobilized with high efficacy from integrase-defective lentiviral vectors in primary human cells. Molecular Therapy, 19, 1499-1510. doi:10.1038/mt.2011.47
- Vink, C.A., Gaspar, H.B., Gabriel, R., Schmidt, M., McIvor, R.S., Thrasher, A.J. and Qasim, W. (2009) Sleeping beauty transposition from nonintegrating lentivirus. Molecular Therapy, 17, 1197-1204. doi:10.1038/mt.2009.94
- Ehrhardt, A., Yant, S.R., Giering, J.C., Xu, H., Engler, J.A. and Kay, M.A. (2007) Somatic integration from an adenoviral hybrid vector into a hot spot in mouse liver results in persistent transgene expression levels in vivo. Molecular Therapy, 15, 146-156. doi:10.1038/sj.mt.6300011
- Yant, S.R., Ehrhardt, A., Mikkelsen, J.G., Meuse, L., Pham, T. and Kay, M.A. (2002) Transposition from a gutless adeno-transposon vector stabilizes transgene expression in vivo. Nature Biotechnology, 20, 999-1005. doi:10.1038/nbt738
- Yant, S.R., Huang, Y., Akache, B. and Kay, M.A. (2007) Site-directed transposon integration in human cells. Nucleic Acids Research, 35, e50. doi:10.1093/nar/gkm089
- Hausl, M., Zhang, W., Voigtlander, R., Muther, N., Rauschhuber, C. and Ehrhardt, A. (2011) Development of adenovirus hybrid vectors for Sleeping Beauty transposetion in large mammals. Current Gene Therapy, 11, 363- 374. doi:10.2174/156652311797415890
- Luo, W.Y., Shih, Y.S., Hung, C.L., Lo, K.W., Chiang, C.S., Lo, W.H., Huang, S.F., Wang, S.C., Yu, C.F., Chien, C.H. and Hu, Y.C. (2012) Development of the hybrid Sleeping Beauty: Baculovirus vector for sustained gene expression and cancer therapy. Gene Therapy, 19, 844- 851. doi:10.1038/gt.2011.129
- Liu, L., Liu, H., Visner, G. and Fletcher, B.S. (2006) Sleeping Beauty-mediated eNOS gene therapy attenuates monocrotaline-induced pulmonary hypertension in rats. FASEB Journal, 20, 2594-2596. doi:10.1096/fj.06-6254fje
- Singh, H., Figliola, M.J., Dawson, M.J., Huls, H., Olivares, S., Switzer, K., Mi, T., Maiti, S., Kebriaei, P., Lee, D.A., Champlin, R.E. and Cooper, L.J. (2011) Reprogramming CD19-specific T cells with IL-21 signaling can improve adoptive immunotherapy of B-lineage malignnancies. Cancer Research, 71, 3516-3527. doi:10.1158/0008-5472.CAN-10-3843
- Staunstrup, N.H., Madsen, J., Primo, M.N., Li, J., Liu, Y., Kragh, P.M., Li, R., Schmidt, M., Purup, S., DagnaesHansen, F., Svensson, L., Petersen, T.K., Callesen, H., Bolund, L. and Mikkelsen, J.G. (2012) Development of transgenic cloned pig models of skin inflammation by DNA transposon-directed ectopic expression of human beta1 and alpha2 integrin. PLoS One, 7, e36658. doi:10.1371/journal.pone.0036658
- Huang, G., Yu, L., Cooper, L.J., Hollomon, M., Huls, H. and Kleinerman, E.S. (2012) Genetically modified T cells targeting interleukin-11 receptor alpha-chain kill human osteosarcoma cells and induce the regression of established osteosarcoma lung metastases. Cancer Research, 72, 271-281. doi:10.1158/0008-5472.CAN-11-2778
- Xiao, J., Meng, X.M., Huang, X.R., Chung, A.C., Feng, Y.L., Hui, D.S., Yu, C.M., Sung, J.J. and Lan, H.Y. (2012) MiR-29 inhibits bleomycin-induced pulmonary fibrosis in mice. Molecular Therapy, 20, 1251-1260. doi:10.1038/mt.2012.36

