American Journal of Molecular Biology
Vol.06 No.04(2016), Article ID:71198,13 pages
10.4236/ajmb.2016.64013
Expression Analysis of WRKY Transcription Factor Genes in Response to Abiotic Stresses in Horsegram (Macrotyloma uniflorum (Lam.) Verdc.)
Kurnool Kiranmai1, Lokanadha Rao Gunupuru2, Ambekar Nareshkumar3, Vennapusa Amaranatha Reddy1, Uppala Lokesh1, Merum Pandurangaiah4, Boya Venkatesh1, Tanguturi Venkata Kirankumar1, Chinta Sudhakar1*
1Department of Botany, Sri Krishnadevaraya University, Anantapur, India
2Molecular Plant-Microbe Interactions Laboratory, University College Dublin, Dublin, Ireland
3Department of Crop Physiology, GKVK, Bangalore, India
4Shenzhen Research Institute, The Chinese University of Hong Kong, Shenzhen, China

Copyright © 2016 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: July 20, 2016; Accepted: October 10, 2016; Published: October 13, 2016
ABSTRACT
Drought and salt stress are two major environmental constraints that limit the productivity of agriculture crops worldwide. WRKY transcription factors are the plant- specific transcription factors that regulate several developmental events and stress responses in plants. The WRKY domain is defined by a 60-amino acid conserved sequence named WRKYGQK at N-terminal and a Zinc Finger-like motif at the C- terminal. WRKY genes are known to respond several stresses which may act as negative or positive regulators. The function of most of the WRKY transcription factors from non-model plants remains poorly understood. This investigation shows the expression levels of eight WRKY transcription factor genes from horsegram plant under drought and salt stress conditions. The increase in mRNA transcript levels of WRKY transcription factor genes was found to be high in drought stressed plants compared to salt-stressed plants. The levels of MDA which indicates the lipid peroxidation were less in drought stress. More ROS is produced in salt stress conditions compared to drought. The results show that the expression of WRKY transcription factors in drought stress conditions is reducing the adverse effect of stress on plants. These results also suggest that, during abiotic stress conditions such as drought and salt stress, WRKY transcription factors are regulated at the transcription level.
Keywords:
Drought, Salt, Lipid Peroxidation, Plant Abiotic Stress, qRT-PCR, Reactive Oxygen Species, WRKY

1. Introduction
Plants in their natural environmental conditions are always subjected to various biotic and abiotic stresses. Abiotic stresses are the primary reason behind the decline of crop quality and yield worldwide [1] . Drought and salt stress are two major abiotic stresses that limit crop productivity. Excessive amounts of salts in the agriculture soil cause both osmotic and ionic stresses [2] . The rapid spread of drought and soil salinity in many regions worldwide may cause serious salt contamination in more than 50% of all arable lands by the year 2050 [3] . Transcription factors (TFs) play a vital role in stress response by regulating their expression in target gene through interaction with specific cis-acting elements present in the promoter of the gene [4] [5] . WRKY TFs are the largest superfamily of TFs specific to plants. They are classified into three groups based on the number of WRKY domains and nature of their zinc-finger motif. Group I contains two WRKY conserved domains and a classical Zinc finger motif. Group II contains single WRKY domain and a classical zinc finger motif. Group II is divided into five or more subgroups based on short conserved structural WRKYGQK motif. Group III protein of WRKY superfamily contains a single WRKY domain and a modified zinc finger motif C2-CH rather than classical C2-H2. Group II WRKY TFs containing WRKYGQK heptapeptide with Zinc finger CX4-5CX22-23HHX1H is the largest group in most of the plants [6] [7] .
A wide range of WRKY TFs has been identified from different plants 72 WRKYs from Arabidopsis and 96 from rice [7] and 52 WRKYs in papaya [8] have been identified. Most of the WRKYs are found to play a role in biotic stress response, some in abiotic stress and few WRKYs are known for their role in both biotic and abiotic stress response. During normal growth conditions, WRKY TFs regulate plant processes like senescence, trichome development [9] biosynthesis of secondary metabolites [6] and dormancy [10] . Many research groups have investigated the role of WRKY TFs in biotic and abiotic stress in various plants. Overexpression of WRKY33 a WRKY TF family protein, providing resistance to necrotrophic fungi species such as Botrytis cinerea and Alterneria brassicicola has been reported in Arabidopsis [11] . WRKY TFs like HvWRKY33 involving in cold and drought tolerance in barley [12] , oxidative stress response WRKY TF like FcWRKY40 from Fortunella crassifolia [13] , and ZmWRKY33 conferring tolerance to salt stress were reported in Arabidopsis [14] . 46 different WRKY TFs were identified from Brassica napus expressing during infection with fungal pathogens Sclerotinia sclerotiorum and Alternaria brassicae, and ABA, JA, SA and ET treatments [7] . However, the function of each WRKY in various stresses is poorly understood. It is important to get further insight and investigate the response of WRKY TFs under abiotic stress conditions.
Membrane damage is taken as a parameter to determine the lipid destruction of the membrane under various stress conditions. Lipids peroxidation is considered as the most desecrate process known to occur in every living organism. During lipid peroxidation, small hydrocarbon fragments namely Malondialdehyde (MDA) is formed from
polyunsaturated precursors [15] . Lipid peroxidation takes place when Reactive oxygen species (ROS) levels are reached above-threshold, directly affecting normal cellular functioning and also aggravating the oxidative stress through the production of lipid-derived radicals [16] . Level of ROS is controlled by plant cells through detoxification of excess ROS during stress. To help in the detoxification of excess ROS, plants have evolved strong antioxidant defence mechanism [17] .
Horsegram belongs to the family Fabaceae and is comparatively a hardy drought tolerant crop which grows well under adverse conditions. The crop accounts for approximately 5% - 10% of the India’s pulses, with annual production of about 0.65 million tonnes. It is considerably used as dry fodder, cattle feed, cover crop and for water conservation in the semi-arid region. Horsegram is widely grown in the semi-arid regions of India and generally considered as protein rich poor man’s crop. It grows well under dry conditions with the need of low rainfall and marginal soil fertility. Depending on its well adaptive growth conditions and life cycle, there is a high possibility that horsegram plant contains a variable number of genes that can be used to provide stress tolerance. Due to the higher similarity in genome among pulse crops, comprehension of the underlying genetic mechanism of various stress related genes from this plant will be of significant advantage to transform these genes to other legume crops.
The present investigation had made an attempt to identify, the abiotic stress responsive role of horsegram WRKY TFs by qRT-PCR expression analysis under drought and salinity stresses. These experimental results, making feasible to use horsegram WRKY TF genes to develop abiotic stress tolerant crop plants in near future.
2. Materials and Methods
2.1. Plant Material and Stress Treatments
Horsegram seeds were sown in pots containing mixed soil (soil: vermiculite in 3:1 ratio) and allowed to grow in a greenhouse under natural photoperiod and temperature conditions for 19 days. To impose salt stress 19 days, old plants were irrigated with 1% and 2 % NaCl solution. The electrical conductivity of the soil saturation extract was 4.0 mScm−2 and care was taken to avoid drainage of solution during the treatment by giving the water slightly less than field capacity; stress was maintained for four days. The electrical conductivity was monitored and adjusted using conductivity bridge (Elico, Hyderabad, India) on alternate days. For drought stress, the plants were withheld from irrigation until to reach 50% and 25% soil moisture level (SML) and maintained at the same level for three days by measuring SML regularly by following standardised gravimetric method. The samples from stressed and non-stressed plants were collected and flash frozen in liquid nitrogen. Each experiment was repeated at least three times.
2.2. Determination of Growth under Stress
After the stress treatments, plants were carefully uprooted from the soil, to avoid any breakage of lateral roots and washed thoroughly under running tap water. Plant growth was determined by measuring the root length, shoot length, and plant fresh weight.
2.3. Histochemical Localization of ROS
ROS are produced as a result of stress which leads to oxidative damage. The levels of ROS species like 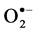 and H2O2, produced during the stress treatments were detected using Diaminobenzidine (DAB) and Nitoblurtetrazolium chloride (NBT) [18] . For detection of
and H2O2, produced during the stress treatments were detected using Diaminobenzidine (DAB) and Nitoblurtetrazolium chloride (NBT) [18] . For detection of  the leaf material was immersed in a solution of 0.1% NBT in 50 mM phosphate buffer (pH 7.8) at room temperature. The immersed leaves were illuminated with two 20-W fluorescent tubes for 2 - 4 h until the dark blue spots appear. For localization of H2O2, excised leaves were immersed in a 0.1% DAB solution in 50 mM phosphate buffer (pH 3.8) and incubated at room temperature for 8 h until brown spots appear. The leaves were then bleached with 50% ethanol to visualize the spots.
the leaf material was immersed in a solution of 0.1% NBT in 50 mM phosphate buffer (pH 7.8) at room temperature. The immersed leaves were illuminated with two 20-W fluorescent tubes for 2 - 4 h until the dark blue spots appear. For localization of H2O2, excised leaves were immersed in a 0.1% DAB solution in 50 mM phosphate buffer (pH 3.8) and incubated at room temperature for 8 h until brown spots appear. The leaves were then bleached with 50% ethanol to visualize the spots.
2.4. Determination of MDA Content
The levels of lipid peroxidation were measured [19] by estimating the MDA content, which is a secondary product of polyunsaturated fatty acids and 2-thiobarbituric acid reactive substances. The MDA content was estimated using 2-thibarbutiric acid at 400, 532 and 600 nm [19] . Five hundred milligrams of leaf tissue was used for the extraction of MDA. One millilitre of the supernatant was mixed with 1 ml of 20% TCA containing 0.65% thiobarbituric acid in a clean glass tube, in another tube 1 ml of supernatant was treated with 1 ml of 20% TCA alone and mixed thoroughly. The mixtures were heated at 95˚C for 30 min and absorbance was recorded at 532 nm (MDA), 440 nm (carbohydrates) and 600 nm (phenylpropanoid pigments). The MDA content was determined by its molar extinction coefficient (155 mM−1∙cm−1) using the formula.
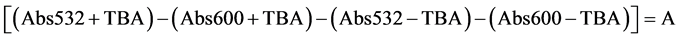
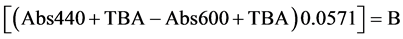
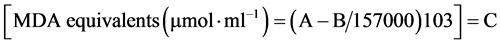

2.5. Isolation of RNA and cDNA Synthesis
Total RNA was isolated from the frozen leaf material using TRIzol reagent. RNA concentration was accurately quantified by the UV-spectrophotometric measurement and 3 µg of total RNA was separated on 0.8% agarose gel containing formaldehyde to check RNA integrity. DNA contamination was removed with TURBO DNA-free kit according to the instructions in the manual. Two hundred nanograms of RNA was used to construct cDNA by Thermo Scientific RevertAid Reverse Transcriptase enzyme and oligodT primers at 42˚C. Obtained cDNA was subsequently used as a template for qRT- PCR analysis [20] .
2.6. Quantitative Real-Time PCR Analysis
Eight WRKY TF genes were selected to study the expression levels under drought and salt stress conditions by the qRT-PCR analysis. For qRT-PCR assay gene, WRKY specific expression primers were designed using the Primer Express 5.0 software at annealing temperature of 56˚C - 60˚C. Primer sequences used for the amplification were listed in Table 1. qRT-PCR was performed using Applied biosystem power SYBR-green mix. The appearance of PCR products was monitored by detecting the increase in fluorescence caused by binding of SYBR green dye. The length of amplification fragment was about 150 bp. Each reaction was repeated at least three times. The expression analysis was conducted in an Applied Biosystem StepOne Realtime PCR-System thermal cycling block. Conditions maintained are, 1min at 95˚C; 1 min at 57˚C and 1 min at 72˚C for 40 cycles, followed by melting curves analysis at 95˚C for 1 min. Expression of MuActin gene was used as internal control for normalization. Relative expression of mRNA was calculated using 2−ΔΔct formula [21] .
2.7. Data Analysis
All physiological and biochemical data were analysed using SPSS version 16.0. Data presented were calculated using One-way ANOVA and statistical analysis was carried out by using Post hoc multiple comparisons Duncan’s test at a significance level of P < 0.05.
Table 1. Primers sequences used for the amplification of gene in qRT-PCR analysis.
3. Results
3.1. Effect of Stress on Plant Growth
Effect of stress is usually perceived as a decrease in photosynthesis and growth of the plant. Growth parameters were calculated for uprooted plants. In general, the overall growth of plant was decreased under both stress conditions. However, the inhibitory effect of salt stress was more on the growth of horsegram when compared to drought. Further, more reduction in the growth of the plant was observed with increased levels of stress. The decrease in root growth was up to 44% in drought and 60% in salt stress. Similarly, the shoot length was decreased up to 22% in drought and 48% under salt stress conditions (Figure 1). There was 37% reduction in the plant total biomass in drought stress conditions and up to 78% under salt stress conditions (Figure 2). The depletion of chlorophyll and leaf drying was also observed at 2% NaCl stress which leads to plant dying. Dying of leaves and eventually entire plant due to high salt concentration in plants was also reported [22] .
Figure 1. Varying root length and shoot length of horsegram plant under control, drought and salt stress conditions (±SD). And letters shown above the bars are significantly different at P < 0.05 (DMR).
Figure 2. Fresh biomass of horsegram plants under control, drought, and salt stress conditions (±SD), and letters shown above the bars are significantly different at P < 0.05 (DMR).
3.2. Effect of Drought and Salt Stress on MDA Levels
MDA, the product of lipid peroxidation is often used as an indicator to check the extent of oxidative stress in plant [23] . The formation of ROS increases the levels of cellular lipid peroxidation, such increase in the production of ROS and peroxidation levels differ from species and severity of stress [24] . Total MDA content was estimated from the leaf tissue of control and stressed plants. The results suggest that the MDA content was significantly increased under both the stresses and an increase in the MDA content was observed proportional to the intensity of stress applied. However, the level of MDA is more in salt stress than in drought stress conditions (Figure 3). An increase in the MDA concentration under abiotic stress conditions like Pb stress was previously reported in groundnut plants by Nareshkumar et al. [25] .
3.3. Histochemical Detection of  and H2O2 in Leaves
and H2O2 in Leaves
In situ histochemical staining assay was employed to detect the accumulation of 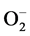 and H2O2, the two important ROS molecules produced during stress. Under control conditions there was no accumulation of free radicals,
and H2O2, the two important ROS molecules produced during stress. Under control conditions there was no accumulation of free radicals, 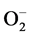 and H2O2 was observed. However, remarkable differences were observed in the accumulation of ROS under stress conditions. Blue spots are the indicator of
and H2O2 was observed. However, remarkable differences were observed in the accumulation of ROS under stress conditions. Blue spots are the indicator of 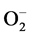 and brown spots indicate the H2O2 accumulation. The results showed that the accumulation of ROS was more under salt stress conditions compared to drought (Figure 4).
and brown spots indicate the H2O2 accumulation. The results showed that the accumulation of ROS was more under salt stress conditions compared to drought (Figure 4).
3.4. Expression Analysis of WRKY TFs under Drought and Salt Stresses
qRT-PCR analysis was conducted to check the expression levels of different WRKY TF genes under drought and salt stress conditions. The results showed the different levels of regulation in the WRKY genes under different abiotic stresses conditions. In drought stress conditions, the highest level of expression was observed in WRKY3 and WRKY 75 was found to be negatively regulated. There was an increase in the transcription levels of different genes from 1.2 fold in WRKY33 to 12 folds in WRKY3 under drought
Figure 3. MDA content in leaf samples of horsegram under control, drought and salt stress conditions. The data represented is the mean of three different experiments ±SD, and the letters shown above the bars are significantly different at P < 0.05 (DMR).

Figure 4. In situ histochemical detection of 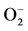 and H2O2 under control, drought and salt stress conditions. (a) Brown spots indicate the H2O2 accumulation which is stained with DAB. (b) Blue colour spots indicate
and H2O2 under control, drought and salt stress conditions. (a) Brown spots indicate the H2O2 accumulation which is stained with DAB. (b) Blue colour spots indicate  accumulation stained with NBT. The intensity of colour spots is seen under salt stress conditions. Control plants did not show any colour due to the absence of ROS.
accumulation stained with NBT. The intensity of colour spots is seen under salt stress conditions. Control plants did not show any colour due to the absence of ROS.
stress conditions at 25% SML. In salt stress, at 2% NaCl, the expression levels of genes were increased up to 6.1 folds in WRKY33. However, the negative regulation was also recorded in WRKY40 and WRKY53 genes whose transcription levels were downregulated by 0.63 fold in WRKY53 and 3.8 folds in WRKY40 genes (Figure 5).
4. Discussion
Abiotic stresses are the major limiting factors that reduce the productivity of crop worldwide. In the present investigation, we are delineated representing the role of WRKY TF gene from horsegram under drought and salt stress conditions. WRKY TFs are the largest superfamily TF that is specific to plants. WRKY recognize and bind to specific TTGAC (C/T) W-box elements found in promoters of a large number of plant defence related genes [6] [27] [28] . The first WRKY cDNAs were cloned from sweet potato, wild oat, parsley and Arabidopsis, based on the ability to bind specifically to the DNA sequence motif TTGACCT, which is known as the W box [29] [30] [31] . Current information available is suggesting that a large number of WRKY TFs plays a pivotal role in pathogen-induced defence mechanism [32] - [34] . However, there are fewer reports on the involvement of WRKY TFs in drought and salt stress response. In earlier studies expression of six WRKY TFs under drought stress have been reported [8] in Carica papaya. AtWRKY57 conferring drought tolerance by elevated ABA levels was reported [35] . A group II WRKY TF named WRKY58 was characterized from maize enhancing the tolerance towards drought and salt stress [36] . There are reports that few WRKY genes can be induced by both biotic and abiotic stress conditions. Zheng et al. [11] have reported that AtWRKY33 is a multifunctional gene that involves both in biotic and abiotic stresses.
In this report, we studied eight WRKY TF genes from horsegram, under drought and salt stress. WRKY3 gene has shown the highest expression level with 12 folds increase in drought stress among eight genes selected. Strong VpWRKY3 transcription levels are
Figure 5. qRT-PCR analysis of WRKY TFs in horsegram plants after imposing drought and salt stress. The relative expression of WRKY TF was more under drought stress conditions.
detected under abiotic stress conditions like high and low temperature, NaCl and drought in transgenic tobacco lines [26] . So we speculate that WRKY3 genes might be involved in stress response and tolerance under drought. All eight WRKY TFs studied are up-regulated in drought stress except the WRKY75 whose mRNA transcript levels are found to be down-regulated by one fold. In salt stress conditions the expression of WRKY33 is more with 6.1 folds increase in the mRNA transcript levels and two genes WRKY 40 and WRKY 53 were found to be down-regulated. In earlier reports, the overexpression of DgWRKY1 from chrysanthemum has shown better tolerance to salt stress in transgenic tobacco plants [37] .
The stress leads to trigger some of the key enzymes of antioxidant defence system. To resist oxidative damage in plants the antioxidant enzymes and certain metabolites; play a vital role leading to adaptation and the ultimate survival under stress [38] . In the present study, we speculate that the expression of WRKY TFs in turn regulating the expression of other stress related antioxidative genes under drought stress conditions. The expression of antioxidative enzymes enhances the scavenging activity in plants and reduces the ROS produced under stress. The low levels of MDA in drought stress conditions also represent the less lipid peroxidation and membrane damage. The overexpression of TaWRKY10 has been reported earlier to enhance the expression level of stress related antioxidant enzyme like SOD, CAT, and GPX in transgenic tobacco plant. The expression of FcWRKY40 altering the expression levels of two ROS scavenging genes like SOD and POD were reported previously [13] . Our results suggest that the up- regulation of WRKY TF genes contributes to plant defence under drought and salt stress conditions. But the genes we studied are upregulated well in drought stress conditions than in salt stress conditions.
5. Conclusion
Based on the result obtained from this study we speculate that horsegram WRKY TF genes are promising to stress responsible gene for modulation and enhancement of abiotic stress tolerance in sensitive crops. In this study, an attempt was made to identify the abiotic stress responsive WRKY TF genes from horsegram plant under various abiotic stress conditions. Furthermore, studies will be carried out to develop abiotic stress tolerant transgenic crops by genetic manipulation of WRKY TF genes and assess the function of these genes in abiotic stress tolerance.
Acknowledgements
Financial support from Council of Scientific and Industrial research (CSIR: 38(1305)/ 11/EMR-11), Department of Science and Technology (DST: SR/SO/PS/0001/2011) and University Grant Commission, New Delhi are greatly acknowledged.
Cite this paper
Kiranmai, K., Gunupuru, L.R., Nareshkumar, A., Reddy, V.A., Lokesh, U., Pandurangaiah, M., Venkatesh, B., Kirankumar, T.V. and Sudhakar, C. (2016) Expression Analysis of WRKY Transcription Factor Genes in Response to Abiotic Stresses in Horsegram (Macrotyloma uniflorum (Lam.) Verdc.). American Journal of Molecular Biology, 6, 125-137. http://dx.doi.org/10.4236/ajmb.2016.64013
References
- 1. Wang, Y. and Frei, M. (2011) Stressed Food—The Impact of Abiotic Environmental Stresses on Crop Quality. Agriculture, Ecosystems & Environment, 141, 271-286.
http://dx.doi.org/10.1016/j.agee.2011.03.017 - 2. Khelil, A., Menu, T. and Ricard, B. (2007) Adaptive Response to Salt Involving Carbohydrate Metabolism in Leaves of a Salt-Sensitive Tomato Cultivar. Plant Physiology and Biochemistry, 45, 551-559.
http://dx.doi.org/10.1016/j.plaphy.2007.05.003 - 3. Wang, W., Vinocur, B. and Altman, A. (2003) Plant Responses to Drought, Salinity and Extreme Temperatures: Towards Genetic Engineering for Stress Tolerance. Planta, 218, 1-14. http://dx.doi.org/10.1007/s00425-003-1105-5
- 4. Tran, L.S.P., Nakashima, K., Sakuma, Y., Simpson, S.D., Fujita, Y., Maruyama, K., Fujita, M., Seki, M., Shinozaki, K. and Yamaguchi-Shinozaki, K. (2004) Isolation and Functional Analysis of Arabidopsis Stress-Inducible NAC Transcription Factors That Bind to a Drought Responsive Cis-Element in the Early Responsive to Dehydration Stress 1 Promoter. Plant Cell, 16, 2481-2498.
http://dx.doi.org/10.1105/tpc.104.022699 - 5. Shinozaki, K., Yamaguchi-Shinozaki, K. and Seki, M. (2003) Regulatory Network of Gene Expression in the Drought and Cold Stress Responses. Current Opinion in Plant Biology, 6, 410-417.
http://dx.doi.org/10.1016/S1369-5266(03)00092-X - 6. Eulgem, T., Rushton, P.J., Robatzek, S. and Somssich, I.E. (2000) The WRKY Superfamily of Plant Transcription Factors. Trends in Plant Science, 5, 199-206.
http://dx.doi.org/10.1016/S1360-1385(00)01600-9 - 7. Yang, B., Jiang, Y.Q., Rahman, M.H., Deyholos, M.K. and Kav, N.N.V. (2009) Identification and Expression Analysis of WRKY Transcription Factor Genes in Canola (Brassica napus L.) in Response to Fungal Pathogens and Hormone Treatments. BMC Plant Biology, 9, 68.
http://dx.doi.org/10.1186/1471-2229-9-68 - 8. Pan, L.J. and Jiang, L. (2014) Identification and Expression of the WRKY Transcription Factors of Carica papaya in Response to Abiotic and Biotic Stresses. Molecular Biology Reports, 41, 1215-1225.
http://dx.doi.org/10.1007/s11033-013-2966-8 - 9. Johnson, C.S., Kolevski, B. and Smyth, D.R. (2002) TRANSPARENT TESTA GLABRA2, a Trichome and Seed Coat Development Gene of Arabidopsis, Encodes a WRKY Transcription Factor. The Plant Cell, 14, 1359-1375.
http://dx.doi.org/10.1105/tpc.001404 - 10. Pnueli, L., Hallak-Herr, E., Rozenberg, M., Cohen, M., Goloubinoff, P., Kaplan, A. and Mittler, R. (2002) Molecular and Biochemical Mechanisms Associated with Dormancy and Drought Tolerance in the Desert Legume Retama raetam. Plant Journal, 31, 319-330.
http://dx.doi.org/10.1046/j.1365-313X.2002.01364.x - 11. Zheng, Z., Qamar, S.A., Chen, Z. and Mengiste, T. (2006) Arabidopsis WRKY33 Transcription Factor Is Required for Resistance to Necrotrophic Fungal Pathogens. Plant Journal, 48, 592-605.
http://dx.doi.org/10.1111/j.1365-313X.2006.02901.x - 12. Mare, C., Mazzucotelli, E., Crosatti, C., Francia, E., Stanca, A.M. and Cattivelli, L. (2004) Hv-WRKY38: A New Transcription Factor Involved in Cold- and Drought-Response in Barley. Plant Molecular Biology, 55, 399-416.
http://dx.doi.org/10.1007/s11103-004-0906-7 - 13. Gong, X., Hu, J. and Liu, J. (2014) Cloning and Characterization of FcWRKY40, A WRKY Transcription Factor from Fortunella crassifolia Linked to Oxidative Stress Tolerance. Plant Cell Tissue and Organ Culture, 119, 197-210.
http://dx.doi.org/10.1007/s11240-014-0526-0 - 14. Li, H., Gao, Y., Xu, H., Dai, H., Deng, D. and Chen, J. (2013) ZmWRKY33, a WRKY Maize Transcription Factor Conferring Enhanced Salt Stress Tolerances in Arabidopsis. Plant Growth Regulation, 70, 207-216.
http://dx.doi.org/10.1007/s10725-013-9792-9 - 15. Garg, N. and Manchanda, G. (2009) ROS Generation in Plants: Boon or Bane? Plant Biosystems, 143, 8-96.
http://dx.doi.org/10.1080/11263500802633626 - 16. Montillet, J.L., Chamnongpol, S., Rusterucci, C., Dat, J., van de Cotte, B., Agnel, J.P., Battesti, C., Inze, D., Van Breusegem, F. and Triantaphylides, C. (2005) Fatty Acid Hydroperoxides and H2O2 in the Execution of Hypersensitive Cell Death in Tobacco Leaves. Plant Physiology, 138, 1516-1526.
http://dx.doi.org/10.1104/pp.105.059907 - 17. Yannarelli, G.G., Noriega, G., Batlle, A. and Tomaro, M. (2006) Heme Oxygenase Up-Regulation in Ultraviolet-B Irradiated Soybean Plants Involves Reactive Oxygen Species. Planta, 224, 1154-1162.
http://dx.doi.org/10.1007/s00425-006-0297-x - 18. Romero-Puertas, M.C., Rodriguez-Serrano, M., Corpas, F.J., Gomez, M., Del Rio, L.A. and Sandalio, L.M. (2004) Chadmium-Induced Subcellular Accumulation of O2- and H2O2 in Pea Leaves. Plant Cell & Environment, 27, 1122-1134.
http://dx.doi.org/10.1111/j.1365-3040.2004.01217.x - 19. Hodges, D.M., Delong, J.M., Forney, C.F. and Prange, R.K. (1999) Improving the Thiobarbitric Acid Reactive Substance Assay for Estimating Lipid Peroxidation in Plant Tissues Containing Anthocyanin and Other Interfering Compounds. Planta, 207, 604-611.
http://dx.doi.org/10.1007/s004250050524 - 20. Hu, W., Yuan, Q., Wang, Y., Cai, R., Deng, X., Wang, J., Zhou, S., Chen, M., Chen, L., Huang, C., Ma, Z., Yang, G. and He, G. (2012) Over-expression of a Wheat Aquaporin Gene, TaAQP8, Enhances Salt Stress Tolerance in Transgenic Tobacco. Plant and Cell Physiology, 53, 2127-2141.
http://dx.doi.org/10.1093/pcp/pcs154 - 21. Livak, K.J. and Schmittgen, T.D. (2001) Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods, 25, 402-408.
http://dx.doi.org/10.1006/meth.2001.1262 - 22. Munns, R. (1993) physiological Process Limiting Plant Growth Saline Soils Some Dogmas and Hypotheses. Plant Cell & Environment, 16, 15-24.
http://dx.doi.org/10.1111/j.1365-3040.1993.tb00840.x - 23. Chen, J., Shiyab, S., Han, F.X., Monts, D.L., Waggoner, A.W. and Su, Z.Y. (2009) Bioaccumulation and Physiological Effects of Mercury in Pteris vittata and Nephrolepis exaltata. Ecotoxicology, 18, 110-121.
http://dx.doi.org/10.1007/s10646-008-0264-3 - 24. Wang, X. and Han, J. (2009) Changes of Proline Content, Activity, and Active Isoforms of Antioxidative Enzymes in Two Alfalfa Cultivars under Salt Stress. Agriculture Sciences in China, 8, 431-440.
http://dx.doi.org/10.1016/S1671-2927(08)60229-1 - 25. Nareshkumar, A., Nagamallaiah, G.V., Pandurangaiah, M., Kiranmai, K., Amaranathareddy, V., Lokesh, U., Venkatesh, B. and Sudhakar, C. (2015) Pb-Stress Induced Oxidative Stress Caused Alterations in Antioxidant Efficacy in Two Groundnut (Arachis hypogaea L.) Cultivars. Agricultural Sciences, 6, 1283-1297.
http://dx.doi.org/10.4236/as.2015.610123 - 26. Zhu, Z., Shi, J., Cao, J., He, M. and Wang, Y. (2012) VpWRKY3, a Biotic and Abiotic Stress-Related Transcription Factor from Chinese Wild Vitis pseudoreticulata. Plant Cell Reports, 11, 2109-2120.
http://dx.doi.org/10.1007/s00299-012-1321-1 - 27. Jing, S.J., Zhou, X., Song, Y. and Yu, D.Q. (2009) Heterogonous Expression of OsWRKY23 Gene Enhances Pathogen Defence and Dark Induced Leaf Senescence in Arabidopsis. Plant Growth Regulation, 58, 181-190.
http://dx.doi.org/10.1007/s10725-009-9366-z - 28. Yang, P., Chen, C., Wang, Z., Fan, B. and Chen, Z. (1999) A Pathogen- and Salicylic Acid-Induced WRKY DNA-Binding Activity Recognizes the Elicitor Response Element of the Tobacco Class I Chitinase Gene Promoter. Plant Journal, 18, 141-149.
http://dx.doi.org/10.1046/j.1365-313X.1999.00437.x - 29. Ishiguro, S. and Nakamura, K. (1994) Characterization of a cDNA Encoding a Novel DNA-Binding Protein, SPF1, That Recognizes SP8 Sequences in the 5’ Upstream Regions of Genes Coding for Sporamin and β-Amylase from Sweet Potato. Molecular and General Genetics, 244, 563-571.
http://dx.doi.org/10.1007/BF00282746 - 30. Rushton, P.J., Torres, J.T., Parniske, M., Wernert, P., Hahlbrock, K. and Somssich, I.E. (1996) Interaction of Elicitor-Induced DNA-Binding Proteins with Elicitor Response Elements in the Promoters of Parsley PR1 Genes. EMBO Journal, 15, 5690-5700.
- 31. De Pater, S., Greco, V., Pham, K., Memelink, J. and Kijne, J. (1996) Characterization of a Zinc-Dependent Transcriptional Activator from Arabidopsis. Nucleic Acids Research, 24, 4624-4631.
http://dx.doi.org/10.1093/nar/24.23.4624 - 32. Oh, S.K., Baek, K.H., Park, J.M., Yi, S.Y., Yu, S.H., Kamoun, S. and Choi, D. (2008) Capsicum Annuum WRKY protein CaWRKY1 Is a Negative Regulator of Pathogen Defence. New Phytologist, 177, 977-989.
http://dx.doi.org/10.1111/j.1469-8137.2007.02310.x - 33. Cai, M., Qiu, D., Yuan, T., Ding, X., Li, H., Duan, L., Xu, C., Li, X. and Wang, S. (2008) Identification of Novel Pathogen-Responsive Cis-Elements and Their Binding Proteins in the Promoter of OsWRKY13, a Gene Regulating Rice Disease Resistance. Plant Cell and Environment, 31, 86-96.
- 34. Yu, F.F., Huaxia, Y.F., Lu, W.J., Wu, C. and Guo, X.Q. (2012) GhWRKY15, a Member of the WRKY Transcription Factor Family Identified from Cotton (Gossypium hirsutum L.), Is Involved in Disease Resistance and Plant Development. BMC Plant Biology, 12, 144.
http://dx.doi.org/10.1186/1471-2229-12-144 - 35. Jiang, Y.J., Liang, G. and Yu, D.Q. (2012) Activated Expression of WRKY57 Confers Drought Tolerance in Arabidopsis. Molecular Plant, 5, 1375-1388.
http://dx.doi.org/10.1093/mp/sss080 - 36. Cai, R., Zhao, Y., Wang, Y., Lin, Y., Peng, X., Li, Q., Chang, Y., Jiang, H., Xiang, Y. and Cheng, B. (2014) Overexpression of a Maize WRKY58 Gene Enhances Drought and Salt Tolerance in Transgenic Rice. Plant Cell Tissue and Organ Culture, 119, 565-577.
http://dx.doi.org/10.1007/s11240-014-0556-7 - 37. Liu, Q., Xu, K., Pan, Y., Jiang, B., Liu, G., Jia, Y. and Zhang, H. (2014) Functional Analysis of a Novel Chrysanthemum WRKY Transcription Factor Gene Involved in Salt Tolerance. Plant Molecular Biology Reports, 32, 282-289.
http://dx.doi.org/10.1007/s11105-013-0639-3 - 38. Zhang, F.Q., Wang, Y.S., Lou, Z.P. and Dong, J.D. (2007) Effect of Heavy Metal Stress on Antioxidative Enzymes and Lipid Peroxidation in Leaves and Roots of Two Mangrove Plant Seedlings (Kandelia candel and Bruguiera gymnorrhiza). Chemosphere, 67, 44-50.
http://dx.doi.org/10.1016/j.chemosphere.2006.10.007






