Advances in Biological Chemistry
Vol. 2 No. 4 (2012) , Article ID: 24535 , 12 pages DOI:10.4236/abc.2012.24040
Insights on the structural characteristics of NDM-1: The journey so far
![]()
Department of Biophysics, Panjab University, Chandigarh, India
Email: *avneet@pu.ac.in
Received 20 August 2012; revised 28 September 2012; accepted 4 October2012
Keywords: NDM-1; Metallo-β-Lactamase; Superbug; β-Lactam; Extended-Spectrum β-Lactamases
ABSTRACT
New Delhi metallo-β-lactamase (NDM-1) has created a medical storm ever since it was first reported; as it is active on virtually all clinically used β-lactam antibiotics. NDM-1 rampancy worldwide is now considered a nightmare scenario, particularly due to its rapid dissemination. An underlying theme in the majority of recent studies is structural characterization as knowledge of the three-dimensional structure of NDM-1 shall help find connections between its structure and function. Moreover, structural details are even critical in order to reveal the resistance mechanism to β-lactam antibiotics. In this perspective, we review structural characteristics of NDM-1 that have been delineated since its first report. We anticipate that these structure-function connections made by its characterization shall further serve as future guidelines for elucidating pathways towards de novo design of functional inhibitors.
1. INTRODUCTION
New Delhi metallo-β-lactamase (NDM-1) is an enzyme that makes bacteria resistant to a broad range of β-lactam antibiotic drugs. Increasing antibiotic resistance has posed a great threat to public health especially multi drug resistance in bacterial strains that have disseminated widely, due to which it has been recognized as one of the greatest challenges for the treatment of clinical infections [1]. Since the first NDM-1 infection reported in 2009, the emergence of New Delhi metallo-β-lactamase (NDM-1) is the latest twist which has made recent headlines for its ability of rapid dissemination worldwide and rendering ineffective the last-resort antibiotics known as carbapenems [2,3]. Following this first case, the past two years have seen a sudden increase in the number of NDM-1 carrying bacteria all over the world [4-14]. Amongst the patients infected by NDM-1 carrying bacteria many different Enterobacteriaceae species were identified, including K. pneumonia, E. coli, E. cloacae, Proteus spp., Citrobacter freundii, K. oxytoca, M. morganii and Providencia spp. [4]. The NDM-1 rampancy worldwide is now considered a potential nightmare scenario, as there is hardly any antibiotic that is effective against the multidrug-resistant “superbug” [3,15].
2. WHERE IT ALL BEGAN?
β-lactams are the most broadly used antibacterials worldwide because of their comparatively high effectiveness, low cost of production, ease of delivery and minimal side effects [16-18]. These antibacterials inhibit the synthesis of the peptidoglycan layer of bacterial cell walls leading to cell lysis [19]. Structurally, all β-lactam antibiotics contain a four-member β-lactam ring that has a nitrogencontaining cyclic amide that is critical for antibacterial activity (Figure 1).
2.1. β-Lactam Antibiotics
The β-lactam antibiotics comprise six different structural subtypes, including penams, cephems, monobactams, clavams, penems and carbapenems (Figure 2). The pe-
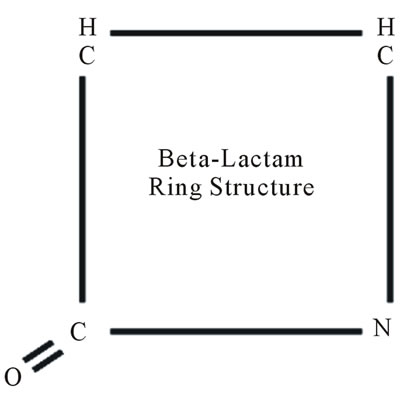
Figure 1. Basic structure of β-lactam ring. IUPAC name of β-lactam is 2-Azetidinone.
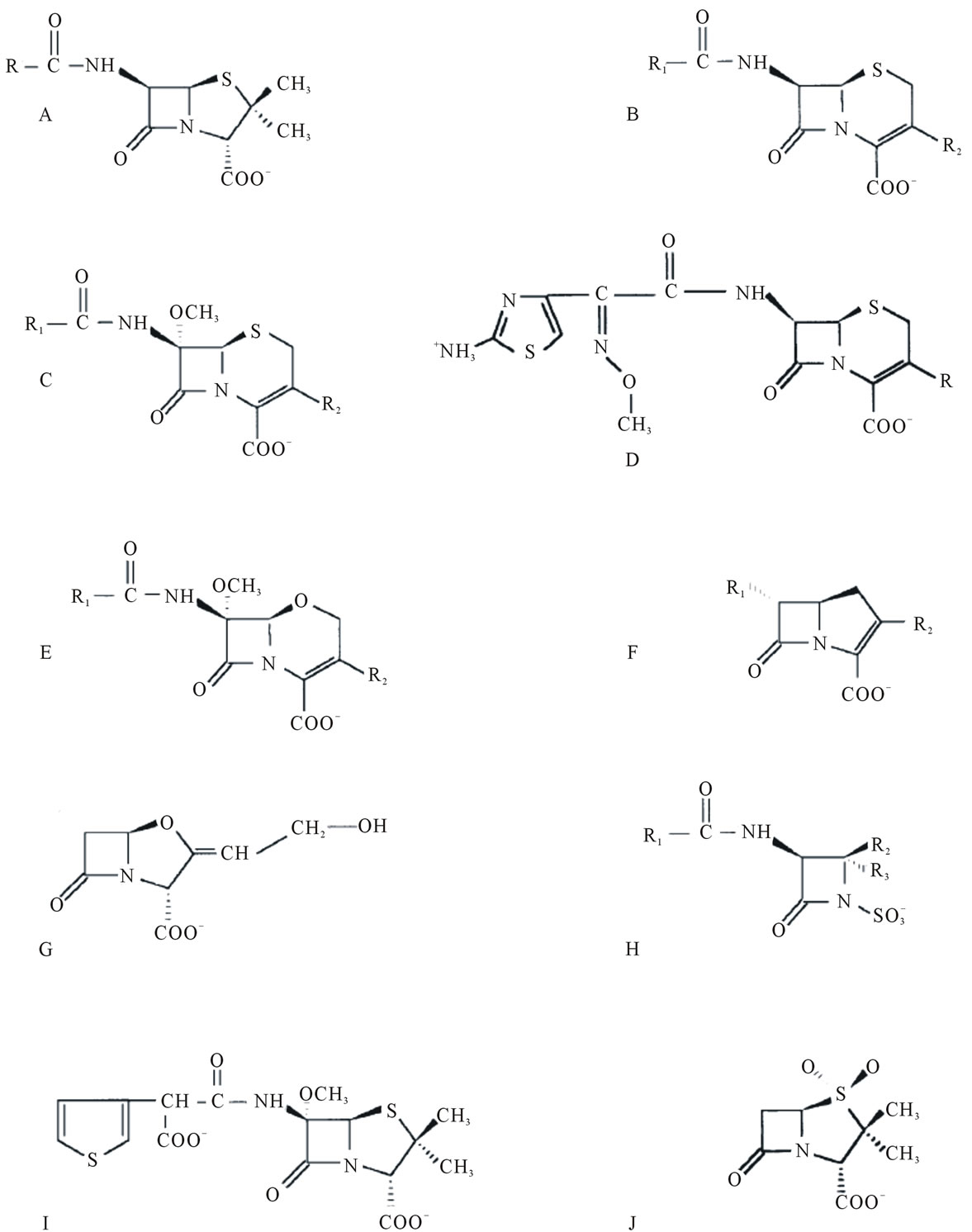
Figure 2. Chemical structures of some β-lactum antibiotics. A) Penams (e.g. benzylpenicillin, ampicillin, amoxycillin); B) Cephems (cephalosporins); C) Cephamycins (e.g. cefoxitin); D) Cefotaxime (oximino cephalosporin ; R = CH2-O-CO-CH3); E) Oxacephamycins (e.g. moxalactam); F) Carbapenems (e.g. imipenem); G) Clavulanate (oxapenam); H) Monobactams (e.g. aztreonam); I) Temocillin (6-α-methoxy penam) and J) Sulbactam (penam sulphone) [22].
nams include benzylpenicillin and ampicillin, while cephems include classical cephalosporins such as cephaloridine, nitrocefin, and cefotaxime, as well as cephamycins (that are 7-α-methoxy-cephalosporins). The monobactams are monocyclic β-lactams and include aztreonam. Penems have a 2, 3-double bond in the fused thiazolidine ring (hence dihydrothiazole), similar to the carbapenems (e.g. imipenem, biapenem), which also have an unsaturated fused five membered ring, with carbon in place of sulfur at the 1-position [19]. These bind to and inhibit the carboxypeptidases and transpeptidases which are the cell wall synthesizing enzymes, also called the penicillin-binding proteins, or PBPs, that catalyze the D-ala D-ala cross linkages of the peptidoglycan wall that surrounds the bacterium. As a result, there is weakening of the cell wall structure, leading to cell lysis [18,20].
The first antibiotic that was discovered by Sir Alexander Fleming in 1927 named penicillin is also a β- lactam. It was not until the early 1940s, through the work of Dr. Florey Chain and Heatley from Oxford University, that penicillin was purified and shown to cure specific bacterial infections. Since that time, many structural derivatives have been developed from penicillin to combat resistance that has arisen in bacteria. These derivatives commonly referred to as the extended spectrum β-lactams, include antibiotics called the cephalosporins, carbapenems and monobactams. Over the last 60 years, the β-lactam class of antibiotics were the most widely used, representing about 60% of all of the antibiotics used (by weight) in human and animal medicine. These antibiotics also contain a β-lactam nucleus in their molecular structure by which they can inhibit the synthesis of the peptidoglycan layer of bacterial cell walls [19].
2.2. β-Lactamase Enzyme
Today, most of the bacteria have developed resistance to the currently available antibiotics which could be the result of spontaneous mutation, β-lactamases, impermeability, efflux and target modification [20]. Hence, bacterial resistance to β-lactam antibiotics can be achieved by 3 broad ways (Figure 3): 1) degrading or modifying the antibiotic before it can reach the site of action by production of enzymes from β-lactamase family; 2) modifying antibiotic target site with β-lactam resistant cell wall transpeptidase (this is major cause of resistance in several pathogens including Gram-positive Staphylococcal and Streptopcoccal species); 3) preventing the access of the antibiotic to the target by way of altered permeability or forced efflux (for example, MexA, BOprM antibiotic efflux pump, which is a major cause of resistance in Pseudomonas and in other pathogenic Gram-negative species) [18]. Out of these, the most common mechanism of bacterial resistance is the production of β-lacatamase enymes that hydrolyze the β-lactam antibiotics by cleaving the amide bond of β-lactam ring in an acylation-deacylation-based process [21,22]. First indentified in an isolate of Escherichia coli in 1940 [23] these enzymes are mainly found in Enterobacteriaceae family [24]. These enzymes are very effective at protecting the organisms against the β-lactam antibiotics, possibly because they are precisely located at optimal positions in the periplasmic space [25] and can be carried on bacterial chromosomes, that is, inherent to the organism, or may be plasmid-mediated with the potential to move between bacterial populations and can be produced in a constitutive or inducible manner [22,26,27].
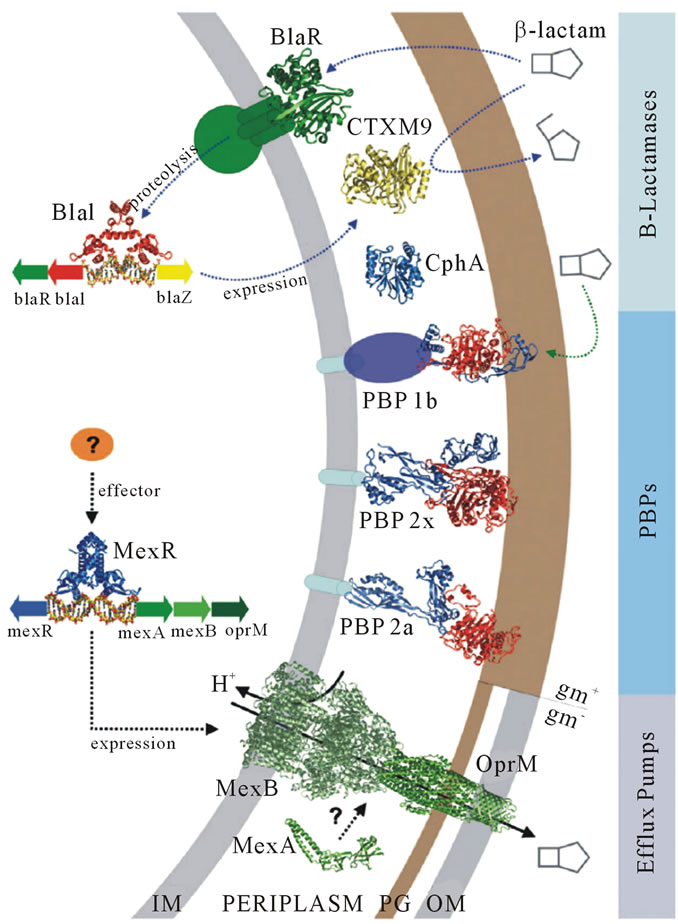
Figure 3. Mechanisms followed by bacteria against β-lactam antibiotics [18].
As the bacteria developed resistance to one type of β- lactam antibiotic, new antibiotic derivatives were made by researchers and were called the cephalosporins, carbapenems and monobactams. Nevertheless, the bacteria continually evolved and changed the existing β-lactamase enzymes in order to break down these new compounds. The enzymes that can break down the newer derivatives are known as the extended-spectrum β-lactamases (ESBLs) and were first observed in the early 1980’s. Thus, it is believed that β-lactamases play an important role in leading to resistance of bacteria to β-lactam antibiotics.
β-lactamases show extensive molecular and functional diversity. Based on the characteristics of the enzymes and their substrate profile, a number of classification schemes have been proposed. Among these the Ambler molecular classification (based on molecular structure) and the Bush-Jacoby-Medeiros classification (based on functional similarities) (Table 1) are the two most widely used classification systems [28-31]. The properties that have been used to differentiate and classify the β-lactamases are: isoelectric point, molecular mass, relative activity towards different β-lactam, interaction with inhibitors and inactivators, the nature of active site, amino acid sequence and three dimensional structures
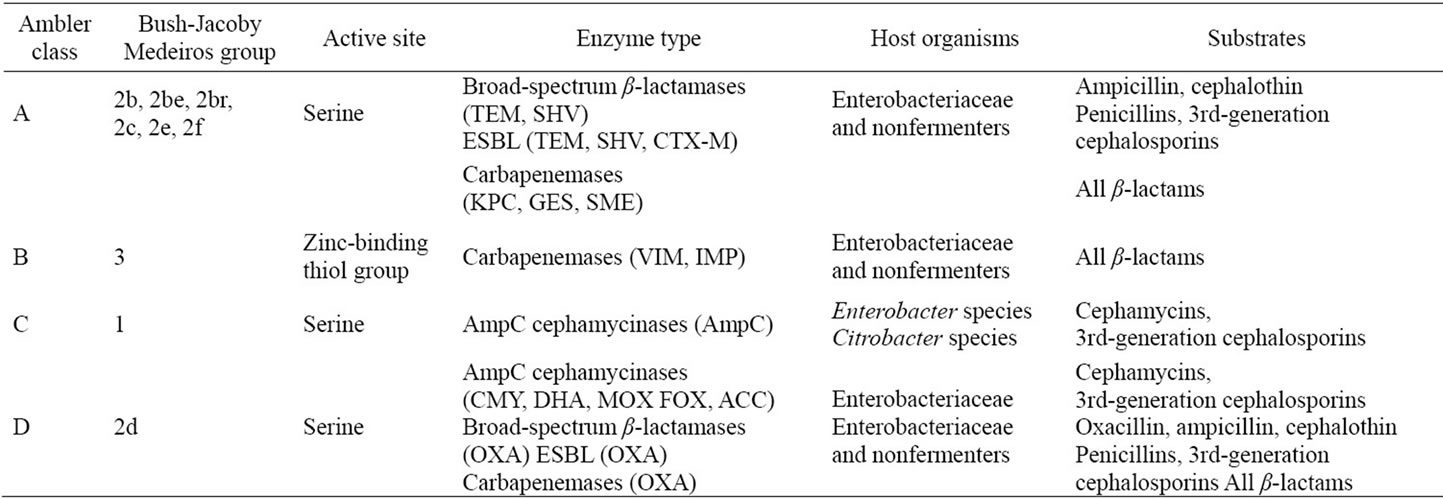
Table 1. Classification of β-lactamase enzymes [28].
[29]. Ambler scheme divides β-lactamases into four major classes A, B, C and D. Enzymes of class A, C and D require serine for deactivation of antibiotics while enzymes from class B, also called Metallo-β-lactamase (MBL), require one or two zinc ions in their mechanism of action. Among all the β-lactamases, metallo-β-lactamases (MBLs) are the major culprits causing bacteria to resist antibiotics as they can degrade all β-lactams except monobactams and that they are special for their constant and efficient carbapenemase activity [32]. The Bush-JacobyMedeiros classification scheme groups β-lactamases according to functional similarities (substrate and inhibitor profile). There are four main groups and multiple subgroups in this system. This classification scheme is of much more immediate relevance to the physician or microbiologist in a diagnostic laboratory because it considers β-lactamase and β-lactam substrates that are clinically relevant.
2.3. Metallo-β-Lactamases
Metallo-β-lactamases (MBLs) constitute the molecular class B of Ambler [29] and group 3 according to the Bush-Jacoby-Medeiros functional classification [30]. BcII, the first MBL discovered was chromosomally encoded by the relatively innocuous Bacillus cereus microbe (PDB ID: 1BMC) and since then, MBLs have grown into a looming global antibiotic resistance threat, most recently highlighted by the spread of NDM-1.
The clinical importance of MBLs is highlighted by the fact that they hydrolyze carbapenems, compounds which most often escape the activity of active-site serine β- lactamases. Metallo-β-lactamases in particular are of global health interest, as many are acquired, capable of traveling across species, and are the most commonly encountered transferable carbapenemases [33].
The MBL super family was defined by Neuwald et al. in 1997 [34]. In addition to metallo-β-lactamases, it includes enzymes which hydrolyze thiol-ester, phosphodiester and sulfuric ester bonds as well as oxydoreductases. Most of the 6000 members of this super family share five conserved motifs [35]: Asp84, His116- X-His118-X-Asp120-His121, His196, Asp221 and His263. Amongst these conserved motifs, Asp84 exhibits a strained secondary conformation and may play a role in maintaining the protein fold. The other conserved residues are involved in metal coordination. For the majority of these enzymes, the first metal ion binding site is composed of His116, His118 and His196, while the second binding site is composed of Asp120, His121 and His263. Daiyasu has further classified these proteins in 17 families on the basis of their biological functions [35]. The MBL superfamily of proteins is quiet diverse in the identity and stoichiometry of its bound metal ions. The best studied examples have two zinc ions bound in a dinuclear active site cluster; however, more exotic examples can include iron or manganese ions, and the number of bound metal ions can range between zero and three, although these are not always bound at the active site [18, 35-40]. Structurally, MBLs are members of a large, diverse, superfamily of proteins that share a similar four-layered αβ/βα structure, composed by two central β-sheets with five solvent-exposed α-helices. The Nterminal and C-terminal parts of the molecule, each of them comprising a β-strand and two a helices, can be superposed by a 180˚ rotation around a central axis, suggesting that the complete structure might have arisen from the duplication of a gene [41]. In all known structures, the active site is located at the external edge of the ββ sandwich.
3. NEW DELHI METALLO-β-LACTAMASE
The recently discovered plasmid-borne New Delhi metallo-β-lactamase (NDM-1), capable of hydrolyzing a broad range of antibiotics, is such a metallo enzyme noted for its ability to confer resistance to all but a small handful of β-lactam antimicrobials. NDM-1 refers not to a single bacterial species but to a transmissible genetic element encoding multiple resistance genes that was initially isolated from a strain of Klebsiella.
NDM-1 has the ability to make bacteria resistant to a wide range of β-lactam antibiotics, including the carbapenem family antibiotics that are a mainstay for the treatment of antibiotic-resistant bacterial infections [15, 42]. Like other MBLs, it also depends on its metal cation cofactor for its catalytic activity and therefore, is a highly potent carbapenems-hydrolyzing zinc dependent MBL [42]. In August 2010, an article in The Lancet Infectious Diseases triggered a media storm and brought the public’s attention to the world’s newest superbug, when 37 strains of NDM-1 were found in the UK and 99 strains were found in India [3]. Since its first report, NDM-1 infections have rapidly spread to five of seven continents. The most common bacteria that make this enzyme are Gram negative bacteria such as Escherichia coli and Klebsiella pneumonia, but the gene for NDM-1 can spread from one strain of bacteria to another by horizontal gene transfer [15]. Further, it is reported that this transmission can be accelerated by “medical tourism”, and by the high level of population exchanges between India and Pakistan and other countries around the globe especially United Kingdom, Australia and Canada. NDM producing Enterobacteriaceae have also recently been isolated from patients residing in the United States (1), the Netherlands (12), Australia (20), Canada (15), France (19), and the Sultanate of Oman (18). Most of the patients received medical care while visiting the subcontinent of Pakistan, India, and Bangadesh [43].
This scenario is of great concern because there are a few new anti-Gram-negative antibiotics in the pharmaceutical pipeline and none that are active against NDM-1 producers [3]. Moreover, the limited structural studies of NDM-1, its active site diversity and mechanism of hydrolysis make the development of broad spectrum inhibitors a great challenge.
Knowledge of the three-dimensional structure of NDM-1 is critical in order to reveal the resistance mechanism to β-lactam antibiotics. The year 2011 saw an exponential increase in the number of structural studies/ reports (both crystallographic and theoretical) on NDM-1 characterization. In this perspective this article reviews the several structural/structure-function studies on NDM- 1 that have been delineated since 2009.
3.1. NDM-1 Gene & Phylogenetic Studies
The blaNDM-1 gene encoding this novel β-lactamase; found on a large 180-kb resistance-conferring genetic element was characterized from Klebsiella pneumonia sequence type 14 from India [2]. It was easily transferred and rapidly disseminated to other Enterobacteriaceae as it is plasmid borne. Moreover, it contained a variety of other resistance determinants, including a gene encoding another broad-spectrum β-lactamases (CMY-4) and genes inactivating erythromycin, ciprofloxacin, rifampicin, and chloramphenicol. In addition, the genetic element encoded an efflux pump capable of causing additional antimicrobial resistance and growth promoters that insured the transcription of the genes contained in the genetic element.
There appears to be a possible transfer of blaNDM-1 in vivo either from K. pneumoniae to E. coli or vice versa, but more interestingly, the plasmids carrying blaNDM-1 in the two species were of two different sizes suggesting that there is rearrangement in vivo which could result from either duplication or insertion (e.g., transposition or rolling circle replication from the smaller plasmid, or deletion from the larger plasmid) [2]. The blaNDM-1 open reading frame encodes a putative protein of 269 amino acids (Table 2) with a molecular mass of approximately 27.5 kDa and also posses N-terminal signal peptide with conserved LXXC like motif [15].
Sequence alignment and similarity studies revealed that NDM-1 shares very little identity with other MBLs and is most closely related to VIM-1/VIM-2, with which it has only 32.4% identity.
Curiously, NDM-1 also has a unique HXHXD motif among the mobile MBLs, as it contains an alanine between the two histidines. Further, NDM-1 possesses a tyrosine at position 222 instead of the universally conserved tryptophan and the theoretical pI of the mature peptide has been reported to be 6.9. NDM-1 has shown tight binding (low Km values) to most cephalosporins and in particular, to cefuroxime, cefotaxime, and cephalothin (cefalotin). It also showed relatively tight binding to penicillins, which is unusual for an MBL [2].
Phylogenetic study to trace the origin of this antibiotic resistant NDM-1 gene using BLAST suggested that NDM-1 gene has closest orthologues in marine bacteria and shared high identity with β-lactamase II from Erythrobacter litoralis (ElBla2), lactamase from Haliangium ochraceum and lactamase from Hirschia baltica with identities of 55%, 49% and 37% respectively. NDM-1 and marine lactamases also share HAHXD motif, principal zinc binding motif, by contrast to other MBLs. Both NDM-1 and ElBla2 have the unique loop at positions 168 to 172, and unique amino acids at positions 195, 229, 246 and 259 [15].
Therefore, it was suggested that NDM-1 might have appeared via horizontal gene transfer from environmental reservoirs. Further suggesting, that that these marine
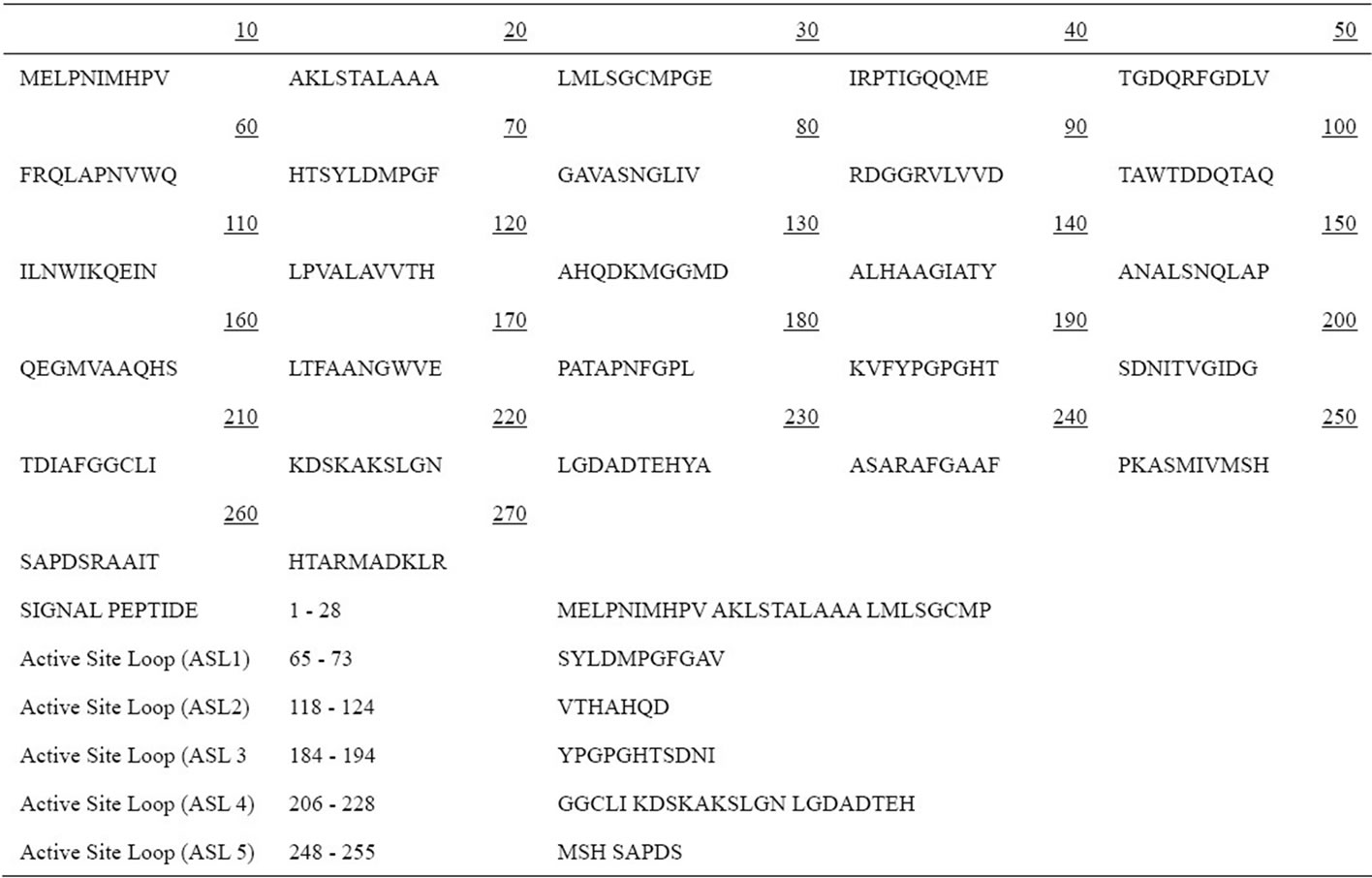
Table 2. Amino acid sequence of NDM-1.
originated β-lactamases act strictly against β-lactam molecules secreted by antibiotic producers sharing the same ecological niche [15].
NDM-1 does show some similarity to the VIM group of MBLs and the active site of NDM-1 is highly homologous to that of VIM-4 [2,44] but at the same time NDM-1 contains additional sequences at positions 226 to 228 indicating that these residues might be playing a unique role in the structure and functioning of NDM-1. In NDM-1 the substitute Arg228Ala and Tyr224Lys are unlikely to serve similar roles [2] due to the completely different amino acid characteristics.
3.2. Crystal Structure & Active Site
Detailed questions regarding the structural architecture of the protein cannot be reliably answered until a resolution of ~2.5 Å or better is achieved. Further, precise calculations of the energetics of ligand binding or intermolecular interfaces requires structure determination carried out at even higher resolution, making possible the mapping of ordered water molecules and an accurate description of hydrogen bonding geometries. Such a set of data mandates a resolution of 2.0 Å or better.
Currently, there are 16 structural hits in PDB database for the molecule “Beta-lactamase NDM-1” (Figure 4), out of which there are two structures with an an X-ray resolution less than 1.3 Å (PDB ID: 4EYB, 4EY2) [45], and another two with a resolution less than 1.5 Å (PDB ID: 3Q6X, 4EXY) [32,45]. Four crystal structures have been reported with a resolution between 1.5 and 2.0 Å (PDB ID: 3ZR9, 4EYF, 4EYL, 1JSZ) [46], while eight of the reported structures have a resolution between 2.0 to 2.5 Å (PDB ID: 3RKK, 3RKJ, 3SBL, 3SPU, 3SFP, 3PG4, 4EXS, 3AGM) [42,44,46]. Further, the source of the protein for all structures is K. pneumoniae.
The core of NDM-1 consists of two β-sheets, one (N-terminal) is composed of seven antiparallel strands (β1 - β7) and the other (C-terminal) is composed of five antiparallel strands (β8 - β12). The seven connecting helices are located below (α1 - α4 and 310 helix 5) and above (α6 - α7) the plane of the β-sandwich. Hydrophobic interactions between helices and β-sheets are critical from the structural point of view but also involve several hydrogen bonds (Gln-96 with carbonyl of Tyr-64, Thr-98 with carbonyl of Ala-92, Tyr-229 with carbonyl of Leu-209 and Ser-232 with carbonyl of Pro-187). Strands and helices are connected through flexible loops with the most prominent loop (residues 206 - 228) located above a 600 Å active site cavity. A reduction in the volume of residues contributing to the active site (Ala- 121 in NDM-1, typically Phe or Trp in other MBLs;
X-ray Resolution
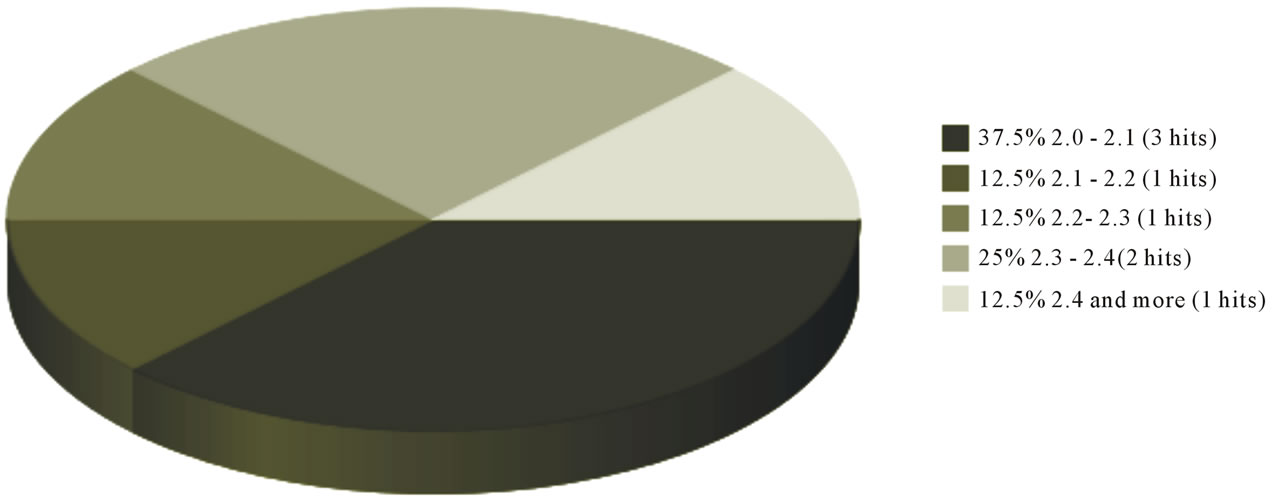
Figure 4. Pie chart depicting the quantity as well as quality of the X-ray crystal structure data for NDM-1. www.pdb.org
Tyr-229 in NDM-1, typically Trp in other MBLs; sequence Ala-72-Val-73 in NDM-1, which is typically Val or Phe in other MBLs; Phe-70 in NDM-1 which is Trp in other MBLs) and also in the volume of the residues contributing to the hydrophobic core was reported. Such changes can contribute to the opening of the groove to the solvent and can also increase the flexibility of the structural elements [42].
The oligomeric state of NDM-1 is reported to be either monomeric [2,47] or (partially) dimeric [44]. Dimerization of NDM- 1 is proposed to occur, in part, through a loop insert (Thr-162 - Gly-167) [44] unique to NDM-1, which contributes to the proposed dimerization interface as visualized in two separate crystal structures [32,44]. In contrast, Thomas et al. [48] described a purification method which produced NDM-1 that elutes from a gel filtration column with an apparent molecular mass near 24 kDa, which is most consistent with classifying NDM- 1 as monomeric and is also consistent with other reports of NDM-1’s monomeric state [2,42,47]. The only apparent difference between the protein studied here and that of dimeric NDM-1 is the length of the N-terminus and its possible lipidation. Thomas et al. also concluded that although the unique NDM-1 loop insert found between residues Thr-162 and Glu-167 may contribute to dimerization, it is not sufficient by itself to enforce dimerization without additional interactions mediated either by the N-terminal extension or by membrane anchoring through lipidation [48]. Future studies are required to address any possible effects of lipidation and dimerization on the activity of NDM-1 and its ability to confer antibiotic resistance.
Similar unique features on primary sequence and unusually large cavity of the active site were also reported for the crystal structure of the 24-kDa truncated NDM-1 by the Molecular Replacement Method (PDB ID: 3S0Z) [47]. Based on similar lines another crystal structure of the apo form of this enzyme solved to a resolution of 2.1 Å using molecular replacement phases generated from a previously determined MBL homolog VIM-4 has been reported [46].
Zhang and Hao’s work on [32] the crystal structure (at 1.3 Å resolution) of NDM-1 enzyme in complex with a hydrolyzed ampicillin at its active site suggested that the overall structure is somewhat different then the other MBLs. The complex structures of hydrolyzed benzylpenicillin-, methicillin-, and oxacillin-bound NDM-1 have been solved to 1.8, 1.2, and 1.2 Å, respectively, and represent the highest-resolution structural data for any metallo-β-lactamase reported to date [45].
Based on these crystal structure data the N-terminus of NDM-1 was suggested to be much longer than that of VIM-2 and IMP-1, forming two more β-strands making NDM-1 more potent to substrate specificity and binding by increasing hydrophobic interactions. The hydrolyzed ampicillin forms coordination bonds to the zinc ions present at the active site, with Zn (I) coordinated by 3 histidine residues (His-120, His-122, and His-189) and Zn (II) coordinated by Asp-124, Cys-208, and His-250, and hydrogen bonds with several critical residues around the active site. In addition to forming a salt bridge by coordinating to Zn (II) and Lys-211, β-lactam ring also coordinated to Zn (I) and Asn-220, which is a conserved residue in subclass B1 and B2 MBLs. The phenyl ring of the hydrolyzed ampicillin was seen forming strong hydrophobic interactions with residues Leu-65 and Met-67 and Trp-93. It was also observed that residue Gln-123 in NDM-1 was replaced by Asp-119 in VIM-2 and Ser-80 in IMP-1, respectively, making the formation of hydrogen bonds less possible in the latter two. NDM-1 shows tighter binding to penicillin and ampicillin compared to VIM-2 and IMP-1 [2] because of strong hydrophobic and hydrogen bond interactions between the nitrogen atom near the phenyl ring of β-lactam ring and residue Gln-123. However, it does not bind to carbepenams as tightly as IMP-1 or VIM-2 and turns over the carbapenems at a rate similar to that of VIM-2 [2]. Structural comparison with other MBLs has revealed that two residues in NDM-1 i.e. Tyr-229 and residue Lys-125 [32] might play critical roles in stabilizing the conformation of the active site. These residues formed extensive hydrophobic and hydrophilic interactions with neighboring residues like Lys-209 both on top and bottom; providing stability and specific orientation and at the same time restricting the flexibility of Asn-220 conferred by Gly-219 [32]. According to a current research, a new weak inhibitor tigecycline has been identified which was previously reported to inhibit the growth of NDM-1 harbored Kleibsiella pneumonia [2].
So far there is only one report on the proposed hydrolysis mechanism of β-lactams by NDM-1 [32]. During hydrolysis of β-lactams, the shared hydroxide between the two zinc ions attacks the carbonyl carbon of β-lactam ring as a nucleophile. The C-N bond breaks immediately on the nucleophile attack resulting in a nitrogen anion intermediate; while, in the second path the breaking of the C-N bond is concerted with the protonation of the nitrogen atom and no nitrogen anion intermediate is generated. In either path, the nitrogen atom of the β- lactam ring gets a proton from the attacking hydroxide. Unlike other MBLs, NDM-1 contains an additional insert between residues 162 and 166 which is present in the opposite side of the active site with distance of around 20 Å, its role in the hydrolysis reaction is still unknown [2] and needs a careful investigation.
NDM-1 is unique among other MBLs due to its enlarged and flexible active site that was revealed by the crystal structures of its metal free apo and monozinc forms. This facilitated NDM-1 to accommodate many β-lactam substrates, hence, explaining the extended β- lactamase catalytic activity [42]. NDM-1 active site is a deep cavity that is formed by the loop regions between β5-α2 and β10-α4, in which the catalytic zinc ions are deeply buried. This cavity is clamped by the loop regions between Thr-119 & Met-126 and Ser-217 & Asp-225 (i.e., the ceiling and floor), while the Met-67 & Gly-71 loop acts as a doorkeeper to close the site during substrate binding and open it for product release via conformational changes (Figure 3) [49]. The active site of NDM-1 is fully accessible to solvent and is located at the bottom of a shallow groove, which is a common characteristic of all MBLs. In the active site Zn2+ (I) is coordinated with three conserved histidine residues 120, 122 and 189, while Zn2+ (II) is coordinated with the conserved residues Asp-124, Cys-208 and His-250. A water molecule or more probably a hydroxide moiety bridges both zinc ions, which has been suggested to act as a nucleophile during the β-lactam hydrolysis [49,50]. Substrates that are most efficiently (kcat/KM) turned over, in general have extended hydrophobic characteristics that complement the linear and hydrophobic nature of the NDM-1 active site cavity (Table 3). The primary hydrophobic arrangement is contributed by residues Leu- 65, Met-67, Pro-68, Val-73, Gly-69, Phe-70 and Val-73, which contour the flexible roof of the active site (Figure 5). In addition, alkyl moieties of Leu-209, Ile-210, Lys- 211, Asp-212, Lys-214, Ala-215, Lys-216, and Asn-220 side chains give the distal region and base of the active site a partially hydrophobic surface area, while simultaneously maintaining hydrogen bond capability. These residues contribute to the “keg” set of side chains that constitute the active site [42].
NDM-1 is classified as a group B1 MBL according to its primary sequence [2]. This group generally contains a dinuclear zinc cluster at the active site but also contains examples for which the affinities of the two zinc ions differ sufficiently to favor the monozinc form [18,41, 51,52]. The structures of NDM-1 that have been reported till date contain a dinuclear metal ion cluster at the active site [32,46,47] with one exception showing a monozinc
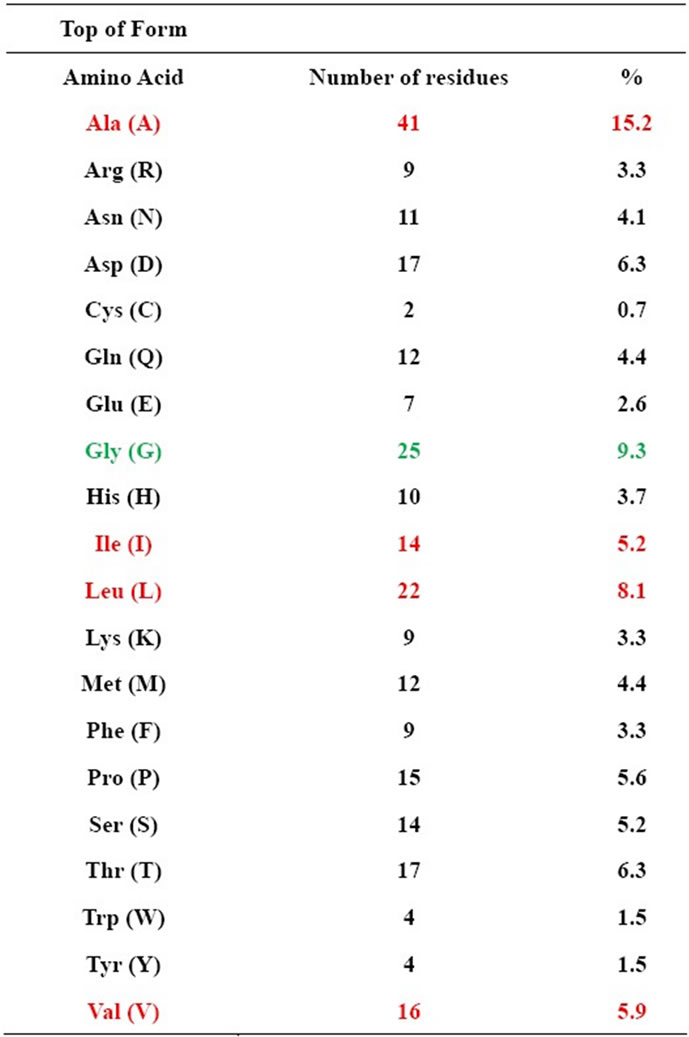
Table 3. Amino acid composition in NDM-1 (hydrophobic amino acidsred).
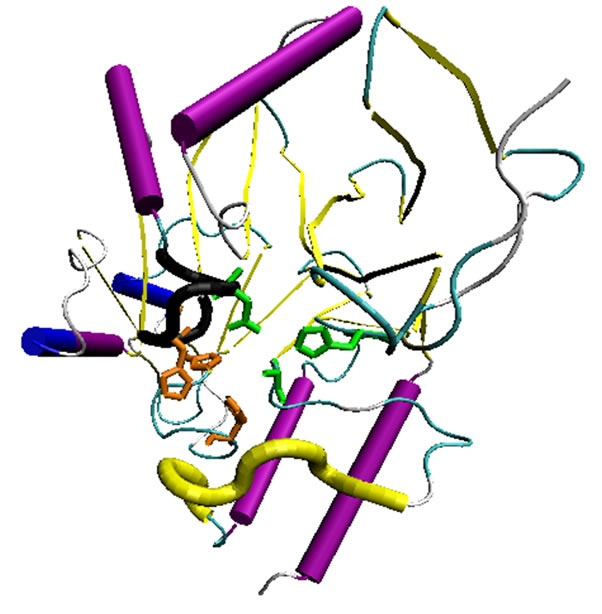
Figure 5. Zn (I) and (II) binding pockets of NDM-1 have been highlighted by the corresponding amino acid side chains in orange and green color respectively. The fragment in black marks the ceiling of the active site while that in yellow shows the floor.
NDM-1 in which the site containing ligands: His-120, His-122, and His-189 is occupied by a zinc (II) ion but the site with Asp-124, Cys-208, and His-250 is not occupied by a metal ion [42]. In a recent study recombinant NDM-1 was released in the culture medium as a monomer and purified as a dizinc protein. However, the two zinc binding sites have different affinities, allowing the preparation of monozinc NDM-1, as well [48].
3.3. Homology Modeling
A few theoretical studies have also been devoted to understanding the structural and mechanistic properties of NDM-1. Homology Modeling has been reported by using VIM-2 [21] and VIM-4 [53,54] crystal structures as templates. In one of the earliest structural reports on NDM-1 the three-dimensional structure was developed using homology modeling using the crystal structure of VIM-2 (PDB ID: 1ko3) [21]. The model thus obtained also showed that like most of the other MBLs, the NDM-1 protein belongs to the α/β structural class [55], with 3 helices and 7 β-strands. The 3 helices were exposed to the solvent. The N-terminal and C-terminal regions of NDM-1 can be superposed by a 180˚ rotation around a central axis, indicating that the entire structure may have arisen from the duplication of a gene [21]. On similar lines, X-ray crystallographic structure of Di-Zinc metallo-β-lactamase Vim-4 from Pseudomonas aeruginosa (PDB ID: 2WHG) was selected as a template protein for homology modeling in another report [44,54]. Two mobile loops in the active site were found to be crucial for substrate recognition, binding and catalysis in MBLs [56,57]. In NDM-1, the loop1, also called the flapping loop, is composed of amino acids LDMPGFGAVA (residues 65 - 74). Compared with the loop1 (QSFDGAVYP) in VIM-2 and VIM-4, the size of the active site cavity is enlarged with less bulky amino acids, suggesting a broader substrate profile. Besides it is worth noting that it is more hydrophobic than others [32]. There is only one benzene ring of Phe-70 in the middle of the loop1 of NDM-1, and the side-chains of Asp-66 and Met-67 pointed to the solvent, the less bulky amino acids, especially for the glycines, are proposed to make the loop1 more flexible for substrate binding [47].
These reports further add to the growing evidences indicating that various tools in structural bioinformatics, such as homology modeling, molecular docking, as well as molecular dynamics simulations can timely provide very useful information and insights for biomedical science and drug development.
3.4. Docking Studies
3.4.1. Enzyme-Ligand Complex
Molecular docking with Monte Carlo simulated annealing was used to predict the favorable binding interaction modes for NDM-1 with Imipenem and Meropenem, two typical β-lactam antibiotic drugs (as it is reported recently that NDM-1 showed a comparatively high resistance to Imipenem and Meropenem [3,21]). Docking studies with Imipenem showed that its binding pocket is formed by 9 residues that have at least one heavy atom with a distance within 5 Å to Imipenem. In addition to the van der Waals interaction of the Imipenem drug with the surrounding binding pocket residues, there are remarkable hydrogen bonds that have tightly tethered the drug to Glu-88 and Thr-126 of the receptor, making NDM-1 able to recognize the antibiotic drug followed by cleaving the amide bond of its β-lactam ring so as to inactivate Imipenem. Similar binding interactions were evident with Meropenem too [21]. In an additional molecular docking study, it was observed that ligands like Ceftriaxone formed three hydrogen bond with active site residues Lys-165, Ser-203 and Asp-177, Cefepime was also found to form three hydrogen bond with residues Lys-165, Ser-203 and Ala-28. Cefoperazone, was observed to dock with residues Ser-205, Lys-165 and His-204, Imepenum formed 5 hydrogen bonds with residues Lys-165, Ser-203, Gly-161, Asp-78 and Ser-205 and Meropenum bounded to Lys-165, Ser-203, Gly-161, Asp-78, Ile-164 and His-204. These studies suggested that carbapenem group of antibiotics are more potent against NDM-1 as compared to cephalosporins [58].
3.4.2. Potent Inhibitors
Currently, there is no inhibitor of NDM-1 that is approved for medical treatment. To the best of our knowledge there are only two reports of this type where a compound library screening approach was used to identify molecules which may acts as potent inhibitors of NDM-1. In one study certain compounds were identified from the ZINC Database in order to characterize NDM-1 inhibitors from a library of active compounds. This study states that the N-arylsulphonyl hydrazones, Sulfonyltriazole and C-6-Mercaptomethyl Penicillinates derivatives have the potency to inhibit the NDM-1 protein [54]. In another study, homology modeling and docking analysis of NDM-1 with flavanoids indicated that Quercetin may acts as a potent inhibitor of NDM-1 [59]. In one of the most recent reports crystal structure of NDM-1 bound to the potent competitive inhibitor l-captopril has been put forward [45].
4. CONCLUSIONS
This perspective has analyzed NDM-1 in light of its sequence homologs, phylogenetics, structural characteristics, active site analysis as well as docking models and inhibitors. These structural elucidations shall facilitate the thorough mechanistic understanding required for a rational design of small molecule inhibitors specific to NDM-1. Still, the inherent diversity in its active site and limited knowledge about the mechanism of hydrolysis make the development of broad spectrum inhibitors a great challenge.
Bacterial resistance to antibiotics highlights the struggle between new drug development and the evolution of resistance in bacteria. Whenever a new modified β-lactam compound is developed that shows activity against bacterial infections, the bacteria fight back and evolve new enzymes to inactivate the new compound. Increased surveillance is necessary to monitor this type of resistance. Continued research is required to develop novel derivatives of this highly successful antibacterial compound and keep one step ahead of the resistant strains. At present, no reports indicate individual inhibitors with submicromolar Ki values against all 3 MBL subclasses, which is a highly desirable property for clinical use.
As, NDM-1 is likely to be more potent and extensive than known MBLs in inactivating β-lactam antibiotics, so unfolding the ligand binding properties and catalytic mechanisms of NDM-1 is an urgent issue and challenge. Currently, there is no drug or inhibitor of NDM-1 that is approved for medical treatment. Therefore, in view of the emerging multidrug-resistant strains carrying NDM-1, the discovery of effective inhibitors is the need of the hour.
![]()
![]()
REFERENCES
- Levy, S.B. and Marshall, B. (2004) Antibiotic resistance worldwide: Causes, challenges and responses. Nature Medicine, 10, S122-S129. doi:10.1038/nm1145
- Yong, D., Toleman, M.A., Giske, C.G., Cho, S., Hyun, S., Sundman, K., Lee, K. and Walsh, T.R. (2009) Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in klebsiella pneumonia sequence type 14 from India. Antimicrobial Agents and Chemotherapy, 53, 5046-5054. doi:10.1128/AAC.00774-09
- Kumarasamy, K.K., Toleman, M.A., Walsh, T.R., Bagaria, J., Butt, F., Balakrishnan, R., Chaudhary, U., Doumith, M., Giske, C.G., Irfan, S., Krishnan, P., Kumar, A.V., Maharajan, S., Mushtaq, S., Noorie, T., Paterson, D.L., Pearson, A., Perry, C., Pike, R., Rao, B., Ray, U., Sarma, J.B., Sharma, M., Sheridan, E., Thirunarayan, M.A., Turton, J., Upadhay, S., Warner, M., Welfare, W., Livermore, D.M. and Woodford, N. (2010) Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. The Lancet Infectious Diseases, 10, 597-602. doi:10.1016/S1473-3099(10)70143-2
- Rolain, J.M., Parola, P. and Cornaglia, G. (2010) New Delhi metallo-beta-lactamase (NDM-1): Towards a new pandemia? Clinical Microbiology and Infection, 12, 1699- 1701. doi:10.1111/j.1469-0691.2010.03385.x
- Bush, K. (2010) Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Current Opinion in Microbiology, 13, 558-564. doi:10.1016/j.mib.2010.09.006
- Huo, T.I. (2010) The first case of multidrug-resistant NDM-1 harboring Enterobacteriaceae in Taiwan: Here comes the superbacteria! Journal of the Chinese Medical Association, 73, 557-558. doi:10.1016/S1726-4901(10)70121-0
- Moellering Jr., R.C. (2010) NDM-1-A cause for worldwide concern. The New England Journal of Medicine, 363, 2377-2379. doi:10.1056/NEJMp1011715
- Poirei, L., Lagrutta, E., Taylor, P., Pham, J. and Nordmann, P. (2010) Emergence of metallo-β-lactamase NDM-1 producing multidrug resistant Escherichia coli in Australia. Antimicrobial Agents and Chemotherapy, 54, 4914-4916. doi:10.1128/AAC.00878-10
- Poirel, L., Al Maskari, Z., Al Rashdi, F., Bernabeu, S. and Nordmann, P. (2011) NDM-1 producing Klebsiella pneumonia isolated in the Sultanate of Oman. The Journal of Antimicrobial Chemotherapy, 66, 304-306. doi:10.1093/jac/dkq428
- Poirel, L., Revathi, G., Bernabeau, S. and Nordamnn, P. (2011) Detection of NDM-1 producing Kleibsiella pneumonia in Kenya. Antimicrobial Agents and Chemotherapy, 55, 934-936. doi:10.1128/AAC.01247-10
- Poirel, L., Ros, A., Crricajo, A., Berthelot, P., Pozzetto, B., Bernabeu, S. and Nordmann, P. (2011) Extremely drug-resistant Citrobacter freundii identified in a patient returning from India and producing NDM-1 and other carbapenemases. Antimicrobial Agents and Chemotherapy, 55, 447-448. doi:10.1128/AAC.01305-10
- Samuelsen, O., Thilesen, C.M., Heggelund, L., Vada, A.N., Kummel, A. and Sundsfjord, A. (2011) Identification of NDM-1 producing Enterobacteriaceae in Norway. The Journal of Antimicrobial Chemotherapy, 66, 670- 672. doi:10.1093/jac/dkq483
- Walsh, T.R. (2010) Emerging carbapenemases: A global perspective. International Journal of Antimicrobal Agents, 36, S8-S14. doi:10.1016/S0924-8579(10)70004-2
- Zhang, X. (2010) Human in check: New threat from superbugs equipped with NDM-1. Protein Cell, 1, 1051- 1052. doi:10.1007/s13238-010-0134-7
- Zheng, B., Tan, S., Gao, J., Han, H., Liu, J., Lu, G., Liu, D., Yi, Y., Zhu, B. and Gao, G.F. (2011) An unexpected similarity between antibiotic-resistant NDM-1 and betalactamase II from Erythrobacter litoralis. Protein Cell, 2, 250-258. doi:10.1007/s13238-011-1027-0
- Llarrull, L.I., Testero, S.A., Fisher, J.F. and Mobashery, S. (2010) The future of β-lactams. Current Opinion in Microbiology, 13, 551-557. doi:10.1016/j.mib.2010.09.008
- Draw, S.M. and Bonomo, R.A. (2010) Three decades of beta-lactamase inhibitors. Clinical Microbiology Reviews, 23, 160-201. doi:10.1128/CMR.00037-09
- Wilke, M.S., Lovering, A.L. and Strynadka, N.C. (2005) Beta-lactam antibiotic resistance: A current structural perspective. Current Opinion in Microbiology, 8, 525- 533. doi:10.1016/j.mib.2005.08.016
- Bebrone, C. (2007) Metallo-β-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochemical Pharmacology, 74, 1686-1701. doi:10.1016/j.bcp.2007.05.021
- Nair, D. and Rawat, D. (2010) Extended-spectrum β- lactamases in Gram Negative Bacteria. Journal of Global Infectious Diseases, 2, 263-274. doi:10.4103/0974-777X.68531
- Wang, J.-F. and Chou, K.C. (2011) Insights from modeling the 3D structure of New Delhi metallo-β-lactamase and its binding interactions with antibiotic drugs. PLoS ONE, 6, e18414. doi:10.1371/journal.pone.0018414
- Matagne, A., Lamotte-Brasseur, J. and Frere, J.M. (1998) Catalytic properties of class A-lactamases: Efficiency and diversity. The Biochemical Journal, 330, 581-598.
- Abraham, E.P. and Chain, E. (1940) An enzyme from bacteria able to destroy penicillin. Nature, 146, 837-837. doi:10.1038/146837a0
- Turner, P.J. (2005) Extended-spectrum-lactamases. Clinical Infectious Diseases, 41, S273-S275. doi:10.1086/430789
- Ambler, R.P. (1980) The structure of beta-lactamases. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 289, 321-331. doi:10.1098/rstb.1980.0049
- Gupta, V. (2007) An update on newer-lactamases. Indian Journal of Medical Research, 126, 417-427.
- Dhillon, R. and Clark, J. (2011) ESBLs: A clear and present danger? Critical Care Research and Practice, 2012, 625170-625181.
- Kanj, S.S. and Kanafani, Z.A. (2011) Current concepts in antimicrobial therapy against resistant gram-negative organisms: Extended-spectrum beta-lactamase-producing Enterobacteriaceae, carbapenem-resistant Enterobacteriaceae, and multidrug-resistant Pseudomonas aeruginosa. Mayo Clinic Proceedings, 86, 250-259. doi:10.4065/mcp.2010.0674
- Ambler, R.P., Coulson, A.F., Frere, J.M., Ghuysen, J.M., Joris, B., Forsman, M., Levesque, R.C., Tiraby, G. and Waley, S.G. (1991) A standard numbering scheme for the class A beta lactamases. The Biochemical Journal, 276, 269-270.
- Bush, K., Jacoby, G.A. and Medeiros, A.A. (1995) A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrobial Agents and Chemotherapy, 39, 1211-1233. doi:10.1128/AAC.39.6.1211
- Rasmussen, B.A. and Bush, K. (1997) Carbapenem-hydrolyzing betalactamases. Antimicrobial Agents and Chemotherapy, 41, 223-232.
- Zhang, H.M. and Hao, Q. (2011) Crystal structure of NDM-1 reveals a common β-lactam hydrolysis mechanism. The FASEB Journal, 25, 2574-2582. doi:10.1096/fj.11-184036
- Lee, K., Yum, J.H., Yong, D., Lee, H.M., Kim, H.D., Docquier, J.D., Rossolini, G.M. and Chong, Y. (2005) Novel acquired metallo-beta-lactamase gene, bla(SIM-1), in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrobial Agents and Chemotherapy, 49, 4485-4491. doi:10.1128/AAC.49.11.4485-4491.2005
- Neuwald, A.F., Liu, J.S., Lipman, D.J. and Lawrence, C.E. (1997) Extracting protein alignment models from the sequence database. Nucleic Acids Research, 25, 1665- 1677. doi:10.1093/nar/25.9.1665
- Daiyasu, H., Osaka, K., Ishino, Y. and Toh, H. (2001) Expansion of the zinc metallohydrolase family of the beta-lactamase fold. FEBS Letters, 503, 1-6. doi:10.1016/S0014-5793(01)02686-2
- Aravind, L. (1999) An evolutionary classification of the metallo-β-lactamase fold proteins. Silico Biology, 1, 69- 91.
- Costello, A.L., Sharma, N.P., Yang, K.W., Crowder, M.W. and Tierney, D.L. (2006) X-ray absorption spectroscopy of the zinc binding sites in the class B2 metallo-β-lactamase ImiS from Aeromonas veronii bv. sobria. Biochemistry, 5, 13650-13658. doi:10.1021/bi061547e
- Li, Y., Chooi, Y.H., Sheng, Y., Valentine, J.S. and Tang, Y. (2011) Comparative characterization of fungal anthraxcenone and naphthacenedione biosynthetic pathways reveals an α-hydroxylation-dependent claisen-like cyclization catalyzed by a dimanganese thioesterase. Journal of the American Chemical Society, 133, 15773-15785. doi:10.1021/ja206906d
- Llarrull, L.I., Tioni, M.F., Kowalski, J., Bennett, B. and Vila, A.J. (2007) Evidence for a dinuclear active site in the metallo-β-lactamase BcII with substoichiometric Co (II). A new model for metal uptake. The Journal of Biological Chemistry, 282, 30586-30595. doi:10.1074/jbc.M704613200
- Gomes, C.M., Giuffre, A., Forte, E., Vicente, J.B., Saraiva, L.M., Brunori, M. and Teixeira, M. (2002) A novel type of nitric-oxide reductase. Escherichia coli flavorubredoxin. The Journal of Biological Chemistry, 277, 25273-25276. doi:10.1074/jbc.M203886200
- Carfi, A., Pares, S., Duee, E., Galleni, M., Duez, C., Frere, J.M. and Dideberg, O. (1995) The 3-D structure of a zinc metallo-β-lactamas from Bacillus cereus reveals a new type of protein fold. The EMBO Journal, 14, 4914-4921.
- Kim, Y., Tesar, C., Mire, J., Jedrzejczak, R., Binkowski, A., Babnigg, G., Saccgettini, J. and Joachimiak A. (2011) Structure of apoand mono-metalated forms of NDM-1: A highly potent carbanepem-hydrolyzing metallo-β-lactamase. PLoS ONE, 6, e24621. doi:10.1371/journal.pone.0024621
- Peirano, G., Schreckenberger, P.C. and Pitout, J.D.D. (2011) Characteristics of NDM-1 producing Escherichia coli isolates that belong to the successful and virulent clone ST131. Antimicrobial Agents and Chemotherapy, 55, 2986-2988. doi:10.1128/AAC.01763-10
- King, D. and Strynadka, N. (2011) Crystal structure of New Delhi metallo-β-lactamase reveals molecular basis for antibiotic resistance. Protein Science, 20, 1484-1491. doi:10.1002/pro.697
- King, D.T., Worrall, L.J., Gruninger, R. and Strynadka, N. (2012) New Delhi metallo-β-lactamase: Structural insights into β-lactam recognition and inhibition. Journal of the American Chemical Society, 134, 11362-11365. doi:10.1021/ja303579d
- Green, V.L., Verma, A., Owens, R.J., Phillips, S.E. and Carr, S.B. (2011) Structure of New Delhi metallo-β-lactamase 1 (NDM-1). Acta Crystallogr, 67, 1160.
- Guo, Y., Wang, J., Niu, G., Shui, W., Sun, Y., Zhou, H., Zhang, Y., Yang, C., Lou, Z. and Rao, Z. (2011) A structural view of the antibiotic degradation enzyme NDM-1 from a superbug. Protein Cell, 2, 384-394. doi:10.1007/s13238-011-1055-9
- Thomas, P.W., Zheng, M., Wu, S., Guo, H., Liu, D., Xu, D. and Fast, W. (2011) Characterization of purified New Delhi metallo-β-lactamase-1. Biochemistry, 50, 10102- 10113. doi:10.1021/bi201449r
- Toleman, M.A., Bennett, P.M. and Walsh, T.R. (2006) Common regions e.g. orf513 and antibiotic resistance: IS91-like elements evolving complex class I integrons. The Journal of Antimicrobial Chemotherapy, 58, 1-6. doi:10.1093/jac/dkl204
- Toleman, M.A., Bennett, P.M. and Walsh, T.R. (2006) ISCR elements: Novel gene-capturing systems of the 21st century? Microbiology and Molecular Biology Review, 70, 296-316. doi:10.1128/MMBR.00048-05
- Heinz, U. and Adolph, H.W. (2004) Metallo-β-lactamases: Two binding sites for one catalytic metal ion? Cellular and Molecular Life Sciences, 61, 2827-2839. doi:10.1007/s00018-004-4214-9
- Orellano, E.G., Girardini, J.E., Cricco, J.A., Ceccarelli, E.A. and Vila, A.J. (1998) Spectroscopic characterization of a binuclear metal site in Bacillus cereus β-lactamase II. Biochemistry, 37, 10173-10180. doi:10.1021/bi980309j
- Liang, Z., Li, L., Wang, Y., Chen, L., Kong, X., Hong, Y., Lan, L., Zheng, M., Yang, C.G., Hong, S.X., Luo, C., Li, K.K., Chen, K. and Jiang, H. (2011) Molecular basis of NDM-1, a new antibiotic resistance determinant. PLoS One, 6, e23606. doi:10.1371/journal.pone.0023606
- Randhawa, V. and Jamwal, R. (2011) Molecular modelling and virtual screening studies of NDM-1 Beta-lactamase for identification of a series of potent inhibitors. International Research Journal of Biochemistry and Bioinformatics, 1, 95-102.
- Chou, K.C. and Zhang, C.T. (1995) Review: Prediction of protein structural classes. Critical Reviews in Biochemistry and Molecular Biology, 30, 275-349. doi:10.3109/10409239509083488
- Moali, C., Anne, C., Lamotte-Brasseur, J., Groslambert, S., Devreese, B., Van Beeumen, J., Galleni, M. and Frère, J.M. (2003) Analysis of the importance of the metallobeta-lactamase active site loop in substrate binding and catalysis. Chemistry and Biology, 10, 319-329. doi:10.1016/S1074-5521(03)00070-X
- Yamaguchi, Y., Jin, W., Matsunaga, K., Ikemizu, S., Yamagata, Y., Wachino, J., Shibata, N., Arakawa, Y. and Kurosaki, H. (2007) Crystallographic investigation of the inhibition mode of a VIM-2 metallo-betalactamase from Pseudomonas aeruginosa by a mercaptocarboxylate inhibitor. Journal of Medicinal Chemistry, 50, 6647-6653. doi:10.1021/jm701031n
- Sowmiya, M., Umashankar, V., Muthukumaran, S., Madhavan, H.N. and Malathi, J. (2012) Studies on New Delhi metallo-beta-lactamse-1 producing Acinetobacter baumannii isolated from donor swab in a tertiary eye care centre, India and structural analysis of its antibiotic binding interactions. Bioinformation, 8, 445-452. doi:10.6026/97320630008445
- Gagupati, J., Mukkavali, S.V., Atiamula, S. and Siva Sai, K.S.R. (2011) In-silico modelling and docking studies of New Delhi beta-lactamase-1 (Superbug). International Journal of Engineering Science and Technology, 3, 2427- 2534.
NOTES
*Corresponding author.

