Journal of Tuberculosis Research
Vol. 1 No. 1 (2013) , Article ID: 33448 , 8 pages DOI:10.4236/jtr.2013.11001
Molecular characterization of the rpoB gene mutations of Mycobacterium tuberculosis isolated from China
![]()
National Tuberculosis Reference Laboratory, National Center for Tuberculosis Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China; *Corresponding Author: zhaoyanlin@chinatb.org
Copyright © 2013 Shengfen Wang et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 3 May 2013; revised 5 June 2013; accepted 15 June 2013
Keywords: Rifampicin; resistance; Beijing genotype
ABSTRACT
Objective: To analyze characterization of the rpoB gene mutations of Mycobacterium tuberculosis isolated from China and to explore the association of specific mutations conferring rifampicin (RIF) resistance with Beijing genotype strains. Methods: Genotypic analysis of 3479 M. tuberculosis isolates including 402 RIF-resistant and 3077 RIF-susceptible isolated from the national drug-resistant tuberculosis baseline survey was performed. Results: DNA sequencing analysis of the 81-bp RIF resistance determining region (RRDR) of the ropB gene revealed that 98.01% of RIF-resistant strains showed rpoB gene mutation, isolates with mutations at codon rpoB531, rpoB526 and rpoB516 were the most frequently. Analysis of the rpoB gene of 3077 RIF-susceptible strains revealed that 98.96% of the strains had no mutation. The distribution of mutation frequency at different critical codons in different regions of China was statistically significant (p = 0.001). There was no significant difference in the occurrence of mutations at critical codons between the rifampicin-resistant Beijing and non-Beijing isolates. Conclusion: About 98% of RIF-resistant strains isolated from China carry mutations in RRDR of rpoB gene. Mutation profiles in RIF-resistant M. tuberculosis clinical isolates are variable depending on the different geographical regions of China. The results provide valuable information in adopting new molecular methods for diagnosis of TB in China.
1. INTRODUCTION
Tuberculosis (TB) especially drug resistance TB is one of the big challenges in China, and nationwide anti-TB drug resistance survey indicated that 34.2% of new cases and 54.5% of previously treated cases had resistance to at least one of the 4 first-line anti-tuberculosis drugs, 5.7% of new cases and 25.6% of previously treated cases had multidrug-resistant (MDR) tuberculosis [1]. High burden of drug resistant TB make a serious problem to the TB control in China. Treatment of TB infection relies primarily on the use of first-line drugs including isoniazid and RIF with ethambutol and pyrazinamide [2]. RIF remains one of the most efficient drugs in the modern short-course regimens due to its excellent bactericidal activity [3]. Resistance to RIF is an indicator of possible multi-resistance as nearly 90% of RIF resistant strains are also isoniazid-resistant [4]. Reliable and timely drug susceptibility testing is critical to ensure patients receive effective drug treatment and reduce TB transmission. At present, the solid culture and culture-based drug susceptibility testing are widely used in most areas of China, these methods are constrained by the slow growth of M. tuberculosis. The use of molecular methods to identify mutations associated with drug resistance can decrease diagnostic delay. In recent years, studies indicating TB drug resistance are associated with mutations in several genes. Resistance to RIF is due to the genetic alterations in the rpoB gene encoding the beta-subunit of the DNA-dependent ribonucleic acid polymerase. It is reported that 95% of RIF resistance isolates are mediated by mutations in an 81 bp hot spot region (codons 507 to 533) of the rpoB gene [5]. Various molecular methods based on rpoB gene sequence have been developed to detect mutations in the sequence, including direct sequencing of PCR products, single stranded confirmation polymorphism analysis and line probe assay.
In this study, we use a large set of M. tuberculosis isolated from the whole country during nationwide antiTB drug resistance survey, representing a variety of RIF resistance patterns, analyze the molecular pattern of mutations conferring resistance to RIF of M. tuberculosis strains and explore the associations of specific mutations with Beijing genotype strains.
2. MATERIALS AND METHODS
2.1. Mycobacterial Strains
From April 2007 to December 2007, the National Tuberculosis Reference Laboratory, Chinese Center for Disease Control and Prevention conducted the first round nationwide anti-TB drug resistance survey in China. According to protocol, 70 clusters covering all national provinces (autonomous regions and municipalities directly under the central government) were randomly chosen according to multistage cluster sampling method. 51 eligible new cases and 17 previously treated smearpositive patients were enrolled into each cluster during the survey [1]. Mycobacterium strains were isolated on solid Löwenstein-Jensen (L-J) medium at county/regional laboratories, Species differentiation and drug susceptibility testing (DST) were performed at the National Tuberculosis Reference Laboratory. DST results were available for 3929 M. tuberculosis strains isolated from 3037 new and 892 previously treated cases, of which 452 isolates were RIF resistant strains and 3477 were RIF susceptible strains. 3479 isolates including 402 RIF resistant strains representing 88.93% of the RIF resistant strains and 3077 RIF susceptible strains representing 88.50% of the RIF susceptible strains were random selected and included in this study.
2.2. Drug Susceptibility Testing
Antimicrobial drug susceptibility testing was performed using the conventional proportional method: RIF (40 μg/ml), isoniazid (0.2 μg/ml), ethambutol (2.0 μg/ml), streptomycin (4.0 μg/ml), kanamycin (30 μg/ml) and Ofloxacin (2.0 μg/ml) were used in slants, H37Rv strain was used as negative control, ongoing external quality control was assured by the Hong Kong Supranational Tuberculosis Reference Laboratory.
2.3. Preparation of DNA
The bacteria were removed from L-J slants, suspended in 400 μl of 1 × TE buffer, heat killed, boiling at 100˚C for 15 min, and then centrifuged at 12,000 g for 5 min to remove cell debris. The supernatant was stored at −20˚C. For each PCR reaction, 2 μl of extracted genomic DNA was used.
2.4. RpoB Gene Amplification and Sequence Analyse
PCR amplification followed by DNA sequencing of the rpoB RIF RRDR region, PCR primers for rpoB were rpoB F (5’-tacggtcggcgagctgatcc-3’) and rpoB R: (5’- tacggcgtttcgatgaacc-3’). Each 25-μl PCR mixture contained 12.5 μl 2 × Taq PCR Star mix (GenStar Biosolutions Co., Ltd.), 0.5 μl of each set of 20 μM primers, 9.5 μl distilled H2O, and 2 μl of genomic DNA. The amplification parameters included an initial denaturation step at 94˚C for 5 min, followed by 30 cycles of denaturation at 94˚C for 30 s, annealing at 58˚C for 30 s, and elongation at 72˚C for 30 s, with a final extension at 72˚C for 7 min. The 411 bp amplicons were purified and sequenced using the primer rpoB F or rpoB R and sequences analysed using DNAstar and BioEdit software.
2.5. Spoligotyping
All isolates were analysed by spoligotyping using a commercial kit (Isogen Bioscience BV, Maarsen, the Netherlands) according to the manufacturer’s instructions. Strains of the Beijing genotype were identified by their characteristic spoligotype pattern with hybridisation to spacers 35 - 43 only.
2.6. Quality Control
92.04% (370/402) of RIF resistant and 4.87% (150/ 3077) of a random sample of RIF susceptible strains were re-tested by another technicians at National TB Reference Laboratory, the results of rpoB gene sequencing were confirmed.
2.7. Ethical Consideration
The study protocol was approved by the ethical review committees of Chinese Center for Disease Control and Prevention. Written informed consent was obtained from the study participants.
3. RESULTS
3.1. Drug Susceptibility and Beijing Genotype Patterns
Phenotypic DST was performed on all study isolates for six drugs: RIF, isoniazid, ethambutol, streptomycin, kanamycin and Ofloxacin. Of the 3479 isolates tested in this study, 2151 were susceptible to all tested drugs, 1328 were drug-resistant to any one of the tested drugs. 402 were RIF-resistant strains, of which, 37 were RIF monoresistant. 138 were isoniazid monoresistant. 342 were MDR-TB, as defined by resistance to both isoniazid and RIF, and of which 22 were extensively drug-resistant (XDR) TB strains, defined as MDR TB that is also resistant to any fluoroquinolones and one of the second-line injectable drugs. 85.07% (342/402) of RIF resistance strains were MDR-TB strains. 578 were resistant to drugs not including RIF or isoniazid. Spoligotyping was performed for 394 of 402 RIF-resistant isolates, of which, 249 (63.2%) were revealed to be Beijing genotype strains, 145 (36.8%) were found to be non-Beijing strains; of 335 of 342 MDR isolates that were spoligotyping tested, 212 (63.3%) were found to be Beijing strains, 123 (36.7%) were found to be non-Beijing strains; of 3023 of 3077 RIF-susceptible isolates that were tested, 1923 (63.6%) were found to be Beijing strains.
3.2. RIF Resistant and rpoB Gene Mutation
402 phenotypically RIF-resistant isolates were tested for mutations in the rpoB gene. DNA sequence analysis revealed that 98.01% (394/402) strains of RIF-resistant showed rpoB gene mutation versus 1.99% (8/402) strains having no mutation. A total of 23 nonsynonymous single mutations, 2 3-bp insertions, 5 deletions, 20 double mutations and a triple mutation were identified among the study isolates. In 359 of 402 RIF-resistant isolates with point mutation, 5 types of mutations were identified in codon 531, 8 types of mutation were found in codon 526, 2 types of mutation were found in codons 516 and 522, respectively. In 24 of the double mutations, 16 isolates with unique pairs, 2 with the Leu511Pro/Met515lle, 2 with the Leu511Pro /Asp516Gly , 2 with the Leu511Pro/ Asp516 Ala , 2 with the Leu511Pro/His526Gln. Mutations were observed in affected codons 508, 509, 510, 511, 512, 513, 514, 515, 516, 517, 518, 519, 522, 523, 525, 526, 529, 531, and 533 in the rpoB fragment.
Of the mutations found within the RRDR, the codons most frequently involved in mutations were codon 531 (50.50%), codon 526 (26.12%) and codon 516 (7.21%). The three most frequently observed mutations accounted for 83.83% of the RIF-resistant isolates in this study. The commonest mutation was found at codon 531 in 191 (47.51%) of isolates with Ser → Leu substitution (TCG → TTG), the second type of mutation was detected at codon 526 in 34 (8.45%) of isolates with His → Tyr substitution (CAC → TAC), the third type of mutation was found at 526 in 31 (7.70%) of isolates with His → Asp (CAC→ GAC), followed by codon 533 in 21 (5.22%) of isolates with Leu → Pro substitution (CTG → CCG) and codon 516 in 17 (4.23%) of isolates with Asp → Val substitution (GAC → GTC). As to our knowledge, new mutations including 4 deletions, 2 insertions and 6 point mutations were not reported in the previous literature (Table 1).
3.3. RIF Susceptible and rpoB Gene Mutation
Analysis of the rpoB gene of the 3077 RIF-susceptible strains revealed that 98.96% (3045/3077) of the strains had no mutation while 1.04% (32/3077) of strains with mutation in the hot spot region of rpoB gene. 31 isolates with single mutations and 1 with Leu511Pro/His526Gln double mutation were found within the rpoB gene. Of the 31 isolates showing single mutations, 10 had the mutation Leu511Pro, 9 the mutation Leu533Pro, 1 the mutation Asp516Val, 2 the mutation Asp516Gly, 3 the mutation Asp516Tyr, 3 the mutation Ser522Leu, 1 the mutation Ser522Gln, 2 the mutation His526Asn. 9 different mutations in five codons were identified with rpoB codons 511 (0.36%), 533 (0.30%) and 516 (0.19%) most frequently affected (Table 1).
3.4. Mutation Profiles of RIF-Resistant Isolates and Geographical Distribution
Considering different geographical regions may have different rpoB gene mutation profiles, we divide China into six different geographical areas, namely North China, Northeast China, East China, Central and South China, Southwest China and Northwest China. The codons most frequently involved in mutations were codon 531 in North China (60.5%) and Northeast China (66.7%), while the commonest mutation was found at codon 526 in Northwest China (43.2%). The distribution of mutation frequency at different codons in different regions of China was statistically significant (p = 0.001) (Table 2).
3.5. Correlation of Genotypes and rpoB Mutation
Among the 394 RIF-resistant strains with spoligotyping results, 249 were Beijing genotype strains, the frequency of mutations in the rpoB gene hot spot region was 98.0%; while among the 145 RIF-resistant non-Beijing strains, the frequency of mutations in the rpoB gene hot spot region was 97.9%, and there was no significant difference in the occurrence of mutation in the rpoB gene hot spot region between the RIF-resistant Beijing and non-Beijing strains (p = 1.00). Also there was no significant difference in the occurrence of mutation in the rpoB gene hot spot region between the MDR Beijing and nonBeijing strains (97.6% vs. 97.6%, p = 1.00).
Of 394 RIF-resistant strains of known genotype, 386 (98.0%) contained mutations in the rpoB gene hot spot region, substitution of serine with leucine at codon 531 was the most frequent mutation found among both Beijing strains (47.1%; 115/244) and non-Beijing strains (48.6%; 69/142 ), the frequency of mutation among Beijing strains and non-Beijing strains was not statistically significant (p = 0.78), and there was no significant difference in the occurrence of mutations at codon 531 between the Beijing and non-Beijing isolates (50.4% vs.
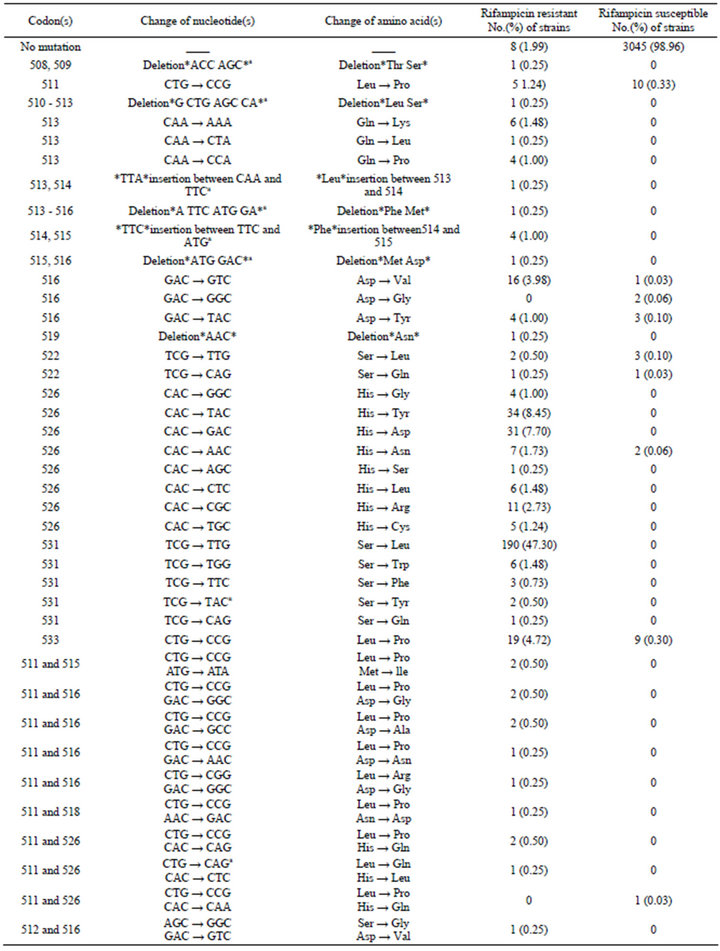
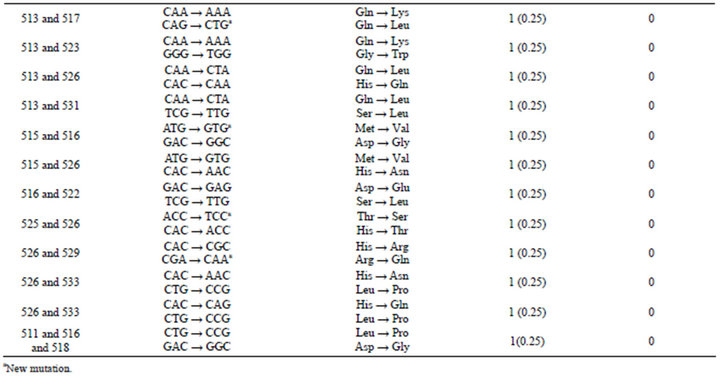
Table1.. Frequency of nucleotide and amino acid changes of different codons in the rpoB gene of M. tuberculosis isolated from China.
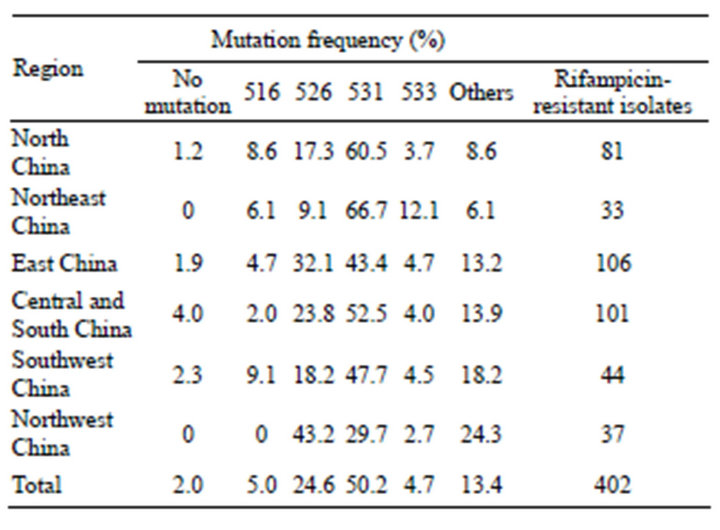
Table 2. The mutation frequency at different locus of the rpoB gene in rifampicin-resistant M. tuberculosis isolated from different areas of China.
51.4%, p = 0.85), also, there were no significant difference in the occurrence of mutation at codon 526, 516 between the Beijing and non-Beijing isolates (23% vs. 30.3%, 6.6% vs. 2.8%, p = 0.11, 0.11, respectively) (Figure 1).
4. DISCUSSION
Analysis of RIF-resistant clinical strains around the world found that about 95% - 98% of resistant strains harbor mutations in an 81 bp region of the rpoB gene [6-11]. Codons 531, 526, and 516 have been reported the most frequent mutations in the rpoB fragment worldwide [12-15]. In this study, DNA sequence analysis revealed
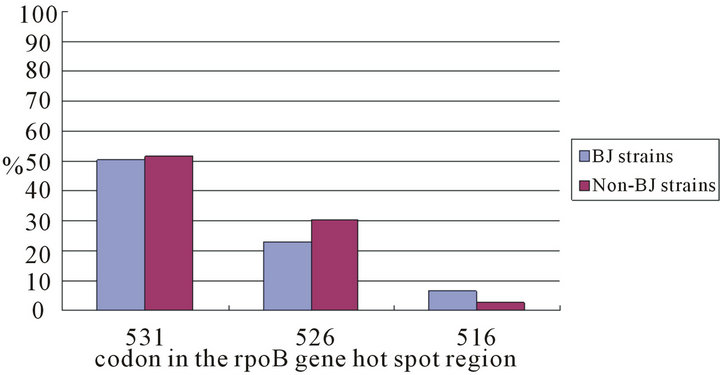
Figure 1. Frequencies of mutations in the rpoB hot spot region among rifampicin resistant Beijing and non Beijing strains.
that 98% RIF resistant strains showed rpoB gene mutation, 83.83% possessed mutations at codons 531, 526, and 516, the codons most frequently involved in mutations were codon 531 (50.50%), codon 526 (26.12%) and codon516 (7.21%). The frequency of mutations in the rpoB gene of RIF-resistant M. tuberculosis isolates varies between different geographical regions of China. The frequency of mutations occurs at the codons encoded Ser 531 (29% to 66%), His 526 (9% to 43%), and Asp 516 (0% to 9%). Previous study from Asia indicated that 47.51% of isolates carried the most common mutation Ser531Leu, while 8.45%, 7.70%, 4.23% of isolates had His526Tyr, His526Asp and Asp516Val mutation [16]. The frequency of Asp516Val mutation was 3.98% in our study, it was found that frequency of Asp516Val mutation occurring less frequently in isolates from China and Hong Kong [17,18].
In this study, some of the rpoB mutations, including Leu511Pro, Leu533Pro, His526Asn, Ser522Leu, Ser522 Gln, Asp516Tyr and Asp516Val were identified in 54 RIF-resistant and 29 sesceptible strains, these results indicate strains harboring these mutations are associated with discordant susceptibility test results, which was in agreement with previous study [19]. Previous study indicated some mutations in codons 511, 516, 518 and 522 could result in a lower resistance to RIF [20,21]. Other authors previously reported the identification of Asp516 Tyr substitutions of rpoB in M. tuberculosis resistance to a high level of RIF [22,23], low level of RIF [20] and in strains sensitive to RIF [24]. Asp516Tyr/Gly substitutions in rpoB were not sufficient to result in RIF resistance of M. tuberculosis [25]. Our results were concordance with previously report, the mutation Leu533Pro was known either to have no effect or to confer weak resistance [26].
Previous study finds that Beijing genotype strains are prevalent in China [27]. Our data suggest that Beijing strains account for about 63% of M. tuberculosis isolated from China, our results did not show any significant difference in the occurrence of mutation in the rpoB gene between the RIF-resistant Beijing and non-Beijing strains, the significant difference in the frequency of mutations at critical codons among Beijing/non-Beijing strains were not detected. These results are in accordance with previously report which tested M. tuberculosis isolated from eastern Asia countries and regions [28]. However, other authors who tested strains from Ukraine [29], Sichuan China [30] found significant difference in the frequency of mutations at codon 531 among Beijing/non Beijing strains. These contradictory reports indicate that the association of particular mutations and Beijing genotype strains is a regional phenomenon. Data from this study and previous reports demonstrate the ropB gene mutation profile in RIF-resistant M. tuberculosis isolated from China differs not only from other countries but also from different parts of China [31-33].
Today, many rpoB gene sequence-based new diagnostic methods such as the MTBDRplus assay (Hain Lifescience) and the Xpert MTB/RIF (Cepheid) have been developed to detect specific mutations associated with RIF drug resistance. The utility of these methods depends on the precise information about the role of any given mutation in RIF resistance, so phenotype and molecular analysis of regional isolates is important. Based on results of this study, the MTBDRplus assay and the Xpert MTB/RIF will detect about 98% of RIF-resistant strains and 99% of RIF-susceptible strains, and for 2% of RIF phenotype resistant strains that had no mutations in the hot spot region of the rpoB gene, these new diagnostic methods will report false negative (sensitive) results, for 1% of RIF-susceptible harboring mutations in the hot spot region, these new methods will give false positive (resistant) results, so if probes covering codons 511, 533, 516 and 522 report resistant results, more attention should be paid to these results, reference results of drug susceptibility test if necessary.
5. CONCLUSION
In summary, we use large set of strains with phenotypic diversity isolated from different areas of China to provide important information about the mutations conferring RIF resistance in M. tuberculosis isolates. Most RIF-resistant strains carried mutations in RRDR of rpoB gene, the mutations were most frequently found at critical codons. Most RIF-susceptible strains didn’t harbor mutations in RRDR of rpoB gene. rpoB gene sequencebased new diagnostic methods are promising for the rapid detection of RIF resistance.
6. ACKNOWLEDGEMENTS
This work was supported by a research grant from the National Science & Technology Major Project (2008ZX10003-009). The sponsor was not involved in study design or the preparation of the manuscript.
REFERENCES
- Zhao, Y., Xu, S., Wang, L., et al. (2012) National survey of drug-resistant tuberculosis in China. The New England Journal of Medicine, 366, 2161-2170. doi:10.1056/NEJMoa1108789
- World Health Organization (2003) Treatment of tuberculosis: Guidelines for national programmes. 3rd Edition, WHO, Geneva, 2003.
- Burman, W.J., Gallicano, K. and Peloquin, C. (2001) Comparative pharmacokinetics and pharmacodynamics of the rifamycin antibacterials. Clinical Pharmacokinetics, 40, 327-341. doi:10.2165/00003088-200140050-00002
- Somoskovi, A.L. Parsons, M. and Salfinger, M. (2001) The molecular basis of resistance to isoniazid, rifampin, and pyrazinamide in Mycobacterium tuberculosis. Respiratory Research, 2, 164-168. doi:10.1186/rr54
- Telenti, A., Imboden, P., Marchesi, F., et al. (1993) Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet, 341, 647-651.
- Bolotin, S., Alexander, D.C., Chedore, P., Drews, S.J. and Jamieson, F. (2009) Molecular characterization of drugresistant Mycobacterium tuberculosis isolates from Ontario, Canada. Journal of Antimicrobial Chemotherapy, 64, 263-266. doi:10.1093/jac/dkp183
- Campbell, P.J., Morlock, G.P., Sikes, R.D., et al. (2011) Molecular detection of mutations associated with firstand second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrobial Agents and Chemotherapy, 55, 2032-2041. doi:10.1128/AAC.01550-10
- Kapur, V., Li, L.L., Iordanescu, S., et al. (1994) Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase beta subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. Journal of Clinical Microbiology, 32, 1095-1098.
- Tan, Y., Hu, Z., Zhao, Y., et al. (2011) The beginning of the rpoB gene in addition to the RRDR might be needed for identifying RIF/Rfb cross resistance in multidrug-resistant Mycobacterium tuberculosis isolates from Southern China. Journal of Clinical Microbiology, 50, 81-85.
- Telenti, A., Imboden, P., Marchesi, F., Schmidheini, T. and Bodmer, T. (1993) Direct, automated detection of rifampin-resistant Mycobacterium tuberculosis by polymerase chain reaction and single-strand conformation polymorphism analysis. Antimicrobial Agents and Chemotherapy, 37, 2054-2058. doi:10.1128/AAC.37.10.2054
- Valim, A.R., Rosetti, M.L., Ribeiro, M.O. and Zaha, A. (2000) Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis isolates from Brazil. Journal of Clinical Microbiology, 38, 3119-3122.
- Ahmad, S., Mokaddas, E. and Fares, E. (2002) Characterization of rpoB mutations in rifampin-resistant clinical Mycobacterium tuberculosis isolates from Kuwait and Dubai. Diagnostic Microbiology & Infectious Disease, 44, 245-252. doi:10.1016/S0732-8893(02)00457-1
- Bakonyte, D., Baranauskaite, A., Cicenaite, J., Sosnovskaja, A. and Stakenas, P. (2005) Mutations in the rpoB gene of rifampin-resistant Mycobacterium tuberculosis clinical isolates from Lithuania. International Journal of Tuberculosis and Lung Disease, 9, 936-938.
- Cavusoglu, C., Hilmioglu, S., Guneri, S. and Bilgic, A. (2002) Characterization of rpoB mutations in rifampinresistant clinical isolates of Mycobacterium tuberculosis from Turkey by DNA sequencing and line probe assay. Journal of Clinical Microbiology, 40, 4435-4438. doi:10.1128/JCM.40.12.4435-4438.2002
- Mokrousov, I., Otten, T., Vyshnevskiy, B. and Narvskaya, O. (2003) Allelespecific rpoB PCR assays for detection of rifampin-resistant Mycobacterium tuberculosis in sputum smears. Antimicrobial Agents and Chemotherapy, 47, 2231-2235. doi:10.1128/AAC.47.7.2231-2235.2003
- Bodmer, T., Zurcher, G., Imboden, P. and Telenti, A. (1995) Mutation position and type of substitution in the beta-subunit of the RNA polymerase influence in vitro activity of rifampin-resistant Mycobacterium tuberculosis. Journal of Antimicrobial Chemotherapy, 35, 345-348. doi:10.1093/jac/35.2.345
- Yue, J., Shi, W., Xie, J., Li, Y., Zeng, E. and Wang, H. (2003) Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis isolates from China. Journal of Clinical Microbiology, 41, 2209-2212. doi:10.1128/JCM.41.5.2209-2212.2003
- Chan, R.C., Hui, M., Chan, E.W., et al. (2007) Genetic and phenotypic characterization of drug-resistant Mycobacterium tuberculosis isolates in Hong Kong. Journal of Antimicrobial Chemotherapy, 59, 866-873. doi:10.1093/jac/dkm054
- Van Deun, A., Barrera, L., Bastian, I., et al. (2009) Mycobacterium tuberculosis strains with highly discordant rifampin susceptibility test results. Journal of Clinical Microbiology, 47, 3501-3506. doi:10.1128/JCM.01209-09
- Williams, D.L., Spring, L., Collins, L., et al. (1998) Contribution of rpoB mutations to development of rifamycin cross-resistance in Mycobacterium tuberculosis. Antimicrobial Agents and Chemotherapy, 42, 1853-1857.
- Moghazeh, S.L., Pan, X., Arain, T., Stover, C.K., Musser J.M. and Kreiswirth, B.N. (1996) Comparative antimycobacterial activities of rifampin, rifapentine, and KRM- 1648 against a collection of rifampin-resistant Mycobacterium tuberculosis isolates with known rpoB mutations. Antimicrobial Agents and Chemotherapy, 40, 2655-2657.
- Yang, B., Koga, H., Ohno, H., et al. (1998) Detection between antimicrobacterial activities of rifampicin, rifabutin and KRM-1648 and rpoB mutations of Mycobacterium tuberculosis. Journal of Antimicrobial Chemotherapy, 42, 621-628. doi:10.1093/jac/42.5.621
- Hwang, H., Chang, C., Chang, L., Chang, S., Chang, Y. and Chen, Y. (2003) Characterisation of rifampicin-resistant Mycobacterium tuberculosis in Taiwan. Journal of Clinical Microbiology, 52, 239-245.
- Somoskovi, A., Dormandy, J., Mitsani, D., Rivenburg, J. and Salfinger, M. (2006) Use of smear-positive samples to assess the PCR-based genotype MTBDR assay for rapid, direct detection of the Mycobacterium tuberculosis complex as well as its resistance to isoniazid and rifampin. Journal of Clinical Microbiology, 44, 4459-4463.
- Zaczek, A., Brzostek, A., Augustynowicz-Kopec, E., Zwolska, Z. and Dziadek, J. (2009) Genetic evaluation of relationship between mutations in rpoB and resistance of Mycobacterium tuberculosis to rifampin. BMC Microbiology, 9, 10. doi:10.1186/1471-2180-9-10
- Ma, X., Wang, H.Y., Deng, Y.F., et al. (2006) rpoB gene mutations and molecular characterization of rifampin-resistant Mycobacterium tuberculosis isolates from Shandong Province, China. Journal of Clinical Microbiology, 44, 3409-3412. doi:10.1128/JCM.00515-06
- Pang, Y., Zhou, Y., Zhao, B., et al. (2012) Spoligotyping and drug resistance analysis of mycobacterium tuberculosis strains from national survey in China. PLoS One, 7, e32976. doi:10.1371/journal.pone.0032976
- Qian, L.S., Abe, C., Lin, T.-P., et al. (2002) rpoB genotypes of Mycobacterium tuberculosis Beijing family isolates from East Asian countries. Journal of Clinical Microbiology, 40, 1091-1094. doi:10.1128/JCM.40.3.1091-1094.2002
- Dymova, M.A., Liashenko, O.O., Poteiko, P.I., Krutko, V.S., Khrapov, E.A. and Filipenko, M.L. (2011) Genetic variation of Mycobacterium tuberculosis circulating in Kharkiv Oblast, Ukraine. BMC Infectious Diseases, 11, 77. doi:10.1186/1471-2334-11-77
- Tang, K., Sun, H., Zhao, Y., et al. (2012) Characterization of rifampin-resistant isolates of Mycobacterium tuberculosis from Sichuan in China. Tuberculosis (Edinburgh, Scotland), 93, 89-95.
- Caws, M., Duy, P.M., Tho, D.Q., Lan, N.T., Hoa, D.V. and Farrar, J. (2006) Mutations prevalent among rifampinand isoniazid-resistant Mycobacterium tuberculosis isolates from a hospital in Vietnam. Journal of Clinical Microbiology, 44, 2333-2337. doi:10.1128/JCM.00330-06
- Mikhailovich, V., Lapa, S., Gryadunov, D., et al. (2001) Identification of rifampin-resistant Mycobacterium tuberculosis strains by hybridization, PCR, and ligase detection reaction on oligonucleotide microchips. Journal of Clinical Microbiology, 39, 2531-2540. doi:10.1128/JCM.39.7.2531-2540.2001
- Fan, X.Y., Hu, Z.Y., Xu, F.H., Yan, Z.Q., Guo, S.Q. and Li, Z.M. (2003) Rapid detection of rpoB gene mutation in rifampin-resistant Mycobacterium tuberculosis isolates in Shanghai by using the amplification refractory mutation system. Journal of Clinical Microbiology, 41, 993-997. doi:10.1128/JCM.41.3.993-997.2003

