American Journal of Molecular Biology
Vol.04 No.03(2014), Article ID:47866,15 pages
10.4236/ajmb.2014.43011
Metallo-β-Lactamases: A Major Threat to Human Health
Emer K. Phelan1,2*, Manfredi Miraula1,2*, Christopher Selleck2, David L. Ollis3, Gerhard Schenk2, Nataša Mitić1#
1Department of Chemistry, National University of Ireland-Maynooth, Maynooth, Ireland
2School of Chemistry and Molecular Biosciences, The University of Queensland, Brisbane, Australia
3Research School of Chemistry, Australian National University, Canberra, Australia
Email: #natasa.mitic@nuim.ie
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 15 April 2014; revised 14 May 2014; accepted 13 June 2014
ABSTRACT
Antibiotic resistance is one of the most significant challenges facing global healthcare. Since the 1940s, antibiotics have been used to fight infections, initially with penicillin and subsequently with various derivatives including cephalosporins, carbapenams and monobactams. A common characteristic of these antibiotics is the four-membered β-lactam ring. Alarmingly, in recent years an increasing number of bacteria have become resistant to these antibiotics. A major strategy em- ployed by these pathogens is to use Zn(II)-dependent enzymes, the metallo-β-lactamases (MBLs), which hydrolyse the β-lactam ring. Clinically useful MBL inhibitors are not yet available. Conse- quently, MBLs remain a major threat to human health. In this review biochemical properties of MBLs are discussed, focusing in particular on the interactions between the enzymes and the func- tionally essential metal ions. The precise role(s) of these metal ions is still debated and may differ between different MBLs. However, since they are required for catalysis, their binding site may present an alternative target for inhibitor design.
Keywords:
Antibiotic Resistance, β-Lactam Antibiotics, Metallo-β-Lactamases, Reaction Mechanism, Metal Ion Binding

1. Introduction
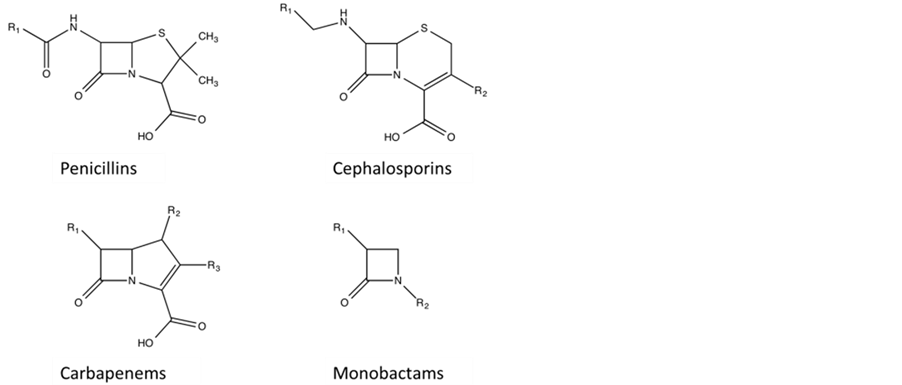
β-lactamases are a family of enzymes that hydrolyse and thus linearise the β-lactam moiety of most of the com- monly used antibiotics, examples of which are shown in Figure 1 [1] . Based on sequence similarity, the
Figure 1 . Chemical Structure of representative antibiotics types containing signature beta lactam ring. The R groups indicate modifications to the core structure of the respective antibiotics.
β-lactamases are subdivided into four groups, A, B, C and D [2] [3] . Subgroups A, C and D are serine-β-lacta- mases SBLs; they employ a serine residue in their active site to initiate hydrolysis of β-lactam substrates and do not require metal ions for their function [2] -[5] . SBLs have been extensively studied and their threat to human health is, at least currently, under control, as clinically useful SBL inhibitors such as clavulanic acid can be co-administered with the antibiotics to maintain their antibacterial effect [4] [5] . In contrast, no such inhibitors are currently available for B-type β-lactamases, termed MBLs. In addition, their threat to human health is further exacerbated by their ability to spread easily between species, mainly through horizontal gene transfer [6] [7] . MBL-encoding genes can be part of either the chromosomal framework of the bacterial species, as observed for instance in Pseudomonas aeruginosa [8] , or are located on mobile genetic elements that can easily be shared among species via horizontal gene transfer (examples include P. aeruginosa, Klebsiella pneumonia and Acine- tobacter baumannii [9] -[11] ). This facile transfer of genetic information amongst pathogenic bacteria, in com- bination with increasing global travel capabilities of the human population provides an ideal framework for the rapid spread of antibiotic resistance [10] .
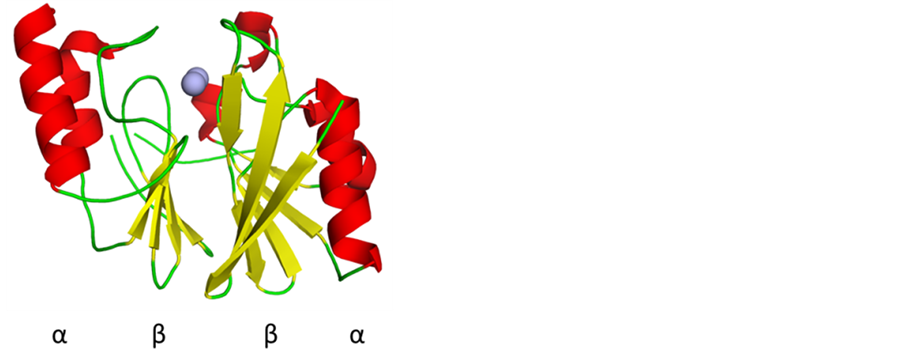
MBLs require at least one, but more commonly two Zn(II) ions in their active sites for catalytic activity [2] [12] -[14] . Based on sequence homology MBLs are divided into as many as four subgroups, labelled B1, B2, B3 and, the most recent addition, B4 [15] . Common to all MBLs is their characteristic 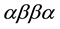 fold (Figure 2).
fold (Figure 2).
While all MBLs require metal ions for their function there are variations with respect to how many metal ions are essential. B1-type MBLs generally require two Zn(II) ions in their active sites [2] [12] -[14] [17] . However, an MBL from Bacillus cereus, BcII, has been shown to be catalytically active in the presence of only one Zn(II) ion (albeit with reduced efficiency) [18] . In contrast, MBLs from the B2 subgroup require only one Zn(II) ion; the binding of a second metal ion in their active sites leads to inhibition of catalysis [19] . Interestingly, B2-type MBLs are more selective in terms of the antibiotics they are able to degrade―their activity against penicillins and cephalosporins is poor, while they have a particular preference for monobactams [2] [20] -[22] . B3-type MBLs resemble B1-type enzymes in their metal ion requirements [23] [24] . The only characterised MBL that may belong to the recently proposed B4 subgroup, SPR-1 from Serratia proteamaculans, may also require two metal ions to be catalytically active. However, unlike other MBLs studied to date SPR-1 may be mononuclear in the absence of substrates; only upon the addition of an antibiotic reactant a binuclear centre is formed [25] . This behaviour may imply the presence of a regulatory mechanism whereby the enzyme is “switched on” only when needed, a mechanism reminiscent of that of the organophosphate-degrading enzyme GpdQ from Enterobacter aerogenes [26] -[33] . While it remains unclear why some MBLs employ such a regulatory mechanism its occur- rence may suggest an additional, as of yet obscure biological function for these enzymes.
Zinc is the naturally occurring metal ion employed by all known MBLs [2] [12] -[14] . In in vitro studies it was demonstrated that catalytic activity can be reconstituted with a range of metal ions, including Co(II), Mn(II)
Figure 2. A representative MBL, the New Dehli M- BL (NDM-1) illustrating the characteristic  fold (PDB: 3S0Z) [16] .
fold (PDB: 3S0Z) [16] .
and Cu(II) [13] [14] [34] . Although not biologically relevant, the MBL derivatives with these metal ions have provided detailed insight into mechanistic aspects of these enzymes.
A series of detailed reviews on the structure, function and clinical relevance of MBLs have been published over the past decade [3] [13] [14] . The aim of this minireview is to briefly summarise the main structural and mechanistic aspects of these diverse enzymes, while focussing on the interactions between metal ions and the ligands in the active site of the MBLs and their role(s) in catalysis. Universal inhibitors for MBLs are currently unavailable but since metal ions are essential for MBL function, a strategy that would interfere with enzyme- metal ion interactions may prove beneficial for the future development of such inhibitors.
2. Overall and Active Site Structures of MBLs
As mentioned above, although MBLs are divided into four subclasses, they all share a common αββα core structure, with eight β strands connected by α helices (Figure 2) [2] [14] . The B1 subclass is the most prevalent and structurally most extensively studied class [2] [8] [17] [35] -[40] . Members include IMP [36] [41] -[48] , VIM [49] -[58] and BcII [14] [15] [59] [60] . More recently, NDM-1 from K. pneumoniae emerged and made global headlines due to its highly pathogenic and dangerous nature because of its ability to degrade most commonly used antibiotics [61] -[66] . Examples of the B2 and B3 subgroups are CphA from A. hydrophila [21] [22] [67] -[71] , ImiS from A. veronii bv. Sobria and Sfh-I from S. fonticola [67] [71] [72] , and L1 from S. maltophilia [24] [73] -[75] , FEZ-1 from F. gormanii [76] , BJP-1 from B. japonicum [77] , MIM-1 from N. pentaromativorans [78] , MIM-2 from S. agarivorans [78] , SMB-1 from S. marcescens [79] , CAR-1 from E carotovora [80] and THIN-B from J. lividum [81] , while the recently proposed B4 subgroup is represented by SPR-1 from S. proteamaculans [25] and CSA-1 from C. sakazaki [15] . B1-type MBLs have two peptide loops, L3 and L8, in the vicinity of the metal ion-containing active site (Figure 3).These loops are believed to be crucial for the determination of the substrate specificity of these enzymes [2] . In contrast, MBLs from the B2 subgroup lack the extended L3 loop. Instead these enzymes have a kinked α-helix positioned directly above the active site cleft [2] [20] . This feature facilitates the formation of a narrow, well defined substrate binding pocket. Consequently, these enzymes display a tighter selectivity for antibiotic substrates than other MBLs, hydrolysing only monobactam substrates with high efficiency [2] . Of the three known representatives from this subgroup (Table 1), CphA is the most extensively studied [19] [21] [22] [67] -[70] . MBLs from the B3 subgroup also lack the extended L3 loop. However, they have an extra loop, located above the active site, which may also influence the substrate specificity of these enzymes. A preference for cephalosporins has been noted for this subgroup [2] [21] . Initially, SPR-1 from S. proteamaculans was also assigned to the B3 subgroup [25] . However, an analysis of its active site structure (see below), together with a homology sequence analysis has indicated that SPR-1 may represent the prototype of the B4 subgroup of MBLs [15] [25] . Its substrate preference is similar to that of the B3-type MBL L1 from S. maltophilia [25] ; no crystallographic data for SPR-1 is currently available.
Figure 3. A representative B1-type MBL illustrating the charachteristic loops L3 and L8 for this subgroup of MBLs. The structure was determined for the enzyme CcrA from B. fragilis. (PDB: 1ZNB) [14] [83] .
Table 1. Geometries and ligands of the two metal ion binding sites in MBLs from different subclasses.
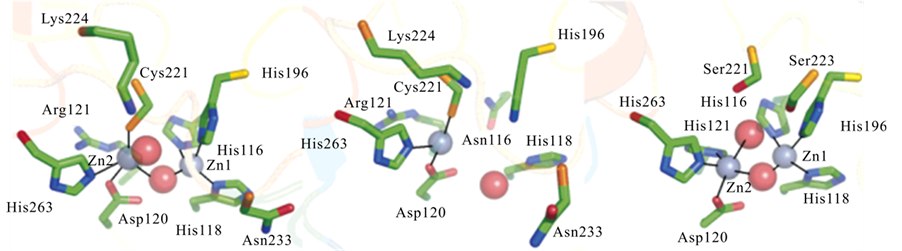
The active sites of MBLs generally accommodate space for two metal ions to bind in close proximity (Figure 4). The two metal ion binding sites are frequently labelled as Zn1 and Zn2, highlighting the fact that in vivo MBLs are Zn(II)-dependent enzymes.
Each metal ion is coordinated by three amino acids; however, the identity of these ligands varies between subgroups. Table 1 lists the relevant amino acids and resulting geometries for the metal ions in the Zn1 and Zn2 sites of MBLs from different subgroups. The numbering of the amino acids is according to a standardised system [3] : water ligands are indicated by W.
In B1- and B3-type MBLs the Zn1 site is formed by three histidine residues-His116, His118 and His196- and is thus referred to as the “histidine site”; the fourth coordination position is occupied by a water molecule that forms a bridge to Zn2. This water molecule is the likely nucleophile that initiates the hydrolysis of the β-lactam substrates [14] .
The overall geometry of the Zn1 site is tetrahedral. In B2-type MBLs one of the histidine ligands, H116, is replaced by an asparagine. This mutation may affect the affinity of the Zn1 site for metal ions. It is important to note that in the catalytically active form of B2-type MBLs this site is not occupied by a metal ion; binding of a metal ion to Zn1 leads to effective inhibition of catalysis [67] . In B4-type MBLs H118 in Zn1 is replaced by an arginine residue [15] [25] . It is not yet known if in these MBLs the Zn1 site is occupied in the resting state. B1- and B2-type MBLs have identical amino acid ligands in their Zn2 sites with D120, C221 and H263-due to the presence of the cysteine ligand this site is frequently also referred to as the “cysteine site”. The metal ion-bridg- ing water completes the coordination sphere of B1-type MBLs. However, in contrast to the Zn1 site the geometry is distorted trigonal bipyramidal, thus providing a possible vacant coordination position for a substrate molecule. In B2-type MBLs a water ligand is also present, but the overall geometry is tetrahedral. In B3 enzymes, the cysteine residue is replaced by H121, but the geometry remains trigonal bipyramidal. More significant alterations appear evident in the Zn2 sites of MBLs belonging to the recently proposed B4 subgroup. Based on sequence analysis and homology modelling it is likely that C221 is replaced either by S221 or Q121, while H263 may be substituted by N262 [15] [25] . These extensive variations in the active site of these enzymes are expected to affect their interactions with the metal ions, and consequently also their reaction mechanism significantly. Although little is currently known about B4-type MBLs they may shed light into the functional diversity of MBLs in general, an insight that may also be exploited in the design of potent inhibitors of clinical relevance.
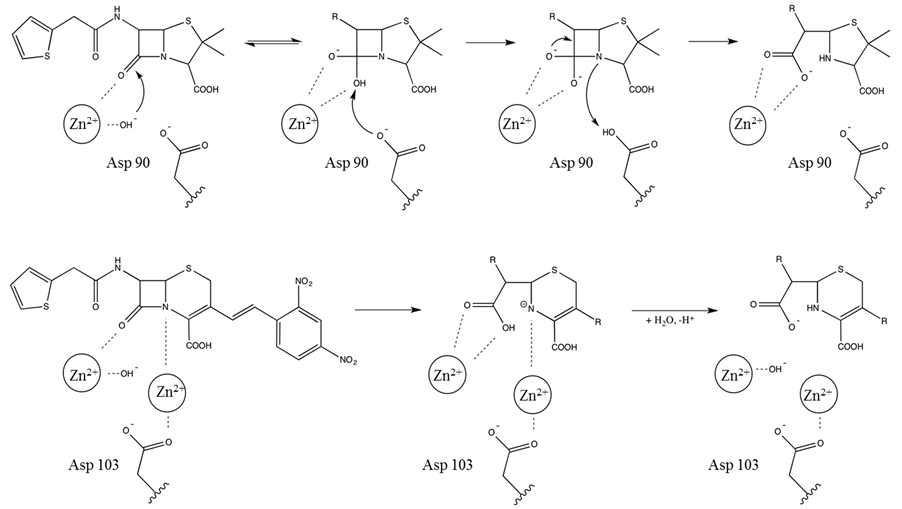
The above discussion illustrates that there is considerable variation in the active sites of MBLs from different subgroups both in terms of the composition of their first coordination spheres and the contribution of the Zn(II) ions to catalysis. Members of the B1 subgroup operate predominantly as binuclear enzymes but may still be functional in mononuclear form, albeit with reduced activity [18] [59] [60] . The B2-type MBLs only operate in mononuclear form and are inhibited if a metal ion occupies their Zn1 site [67] while the known B3-type MBLs appear to require binuclear centres [23] [24] [74] [75] . The least characterised B4-type MBLs may employ yet another mechanistic strategy where the mononuclear form is the resting, inactive form, and the catalytically relevant binuclear form is only assembled once a substrate is present [25] . Consequently, both mono- and binuclear reaction mechanisms have been proposed for MBLs [2] [8] [13] [14] [18] [19] [60] [67] [82] -[84] . Common to the various mechanistic proposals is the involvement of the metal ions in catalysis by coordinating the nucleophilic hydroxide group, which is either bridging the metal ions in binuclear centres or is coordinated to Zn2 in mononuclear MBLs (Figure 5). Despite these mechanistic variations the central and essential role of
Figure 4. Active site structures of B. cereus BcII (B1, left), A. hydrophila CphA (B2, center), and S. maltophilia L1 (B3, right) [14] .
Figure 5. Proposed reaction mechanisms for mono (upper panel) and binuclear (bottom panel) MBLs.
the metal ions in catalysis may facilitate a much needed alternative strategy to develop universal MBL inhibitors for clinical applications. Currently available MBL inhibitors mainly compete with the antibiotic substrates, coordinating either directly to the metal ions in the active site or at least in close vicinity (e.g. Figure 6) [25] [86] - [92] . However, as discussed above, the substrate binding sites reveal large structural variations between MBL subgroups, exemplified by differences in substrate preferences. Compounds that interfere with metal ion binding to Zn1, Zn2 or both sides may thus be more promising as universal MBL inhibitors. In order to exploit the potential of the metal ion binding sites for inhibitor design and development the interactions between the active site and the metal ions need to be investigated. Relatively few studies to date have focused on providing quantitative data that characterise such interactions in MBLs. It is thus a major aim of this minireview to summarise available information with a view to highlight current limitations in our understanding of metal ion binding in MBLs.
3. Interactions between MBLs and Metal Ions
The discussion in the section above highlighted the fact that the metal ion requirement and quite likely also the metal ion binding in the four MBL subgroups may vary. Insight into the role of the metal ions in MBL function is obscured not only by the fact that there is limited quantitative information available, but also by the significant variations in reported metal ion binding affinities. While some of these variations may be due to structural differences in the active sites of MBLs, some may be associated with different methodologies that were employed to obtain binding constants. An example of reported binding constants for Zn(II) to the MBL BcII illustrates the conundrum. The affinities of the two metal binding sites in the catalytic centre of BcII differ greatly, but estimates for the Kd values for each of the two sites also vary largely. For the site with the tighter affinity Kd values range from the low nM scale to 120 μM, and for the lower affinity site from 1.5 μM to 24 mM [18] [60] [93] . A more recent structural and spectroscopic study with BcII suggested positive cooperative binding of the two zinc ions [94] . Thus, it was proposed that the di-zinc form is the biologically relevant one [94] . Positive cooperativity for metal ion binding has also been reported for other B1-type MBLs, i.e. the enzymes CcrA [95] -[99] and IMP-1 [36] [41] -[46] . In contrast, the B1 MBL Bla2 from B. anthracis appears to bind the two Zn(II) in sequential order, leading to the speculation that this enzyme may be active in its mononuclear form under physiological conditions [100] . In the following paragraphs reported metal ion binding constants and their relevance to MBL function are discussed in order to illustrate the significance of the metal ions to MBL function but also to highlight current limitations in our understanding of how precisely the metal ions contribute to these enzymes’ mode of action.
Methods used to measure binding affinities include competition-type assays, spectroscopic and thermodynamic measurements, as well as catalytic assays. Competition-based assays rely on the direct competition of at least two compounds for binding to a target such as a metal ion. In order to study the binding of metal ions, in
Figure 6. Crystal structure of the inhibitor D-captopril bound in the active site of the B3-type MBL L1 [85] . Zn ions are indicated as spheres in magenta. The Fo-Fc difference electron density map of the inhibitor is shown as purple mesh.
particular transition metal ions, frequently used compounds with convenient colorimetric and fluorimetric prop-
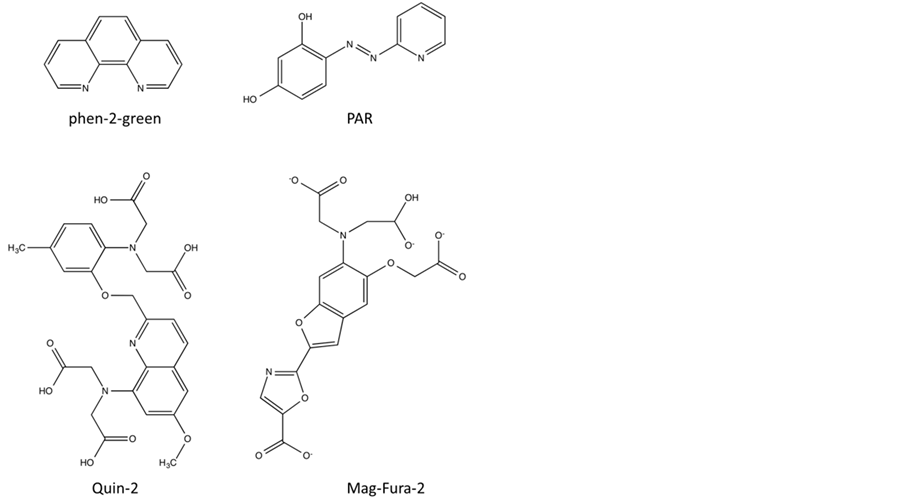
erties are 4-(2-pyridylazo) resorcinol (PAR) and 2-[6-[bis(carboxymethyl)amino]-5(carboxymethoxy)-2-benzof- uranyl], abbreviated here to Mag-Fura, respectively (Figure 7); both compounds have high affinities for Co(II), Ni(II), Cu(II), Zn(II) and Cd(II) [101] -[105] . For a detailed description of the data analysis refer to published literature [18] [60] [106] [107] . A competition assay using Mag-Fura as chelator was employed in order to estimate the binding constants of Zn(II) and Cd(II) to the active site in the B1-type MBL BcII, including several mutants of this enzyme (i.e. H86S, H88S and H149S, point mutations in the Zn1 site, and D90N, C168S and H210S, point mutations in the Zn2 site) [60] . Two binding events are observed, characterised by Kd1 and Kd2; relevant values are summarised in Table 2, and indicate that (a) the affinity of different metal ions for a particular site varies significantly and (b) the two metal ion binding sites also display a large difference in metal ion affinity (note that the binding affinities of Co(II), which were determined spectrophotmetrically by recording the UV-Vis spectrum of BcII as a function of increasing Co(II) concentrations, were included for comparative purposes). For example, in the wild-type enzyme the estimated Kd value of Zn(II) to the tighter binding site (i.e. Kd1) is ~0.6 nM, but only 1.5 µM to the weaker site (i.e. Kd2). Similar values were obtained in competition assays using PAR instead of Mag-Fura [108] . Zn(II) is clearly the preferred metal ion for both binding sites, followed by Cd(II) and Co(II), although catalytic activity can be reconstituted with each of these metal ions [60] .
Interestingly, mutating each of the six metal ion-coordinating amino acid side chains appears to have only a modest effect on the affinities of Zn1 and Zn2; the effect appears stronger for the Co(II) derivatives than the Zn(II) derivatives of the enzyme (Table 2) [18] [60] . Using the same approach it was also shown that the addition of a substrate (imipenem) greatly enhances the affinity of the tighter side (Kd1 ~10 pM); the affinity of the weaker side is only marginally altered (Kd2 ~1.8 µM and 0.8 µM in the absence or presence of substrate, respectively) [60] .
Improved binding to the tighter side was also observed for another B1-type MBL, BlaB from Chryseobacterium meningosepticum, the B2-type MBL CphA from A. hydrophila and the B3-type MBL L1 from S. maltophilia [24] [70] [74] [109] . The effect of the presence of a substrate on binding of Zn(II) to the weaker bound side does not appear to display a trend. For BcII and CphA a ~2- and ~25-fold increase in binding affinity is reported, while a ~3- and 20-fold reduction, respectively, is measured for BlaB and L1 [24] [59] [60] [70] [98] [109] . Based on the very tight binding of only one of the metal ions in the active site it was suggested that MBLs may only require one Zn(II) for catalysis under physiological conditions, especially since it is estimated that the concentration of free Zn(II) in cells may be in the picomolar range or lower [12] . However, this inter-
Figure 7. Chemical structures for PAR, Mag-Fura-2, quin-2 and phen-2-green.
Table 2. Binding constants for several metal ions (i.e. Zn(II), Cd(II) and Co(II)) to the wild-type form of BcII and several of this MBLs mutants [60] .
pretation raises a serious issue in terms of why all known MBLs have two closely spaced metal ion binding sites in their catalytic centres-as already mentioned in a previous report it appears unlikely that such a structural arrangement would be conserved throughout evolution if there was no functional benefit for the enzyme [110] . It should also be noted that the binuclear form of BcII is at least twice as reactive as its mononuclear counterpart, and that the Kd values of Zn(II), especially that for the tighter site, may not be as low as estimated by the abovementioned competition-based assays. Using equilibrium dialysis [108] or catalytic activity measurements as a function of Zn(II) concentration [18] Kd1 values of ~0.3 µM and 0.66 µM, and Kd2 values of ~3 µM and 890 µM, respectively, were recorded. To complicate matters further, using isothermal titration calorimetry (ITC) only one binding event was observed when metal ion-free BcII was titrated with Zn(II). Since the associated stoichiometry was, however, close to 2 it was suggested that two Zn(II) ions with comparable affinities (with an estimated Kd value of ~30 µM) are bound to the enzyme [111] . Similar metal ion affinities for the two binding sites were also recorded for the B1-type MBL Ccr from Bacteroides fragilis with estimated Kd values ≤ 10 µM [97] ; only the binuclear enzyme is believed to be active [98] . In a more recent study with BcII the competition assay using Mag-Fura as chelator was repeated as part of an extended study to investigate the process of Zn(II) binding in this enzyme [109] . The authors concluded that metal ion binding is a positively cooperative process and estimated metal ion affinities similar to the values reported by Badarau and Page, using ITC [111] . The reason for the largely differing Kd values obtained from competition-based assays remains unclear, but it appears increasingly likely that the binuclear form of the B1-type MBLs (and likely also the B3-type MBLs) is the physiologically relevant one [74] , especially since extracellular Zn(II) concentration may be significantly larger than within cells. While in B1- and possibly also B3-type MBLs the binding of a second metal ion is driven by positive cooperativity and leads to an increase in reactivity, in B2-type MBLs it leads to inhibition (with a Ki of ~50 µM) [71] . Metal ion affinity constants for the B2-type MBL CphA from A. hydrophila were also estimated in competition experiments using quin-2 as chelator for Zn(II) and Cd(II), and phen-green for Cu(II) (Figure 7). Co(II) affinities were estimated spectrophotmetrically by recording the UV-Vis spectrum of CphA as a function of increasing Co(II) concentrations [19] . Kd values ranged from the low picomolar range for Zn(II) to ~600 nM for Cu(II) for the tighter metal ion site to micromolar range for the weaker one. These values are of a similar magnitude as those reported for the B1-type MBL BcII (see above).
At this point it may be necessary to briefly discuss the connection between the tighter and weaker metal ion binding sites (characterised by Kd1 and Kd2) and the two available binding sites in the catalytic centre, labelled Zn1 and Zn2 (Figure 4). In B1- and B3-type MBLs Zn1 is likely to be associated with Kd1, supported, for instance, by crystallographic data that indicate a higher metal ion occupancy for this site than Zn2 [35] [36] [46] [59] [64] . An exception appears to be the B3-type MBL GOB from Elizabethkingia meningoseptica, where H116 in the Zn1 position (Figure 4) is replaced by a glutamine; a combination of kinetic and spectroscopic data indicate that this enzyme operates in mononuclear form with the metal ion bound to Zn2 (i.e. Kd1 is associated with Zn2; its magnitude has not been reported) [112] . Hence, GOB resembles B2-type MBLs where available structural and functional information also indicates that the Zn2 site is catalytically relevant and associated with Kd1 [112] . The surprising outcome of these comparisons is that the Zn2 site in B2-type MBLs and GOB is the preferred binding site despite the fact that in GOB the ligand environment of Zn2 is identical to that in B3-type MBLs, enzymes with a distinct preference for metal ion binding to the Zn1 site. Common to both B2-type MBLs and GOB is a single mutation in position 116 in the Zn1 site, where a histidine present in B1-type and most B3-type MBLs is replaced by glutamine. Furthermore, the Zn2 site in B1-type-MBLs is identical to that in B2-type enzymes (Table 1), yet the preferred binding site in the former is Zn1. Hence, relatively small changes in terms of amino acid substitutions may have significant effects in metal ion binding in this family of enzymes. In this respect the recent identification of a novel MBL that may be part of a new subgroup, labelled B4, may be an evolutionary intermediate. SPR-1 from S. proteamaculans also has three mutations in the metal ion binding site, one in Zn1 and two in Zn2 (Figure 4). It has been proposed that in the absence of substrate the enzyme may be in a mononuclear, catalytically inactive state; upon addition of substrate a catalytically active binuclear centre is formed [25] . Although no metal ion binding or crystallographic studies with SPR-1 have been reported it appears likely that in its mononuclear form the Zn2 site is occupied since the Zn1 site resembles that of both GOB and B2-type MBLs. It should be noted that the proposed substrate-promoted metal ion assembly mechanism in SPR-1 has also been observed in other binuclear metalloenzymes, including a glycerophosphatediesterase (GpdQ) from E. aerogenes [26] -[33] .
The above discussion highlighted the considerable difficulties encountered in the study of the contribution of the metal ions to catalysis in the different subgroups of MBLs. While there is clear evidence for functional differences between members from different subgroups, it has also become evident that different methodologies used to estimate metal ion binding may have led to different conclusions. It thus seems essential that metal ion interactions are carried out under well defined, identical conditions in order to compare different MBLs (including different metal ion derivatives). An ideal, universally applicable method may thus be ITC since it is not dependent on particular chelating agents or spectroscopic properties. It is somewhat surprising that this methodology has rarely been used in studies with MBLs. As mentioned above, Badarau and Page reported Kd values for Zn1 and Zn2 in BcII [111] , and in a more recent report Horton et al. employed ITC to measure Kd2 (i.e. the Kd of the Zn2 site) of the C221G mutant of IMP-1, which was purified with the Zn1 site occupied by a zinc ion [48] . The binding constant was estimated to be 17 μM, indicating that the cysteine ligand may not play a major role in binding the metal ion in Zn2 in B1-type MBLs, an observation in agreement with the proposed positive cooperativity for metal ion binding in the subgroup.
4. Conclusions
MBLs have emerged as a major threat to global health. They inactivate an increasing number of commonly used antibiotics and spread easily among various pathogens on mobile genetic elements. Crystal structures for several MBLs have been determined and an extensive amount of information about their biochemical properties has been accumulated. Some potent in vitro inhibitors of MBLs have also been detected. However, to date none of the available MBL inhibitors are of clinical use. The search for universal and clinically applicable MBL antagonists is still very much at the beginning.
This search is complicated further by the fact that MBLs are able to mutate rapidly and thus evade inhibition. This large mutational space is illustrated by the small degree of sequence and structure conservation in the substrate binding pockets of various MBLs; accordingly, their substrate preference and response to potential inhibitors can vary considerably. Thus, new strategies to comprehensively inhibit MBLs are needed. The main common aspect of their function is their requirement for metal ions, one (in the Zn2 site) for B2-type MBLs, and mostly two in the remaining ones (see discussion above). It is thus surprising that the precise role(s) of metal ions in the catalytic mechanism of MBLs, and in particular their binding interactions in the active sites are still obscure. It appears unlikely that the metal ion binding site can afford a large mutational degree of freedom― metal ion affinities are expected to be severely affected by most changes in their coordination environment. Hence, we propose that universal MBL inhibitors that may retain their effect long-term should target the metal ion binding site. It is thus essential to investigate and compare metal binding interactions among different MBLs under experimentally well-defined and conserved conditions. Since the early metal ion binding studies by de Seny, Wommer and coworkers [56] [103] ITC has emerged as a method of choice to assess binding affinities under physiologically relevant conditions. A detailed characterisation of comparative metal ion affinities in various MBLs will provide essential information to design and develop compounds that effectively interfere with metal ion binding in these enzymes. Such compounds are not expected to be affected by mutations as significantly as molecules that compete with substrates, and hence they may prove to be highly useful as clinical chemotherapeutics in the fight against antibiotic resistance.
Acknowledgements
N. M. thanks the Science Foundation Ireland (SFI) for financial support in form of a President of Ireland Young Researcher Award (PIYRA) and G. S. acknowledges the award of a Future Fellowship from the Australian Research Council (FT120100694) and is grateful to the National Health and Medical Research Council of Australia for funding.
References
- Davies, J. (1994) Inactivation of Antibiotics and the Dissemination of Resistance Genes. Science, 264, 375-382. http://dx.doi.org/10.1126/science.8153624
- Bebrone, C. (2007) Metallo-β-Lactamases (Classification, Activity, Genetic Organization, Structure, Zinc Coordination) and Their Superfamily.Biochemical Pharmacology, 74, 1686-1701. http://dx.doi.org/10.1016/j.bcp.2007.05.021
- Galleni, M., Lamotte-Brasseur, J., Rossolini, G.M., Spencer, J., Dideberg, O. and Frère, J.M. (2001) Standard Numbering Scheme for Class B β-Lactamases.Antimicrobial agents and chemotherapy, 45, 660-663. http://dx.doi.org/10.1128/AAC.45.3.660-663.2001
- Bebrone, C., Lassaux, P., Vercheval, L., Sohier, J.S., Jehaes, A., Sauvage, E. and Galleni, M. (2010) Current Challenges in Antimicrobial Chemotherapy Focus on β-Lactamase Inhibition. Drugs, 70, 651-679. http://dx.doi.org/10.2165/11318430-000000000-00000
- Pérez-Llarena, F.J. and Bou, G. (2009) β-Lactamase Inhibitors: The Story so Far. Current Medicinal Chemistry, 16, 3740-3765. http://dx.doi.org/10.2174/092986709789104957
- Maltezou, H.C. (2009) Metallo-β-Lactamases in Gram-Negative Bacteria: Introducing the Era of Pan-Resistance? International Journal of Antimicrobial Agents, 33, 405.e1-405.e7. http://dx.doi.org/10.1016/j.ijantimicag.2008.09.003
- Perez, F., Hujer, A.M., Hujer, K.M., Decker, B.K., Rather, P.N. and Bonomo, R.A. (2007) Global Challenge of Multidrug-Resistant. Acinetobacter baumannii Antimicrobial Agents and Chemotherapy, 51, 3471-3484. http://dx.doi.org/10.1128/AAC.01464-06
- Pollini. S., Maradei, S., Pecile, P., Olivo, G., Luzzaro, F., Docquier, J.D. and Rossolini, G.M. (2013) FIM-1, a New Acquired Metallo―Lactamase from a Pseudomonas aeruginosa Clinical Isolate from Italy. Antimicrobial Agents and Chemotherapy, 57, 410-416. http://dx.doi.org/10.1128/AAC.01953-12
- Nordmann, P. and Poirel, L. (2012) Strategies for Identification of Carbapenemase-Producing Enterobacteriaceae. Journal of Antimicrobial Chemotherapy, 68, 487-489. http://dx.doi.org/10.1093/jac/dks426
- Livermore, D.M. and Woodford, N. (2006) The b-Lactamase Threat in Enterobacteriaceae, Pseudomonas and Acinetobacter. Trends in Microbiology, 14, 413-420. http://dx.doi.org/10.1016/j.tim.2006.07.008
- Lee, K., Yum, J. H., Yong, D., Lee, H.M., Kim, H.D., Docquier, J.-D., Rossolini, G.M. and Chong, Y. (2005) Novel Acquired Metallo-β-Lactamase Gene, blaSIM-1, in a Class 1 Integron from Acinetobacter baumannii Clinical Isolates from Korea. Antimicrobial Agents and Chemotherapy, 49, 4485-4491.
- Heinz, U. and Adolph, H.W. (2004) Metallo-β-Lactamases: Two Binding Sites for One Catalytic Metal Ion? Cellular and Molecular Life Sciences, 61, 2827-2839. http://dx.doi.org/10.1007/s00018-004-4214-9
- Page, M. I. and Badarau, A. (2008) Loss of Enzyme Activity during Turnover of the Bacillus cereus β-Lactamase Catalysed Hydrolysis of b-Lactams Due to Loss of Zinc Ion. Journal of Bioilogical Inorganic Chemistry, 13, 919-928. http://dx.doi.org/10.1007/s00775-008-0379-2
- Crowder, M.W., Spencer, J. and Vila, A.J. (2006) Metallo-β-Lactamases: Novel Weaponry for Antibiotic Resistance in Bacteria. Accounts of Chemical Research, 39, 721-728. http://dx.doi.org/10.1021/ar0400241
- Hou, C.F., Phelan E.K., Miraula, M., Ollis, D.L., Schenk, G. and Mitic, N. (2014) Unusual Metallo-β-Lactamases May Constitute a New Subgroup in This Family of Enzymes. American Journal of Molecular Biology, 4, 11-15. http://dx.doi.org/10.4236/ajmb.2014.41002
- Guo, Y., Wang, J., Niu, G.J., Shui, W.Q., Sun, Y.N., Zhou, H.G., Zhang, Y.Z., Yang, C., Lou, Z.Y. and Rao, Z.H. (2011) A Structural View of the Antibiotic Degradation Enzyme NDM-1 from a Superbug. Protein & Cell, 2, 384-395. http://dx.doi.org/10.1007/s13238-011-1055-9
- Llarrull, L.I., Tioni, M.F. and Vila, A.J. (2008) Metal Content and Localization during Turnover in B. cereus Metallo- β-Lactamase. Journal of the American Chemical Society, 130, 15842-15851. http://dx.doi.org/10.1021/ja801168r
- Rasia, R.M. and Vila, A.J. (2002) Exploring the Role and the Binding Affinity of a Second Zinc Equivalent in B. cereus Metallo-β-Lactamase. Biochemistry, 41, 1853-1860. http://dx.doi.org/10.1021/bi010933n
- Valladares, M.H., Kiefer, M., Heinz, U., Soto, R.P., Meyer-Klaucke, W., Nolting, H.F., Zeppezauer, M., Galleni, M., Frere, J.M., Rossolini, G.M., Amicosante, G. and Adolph, H.W. (2000) Kinetic and Spectroscopic Characterization of Native and Metal-Substituted β-Lactamase from Aeromonas hydrophila AE036. FEBS Letters, 467, 221-225. http://dx.doi.org/10.1016/S0014-5793(00)01102-9
- Massida, O., Rossolini, G.M. and Satta, G. (1991) The Aeromonas hydrophila cphA Gene: Molecular Heterogeneity among Class B Metallo-β-Lactamases. Journal of Bacteriology, 173, 4611-4617.
- Segatore, B., Massidda, O., Satta, G., Setacci, D. and Amicosante, G. (1993) High Specificity of cphA-Encoded Metallo-β-Lactamase from Aeromonas hydrophila AE036 for Carbapenems and Its Contribution to β-Lactam Resistance. Antimicrobial Agents and Chemotherapy, 37, 1324-1328. http://dx.doi.org/10.1128/AAC.37.6.1324
- Bebrone, C., Anne, C., De Vriendt, K., Devreese, B., Rossolini, G.M., Van Beeumen, J., Frere, J.M. and Galleni, M. (2005) Dramatic Broadening of the Substrate Profile of the Aeromonas hydrophila CphA Metallo-β-Lactamase by Site-Directed Mutagenesis. The Journal of Biological Chemistry, 280, 28195-28202. http://dx.doi.org/10.1074/jbc.M414052200
- Leiros, H.L.S., Borra, P.S., Brandsdal, B.O., Edvardsen, K.S.W., Spencer, J., Walsh, T.R. and Samuelson, O. (2012) Crystal Structure of the Mobile Metallo-β-Lactamase AIM-1 from Pseudomonas aeruginosa: Insights into Antibiotic Binding and the Role of Gln157. Antimicrobial Agents and Chemotherapy, 56, 4341-4353. http://dx.doi.org/10.1128/AAC.00448-12
- Costello, A., Periyannan, G., Yang, K.W., Crowder, M.W. and Tierney, D.L. (2006) Site-Selective Binding of Zn(II) to Metallo-β-Lactamase L1 from Stenotrophomonas maltophilia. Journal of Biological Inorganic Chemistry, 11, 351-358. http://dx.doi.org/10.1007/s00775-006-0083-z
- Vella, P., Miraula, M., Phelan, E.K., Leung, E.W., Ely, F., Ollis, D.L., McGeary, R.P., Schenk, G. and Mitic, N. (2013) Identification and Characterization of an Unusual Metallo-β-Lactamase from Serratia proteamaculans. Journal of Biological Inorganic Chemistry, 18, 855-863. http://dx.doi.org/10.1007/s00775-013-1035-z
- Jackson, C.J., Hadler, K.S., Carr, P.D., Oakley, A.J., Yip, S., Schenk, G. and Ollis, D.L. (2008) Malonate-Bound Structure of the Glycerophosphodiesterase from Enterobacter aerogenes (GpdQ) and Characterization of the Native Fe2+ Metal-Ion Preference. Acta Crystallographica Section F: Structural Biology and Crystallization Communications, 64, 681-685. http://dx.doi.org/10.1107/S1744309108017600
- Hadler, K.S., Tanifum, E.A., Yip, S.H.C., Mitìc, N., Guddat, L.W., Jackson, C.J., Gahan, L.R., Nguyen, K., Carr, P.D., Ollis, D.L., Hengge, A.C., Larrabee, J.A. and Schenk, G. (2008) Substrate-Promoted Formation of a Catalytically Competent Binuclear Center and Regulation of Reactivity in a Glycerophosphodiesterase from Enterobacter aerogenes. Journal of the American Chemical Society, 130, 14129-14138. http://dx.doi.org/10.1021/ja803346w
- Hadler, K.S., Mitic, N., Ely, F., Hanson, G.R., Gahan, L.R., Larrabee, J.A., Ollis, D.L. and Schenk, G. (2009) Structural Flexibility Enhances the Reactivity of the Bioremediator Glycerophosphodiesterase by Fine-Tuning Its Mechanism of Hydrolysis. Journal of the American Chemical Society, 131, 11900-11908. http://dx.doi.org/10.1021/ja903534f
- Hadler, K.S., Mitić, N., Yip, S.H., Gahan, L.R., Ollis, D.L., Schenk, G. and Larrabee, J.A. (2010) Electronic Structure Analysis of the Dinuclear Metal Center in the Bioremediator Glycerophosphodiesterase (GpdQ) from Enterobacter aerogenes. Inorganic Chemistry, 49, 2727-2734. http://dx.doi.org/10.1021/ic901950c
- Hadler, K.S., Gahan, L.R., Ollis, D.L. and Schenk, G. (2010) The Bioremediator Glycerophosphodiesterase Employs a Non-Processive Mechanism for Hydrolysis. Journal of Inorganic Biochemistry, 104, 211-213. http://dx.doi.org/10.1016/j.jinorgbio.2009.10.012
- Yip, S.H.C., Foo, J.L., Schenk, G., Gahan, L.R., Carr, P.D. and Ollis, D.L. (2011) Directed Evolution Combined with Rational Design Increases Activity of GpdQ toward a Non-Physiological Substrate and Alters the Oligomeric Structure of the Enzyme. Protein Engineering Design and Selection, 24, 861-872. http://dx.doi.org/10.1093/protein/gzr048
- Schenk, G., Mitic, N., Gahan, L.R., Ollis, D.L., McGeary, R.P. and Guddat, L.W. (2012) Binuclear Metallohydrolases: Complex Mechanistic Strategies for a Simple Chemical Reaction. Accounts of Chemical Research, 45, 1593-1603. http://dx.doi.org/10.1021/ar300067g
- Daumann, L.J., McCarthy, B.Y., Hadler, K.S., Murray, T.P., Gahan, L.R., Larrabee, J.A., Ollis, D.L. and Schenk, G. (2013) Promiscuity Comes at a Price: Catalytic Versatility vs Efficiency in Different Metal Ion Derivatives of the Potential Bioremediator GpdQ. Biochimica et Biophysica Acta, 1834, 425-432. http://dx.doi.org/10.1016/j.bbapap.2012.02.004
- Badarau, A. and Page, M.I. (2006) The Variation of Catalytic Efficiency of Bacillus cereus Metallo-β-Lactamase with Different Active Site Metal Ions. Biochemistry, 45, 10654-10666. http://dx.doi.org/10.1021/bi060934l
- Carfi, A., Pares, S., Duee, E., Galleni, M., Duez, C., Frere, J.M. and Dideberg, O. (1995) Spectroscopic Characteriza- tion of a Binuclear Metal Site in Bacillus cereus β-Lactamase II. EMBO Journal, 14, 4914-4921. http://dx.doi.org/10.1021/bi980309j
- Concha, N.O., Janson, C.A., Rowling, P., Pearson, S., Cheever, C.A., Clarke, B.P., Lewis, C., Galleni, M., Frere, J.M., Payne, D.J., Bateson, J.H. and Abdel-Meguid, S.S. (2000) Crystal Structure of the IMP-1 Metallo β-Lactamase from Pseudomonas aeruginosa and Its Complex with a Mercaptocarboxylate Inhibitor: Binding Determinants of a Potent, Broad-Spectrum Inhibitor. Biochemistry, 39, 4288-4298. http://dx.doi.org/10.1021/bi992569m
- Bellais, S., Girlich, D., Karim, A. and Nordmann, P. (2002) EBR-1, a Novel Ambler Subclass B1 β-Lactamase from Empedobacter brevis. Antimicrobial Agents and Chemotherapy, 46, 3223-3227. http://dx.doi.org/10.1128/AAC.46.10.3223-3227.2002
- Yamaguchi, Y., Takashio, N., Wachino, J., Yamagata, Y., Arakawa, Y., Matsuda, K. and Kurosaki, H. (2010) Structure of Metallo-β-Lactamase IND-7 from a Chryseobacterium indologenes Clinical Isolate at 1.65―A Resolution. Journal of Biochemistry, 147, 905-915. http://dx.doi.org/10.1093/jb/mvq029
- Borra, P.S., Samuelsen, O., Spencer, J., Walsh, T.R., Lorentzen, M.S. and Leiros, H.K. (2013) Crystal Structures of Pseudomonas aeruginosa GIM-1: Active-Site Plasticity in Metallo-β-Lactamases. Antimicrobial Agents and Chemo- therapy, 57, 848-854. http://dx.doi.org/10.1128/AAC.02227-12
- Poirel, L., Héritier, C. and Nordmann, P. (2005) Genetic and Biochemical Characterization of the Chromosome-En- coded Class B β-Lactamases from Shewanella livingstonensis (SLB-1) and Shewanella frigidimarina (SFB-1). Journal of Antimicrobial Chemotherapy, 55, 680-685. http://dx.doi.org/10.1093/jac/dki065
- Laraki, N., Franceschini, N., Rossolini, G.M., Santucci, P., Meunier, C., de Pauw, E., Amicosante, G., Frere, J.M. and Galleni, M. (1999) Biochemical Characterization of the Pseudomonas aeruginosa 101/1477 Metallo-β-Lactamase IMP-1 Produced by Escherichia coli. Antimicrobial Agents and Chemotherapy, 43, 902-906.
- Iyobe, S., Kusadokoro, H., Ozaki, J., Matsumura, N., Minami, S., Haruta, S., Sawai, T. and O’Hara, K. (2000) Amino Acid Substitutions in a Variant of IMP-1 Metallo-β-Lactamase. Antimicrobial Agents and Chemotherapy, 44, 2023- 2027. http://dx.doi.org/10.1128/AAC.44.8.2023-2027.2000
- Riccio, M.L., Franceschini, N., Boschi, L., Caravelli, B., Cornaglia, G., Fontana, R., Amicosante, G. and Rossolini, G.M. (2000) Characterization of the Metallo-β-Lactamase Determinant of Acinetobacter baumannii AC-54/97 Reveals the Existence of bla(IMP) Allelic Variants Carried by Gene Cassettes of Different Phylogeny. Antimicrobial Agents and Chemotherapy, 44, 1229-1235. http://dx.doi.org/10.1128/AAC.44.5.1229-1235.2000
- Oelschlaeger, P. and Mayo, S. (2005) Hydroxyl Groups in the ββ Sandwich of Metallo-β-Lactamases Favor Enzyme Activity: A Computational Protein Design Study. Journal of Molecular Biochemistry, 350, 395-401. http://dx.doi.org/10.1016/j.jmb.2005.04.044
- Yamaguchi, Y., Ding, S., Murakami, E., Imamura, K., Fuchigami, S., Hashiguchi, R., Yutani, K., Mori, H., Suzuki, S., Arakawa, Y. and Kurosaki, H. (2011) A Demetallation Method for IMP-1 Metallo-β-Lactamase with Restored Enzymatic Activity upon Addition of Metal Ion(s). ChemBiochem, 12, 1979-1983. http://dx.doi.org/10.1002/cbic.201100342
- Griffin, D.H., Richmond, T.K., Sanchez, C., Moller, A.J., Breece, R.M., Tierney, D.L., Bennett, B. and Crowder, M.W. (2011) Structural and Kinetic Studies on Metallo-β-Lactamase IMP-1. Biochemistry, 50, 9125-9134. http://dx.doi.org/10.1021/bi200839h
- Vella, P., Hussein, W.M., Leung, E.W., Clayton, D., Ollis, D.L., Mitic, N., Schenk, G. and McGeary, R.P. (2011) The Identification of New Metallo-β-Lactamase Inhibitor Leads from Fragment-Based Screening. Bioorganic & Medicinal Chemistry Letters, 21, 3282-3285. http://dx.doi.org/10.1016/j.bmcl.2011.04.027
- Horton, L.B., Shanker, S., Mikulski, R., Brown, N.G., Phillips, K.J., Lykissa, E., Venkataram Prasad, B.V. and Palzkill, T. (2012) Mutagenesis of Zinc Ligand Residue Cys221 Reveals Plasticity in the IMP-1 Metallo-β-Lactamase Active Site. Antimicrobial Agents and Chemotherapy, 56, 5667-5677. http://dx.doi.org/10.1128/AAC.01276-12
- Lauretti, L., Riccio, M.L., Mazzariol, A., Cornaglia, G., Amicosante, G., Fontana, R. and Rossolini, G.M. (1999) Cloning and Characterization of blaVIM, a New Integron-Borne Metallo-β-Lactamase Gene from a Pseudomonas aeruginosa Clinical Isolate. Antimicrobial Agents and Chemotherapy, 43, 1584-1590.
- Cornaglia, G., Mazzariol, A., Lauretti, L., Rossolini, G.M. and Fontana, R. (2000) Hospital Outbreak of Carbapenem- Resistant Pseudomonas aeruginosa Producing VIM-1, a Novel Transferable Metallo-β-Lactamase. Clinical Infectious Diseases, 31, 1119-1125. http://dx.doi.org/10.1086/317448
- Tsakris, A., Pournaras, S., Woodford, N., Palepou, M.F., Babini, G.S., Douboyas, J. and Livermore, D.M. (2000) Out- break of Infections Caused by Pseudomonas aeruginosa Producing VIM-1 Carbapenemase in Greece. Journal of Clinical Microbiology, 38, 1290-1292.
- Giakkoupi, P., Xanthaki, A., Kanelopoulou, M., Vlahaki, A., Miriagou, V., Kontou, S., Papafraggas, E., Malamou- Lada, H., Tzouvelekis, L., Legakis, N. and Vatopoulos, A.C. (2003) VIM-1 Metallo-β-Lactamase-Producing Klebsiella pneumoniae Strains in Greek Hospitals. Journal of Clinical Microbiology, 41, 3893-3896. http://dx.doi.org/10.1128/JCM.41.8.3893-3896.2003
- Pournaras, S., Maniati, M., Petinaki, E., Tzouvelekis, L., Tsakris, A., Legakis, N. and Maniatis, A. (2003) Hospital Outbreak of Multiple Clones of Pseudomonas aeruginosa Carrying the Unrelated Metallo-β-Lactamase Gene Variants blaVIM-2 and blaVIM-4. Journal of Antimicrobial Chemotherapy, 51, 1409-1414. http://dx.doi.org/10.1093/jac/dkg239
- Miriagou, V., Tzelepi, E., Gianneli, D. and Tzouvelekis, L.S. (2003) Escherichia coli with a Self-Transferable, Multiresistant Plasmid Coding for Metallo-β-Lactamase VIM-1. Antimicrobial Agents and Chemotherapy, 47, 395-397. http://dx.doi.org/10.1128/AAC.47.1.395-397.2003
- Giske, C.G., Rylander, M. and Kronvall, G. (2003) VIM-4 in a Carbapenem-Resistant Strain of Pseudomonas aeruginosa Isolated in Sweden. Antimicrobial Agents and Chemotherapy, 47, 3034-3035. http://dx.doi.org/10.1128/AAC.47.9.3034-3035.2003
- Luzzaro, F., Docquier, J.D., Colinon, C., Endimiani, A., Lombardi, G., Amicosante, G., Rossolini, G.M. and Toniolo, A. (2004) Emergence in Klebsiella pneumoniae and Enterobacter cloacae Clinical Isolates of the VIM-4 Metallo- β-Lactamase Encoded by a Conjugative Plasmid. Antimicrobial Agents and Chemotherapy, 48, 648-650. http://dx.doi.org/10.1128/AAC.48.2.648-650.2004
- Ktari, S., Arlet, G., Mnif, B., Gautier, V., Mahjoubi, F., Jmeaa, M.B., Bouaziz, M. and Hammami, A. (2006) Emergence of Multidrug-Resistant Klebsiella pneumoniae Isolates Producing VIM-4 Metallo-β-Lactamase, CTX-M-15 Extended-Spectrum β-Lactamase, and CMY-4 AmpC β-Lactamase in a Tunisian University Hospital. Antimicrobial Agents and Chemotherapy, 50, 4198-4201. http://dx.doi.org/10.1128/AAC.00663-06
- Garcia-Saez, I., Docquier, J.D., Rossolini, G.M. and Dideberg, O. (2008) The Three-Dimensional Structure of VIM-2, a Zn-β-Lactamase from Pseudomonas aeruginosa in Its Reduced and Oxidised Form. Journal of Molecular Biology, 375, 604-611. http://dx.doi.org/10.1016/j.jmb.2007.11.012
- de Seny, D., Heinz, U., Wommer, S., Kiefer, M., Meyer-Klaucke, W., Galleni, M., Frere, J.M., Bauer, R. and Adolph, H.W. (2001) Metal Ion Binding and Coordination Geometry for Wild Type and Mutants of Metallo-β-Lactamase from Bacillus cereus 569/H/9 (BcII): A Combined Thermodynamic, Kinetic, and Spectroscopic Approach. Journal of Bio- logical Chemistry, 276, 45065-45078. http://dx.doi.org/10.1074/jbc.M106447200
- Abriata, L.A., González, L.J., Llarrull, L.I., Tomatis, P.E., Myers, W.K., Costello, A.L., Tierney, D.L. and Vila, A.J. (2008) Engineered Mononuclear Variants in Bacillus cereus Metallo-β-Lactamase BcII Are Inactive. Biochemistry, 47, 8590-8599. http://dx.doi.org/10.1021/bi8006912
- Yong, D., Toleman, M.A., Giske, C.G., Cho, H.S., Sundman, K., Lee, K. and Walsh, T.R. (2009) Characterization of a New Metallo-β-Lactamase Gene, blaNDM-1, and a Novel Erythromycin Esterase Gene Carried on a Unique Genetic Structure in Klebsiella pneumoniae Sequence Type 14 from India. Antimicrobial Agents and Chemotherapy, 53, 5046- 5054. http://dx.doi.org/10.1128/AAC.00774-09
- Poirel, L., Hombrouck-Alet, C., Freneaux, C., Bernabeu, S. and Nordmann, P. (2010) Global Spread of New Delhi Metallo-β-Lactamase 1. The Lancet Infectious Diseases, 10, 832. http://dx.doi.org/10.1016/S1473-3099(10)70279-6
- Rolain, J.M., Parola, P. and Cornaglia, G. (2010) New Delhi Metallo-β-Lactamase (NDM-1): Towards a New Pandemia? Clinical Microbiology and Infection, 16, 1699-1701. http://dx.doi.org/10.1111/j.1469-0691.2010.03385.x
- Zhang, H.M. and Hao, Q. (2011) Crystal Structure of NDM-1 Reveals a Common β-Lactam Hydrolysis Mechanism. The FASEB Journal, 25, 2574-2582. http://dx.doi.org/10.1096/fj.11-184036
- Walsh, T.R., Weeks, J., Livermore, D.M. and Toleman, M.A. (2011) Dissemination of NDM-1 Positive Bacteria in the New Delhi Environment and Its Implications for Human Health: An Environmental Point Prevalence Study. The Lancet Infectious Diseases, 11, 355-362. http://dx.doi.org/10.1016/S1473-3099(11)70059-7
- Li, N., Xu, Y., Xia, Q., Bai, C., Wang, T., Wang, L., He, D., Xie, N., Li, L., Wang, J., et al. (2014) Simplified Capto- pril Analogues as NDM-1 Inhibitors. Bioinorganic and Medicinal Chemistry Letters, 24, 386-389. http://dx.doi.org/10.1016/j.bmcl.2013.10.068
- Crawford, P.A., Sharma, N., Chandrasekar, S., Sigdel, T., Walsh, T.R., Spencer, J. and Crowder, M.W. (2004) Over- Expression, Purification, and Characterization of Metallo-β-Lactamase ImiS from Aeromonas veronii bv. sobria. Pro- tein Expression and Purification, 36, 272-279. http://dx.doi.org/10.1016/j.pep.2004.04.017
- Garau, G., Bebrone, C., Anne, C., Galleni, M., Frere, J.M. and Dideberg, O. (2005) A Metallo-β-Lactamase Enzyme in Action: Crystal Structures of the Monozinc Carbapenemase CphA and Its Complex with Biapenem. Journal of Molecular Biology, 345, 785-795. http://dx.doi.org/10.1016/j.jmb.2004.10.070
- Xu, D.G., Zhou, Y.Z., Xie, D.Q. and Guo, H. (2005) Antibiotic Binding to Monozinc CphA β-Lactamase from Aeromonas hydropila: Quantum Mechanical/Molecular Mechanical and Density Functional Theory Studies. Journal of Medicinal Chemistry, 48, 6679-6689. http://dx.doi.org/10.1021/jm0505112
- Simona, F., Magistrato, A., Dal Peraro, M., Cavalli, A., Vila, A.J. and Carloni, P. (2009) Common Mechanistic Features among Metallo-β-Lactamases: A Computational Study of Aeromonas hydrophila CphA Enzyme. Journal of Biological Chemistry, 284, 28164-28171. http://dx.doi.org/10.1074/jbc.M109.049502
- Bebrone, C., Delbrück, H., Kupper, M.B., Schlömer, P., Willmann, C., Frère, J.M., Fischer, R., Galleni, M. and Hoffmann, K.M. (2009) The Structure of the Dizinc Subclass B2 Metallo-β-Lactamase CphA Reveals that the Second Inhibitory Zinc Ion Binds in the Histidine Site. Antimicrobial Agents and Chemotherapy, 53, 4464-4471. http://dx.doi.org/10.1128/AAC.00288-09
- Fonseca, F., Bromley, E.H.C., Saavedra, M.J., Correia, A. and Spencer, J. (2011) Crystal Structure of Serratia fonticola Sfh-I: Activation of the Nucleophile in Mono-Zinc Metallo-β-Lactamases. Journal of Molecular Biology, 411, 951-959. http://dx.doi.org/10.1016/j.jmb.2011.06.043
- Ullah, J.H., Walsh, T.R., Taylor, I.A., Emery, D.C., Verma, C.S., Gamblin, S.J. and Spencer, J. (1998) The Crystal Structure of the L1 Metallo-β-Lactamase from Stenotrophomonas maltophilia at 1.7 A Resolution. Journal of Mole- cular Biology, 284, 125-136. http://dx.doi.org/10.1006/jmbi.1998.2148
- Hu, Z., Periyannan, G., Bennett, B. and Crowder, M.W. (2008) Role of the Zn1 and Zn2 Sites in Metallo-β-Lactamase L1. Journal of the American Chemical Society, 130, 14207-14216. http://dx.doi.org/10.1021/ja8035916
- Hu, Z., Periyannan, G.R. and Crowder, M.W. (2008) Folding Strategy to Prepare Co(II)-Substituted Metallo-β-Lacta- mase L1. Analytical Biochemistry, 378, 177-183. http://dx.doi.org/10.1016/j.ab.2008.04.007
- Garcia-Saez, I., Mercuri, P.S., Papamicael, C., Kahn, R., Frere, J.M., Galleni, M., Rossolini, G.M. and Dideberg, O. (2003) Three-Dimensional Structure of FEZ-1, a Monomeric Subclass B3 Metallo-β-Lactamase from Fluoribacter gormanii, in Native Form and in Complex with D-Captopril. Journal of Molecular Biology, 325, 651-660. http://dx.doi.org/10.1016/S0022-2836(02)01271-8
- Docquier, J.D., Benvenuti, M., Calderone, V., Stoczko, M., Menciassi, N., Rossolini, G.M. and Mangani, S. (2010) High-Resolution Crystal Structure of the Subclass B3 Metallo-β-Lactamase BJP-1: Rational Basis for Substrate Speci- ficity and Interaction with Sulfonamides. Antimicrobial Agents and Chemotherapy, 54, 4343-4351. http://dx.doi.org/10.1128/AAC.00409-10
- Miraula, M., Brunton, C.S., Schenk, G. and Mitic, N. (2013) Identification and Preliminary Characterization of Novel B3-Type Metallo-β-Lactamases. American Journal of Molecular Biology, 3, 198-203. http://dx.doi.org/10.4236/ajmb.2013.34026
- Wachino, J.I., Yoshida, H., Yamane, K., Suzuki, S., Matsui, M., Yamagishi, T., Tsutsui, A., Konda, T., Shibayama, K. and Arakawa, Y. (2011) SMB-1, a Novel Subclass B3 Metallo-β-Lactamase, Associated with ISCR1 and a Class 1 Integron, from a Carbapenem-Resistant Serratia marcescens Clinical Isolate. Antimicrobial Agents and Chemotherapy, 55, 5143-5149. http://dx.doi.org/10.1128/AAC.05045-11
- Stoczko, M., Frère, J.M., Rossolini, G.M. and Docquier, J.D. (2008) Functional Diversity among Metallo-β-Lacta- mases: Characterization of the CAR-1 Enzyme of Erwinia carotovora. Antimicrobial Agents and Chemotherapy, 52, 2473-2479. http://dx.doi.org/10.1128/AAC.01062-07
- Docquier, J.D., Lopizzo, T., Liberatori, S., Prenna, M., Thaller, M.C., Frere, J.M. and Rossolini, G.M. (2004) Biochemical Characterization of the THIN-B Metallo-β-Lactamase of Janthinobacterium lividum. Antimicrobial Agents and Chemotherapy, 48, 4778-4783. http://dx.doi.org/10.1128/AAC.48.12.4778-4783.2004
- Concha, N.O., Rasmussen, B.A., Bush, K. and Herzberg, O. (1996) Crystal Structure of the Wide-Spectrum Binuclear Zinc β-Lactamase from Bacteroides fragilis. Structure, 4, 823-836. http://dx.doi.org/10.1016/S0969-2126(96)00089-5
- Wang, Z., Fast, W., Valentine, A.M. and Benkovic, S.J. (1999) Metallo-β-Lactamase: Structure and Mechanism. Current Opinion in Chemical Biology, 3, 614-622. http://dx.doi.org/10.1016/S1367-5931(99)00017-4
- Paul-Soto, R., Bauer, R., Frere, J.M., Galleni, M., Meyer-Klaucke, W., Nolting, H., Rossolini, G.M., de Seny, D., Valladares, M., Zeppezauer, M. and Adolf, H.W. (1999) Mono- and Binuclear Zn2+-β-Lactamase. Role of the Conserved Cysteine in the Catalytic Mechanism. Journal of Biological Chemistry, 274, 13242-13249. http://dx.doi.org/10.1074/jbc.274.19.13242
- Nauton, L., Khan, R., Garau, G., Hernandez, J.F. and Dideberg, O. (2008) Structural Insights into the Design of Inhi- bitors for the L1 Metallo-β-Lactamase from Stenotrophomonas maltophilia. Journal of Molecular Biology, 375, 257- 269. http://dx.doi.org/10.1016/j.jmb.2007.10.036
- Heinz, U., Bauer, R., Wommer, S., Meyer-Klaucke, W., Papamichaels, C., Bateson, J. and Adolph, H.W. (2003) Coor- dination Geometries of Metal Ions in D- or l-Captopril-Inhibited Metallo-β-Lactamases. Journal of Biological Chemi- stry, 278, 20659-20666. http://dx.doi.org/10.1074/jbc.M212581200
- Garcia-Saez, I., Hopkins, J., Papamicael, C., Franceschini, N., Amicosante, G., Rossolini, G.M., Galleni, M., Frere, J.M. and Dideberg, O. (2003) The 1.5-A Structure of Chryseobacterium meningosepticum Zinc β-Lactamase in Complex with the Inhibitor, D-Captopril. Journal of Biological Chemistry, 278, 23868-23873. http://dx.doi.org/10.1074/jbc.M301062200
- Wachino, J., Yamaguchi, Y., Mori, S., Kurosaki, H., Arakawa, Y. and Shibayama, K. (2013) Structural Insights into the Subclass B3 Metallo-β-Lactamase SMB-1 and the Mode of Inhibition by the Common Metallo-β-Lactamase Inhibitor Mercaptoacetate. Antimicrobial Agents and Chemotherapy, 57, 101-109. http://dx.doi.org/10.1128/AAC.01264-12
- Mohamed, M.S., Hussein, W.M., McGeary, R.P., Vella, P., Schenk, G. and Abd El-hameed, R.H. (2011) Synthesis and Kinetic Testing of New Inhibitors for a Metallo-β-Lactamase from Klebsiella pneumonia and Pseudomonas aeruginosa. European Journal of Medicinal Chemistry, 46, 6075-6082. http://dx.doi.org/10.1016/j.ejmech.2011.10.030
- Faridoon, F.F., Hussein, W.M., Vella, P., Islam, N.U., Ollis, D.L., Schenk, G. and McGeary, R.P. (2012) 3-Mercapto- 1,2,4-Triazoles and N-Acylated Thiosemicarbazides as Metallo-β-Lactamase Inhibitors. Bioorganic and Medicinal Chemistry Letters, 22, 380-386. http://dx.doi.org/10.1016/j.bmcl.2011.10.116
- Hussein, W.M., Fatahala, S.S., Mohamed, Z.M., McGeary, R.P., Schenk, G., Ollis, D.L. and Mohamed, M.S. (2012) Synthesis and Kinetic Testing of Tetrahydropyrimidine-2-Thione and Pyrrole Derivatives as Inhibitors of the Metallo- β-Lactamase from Klebsiella pneumonia and Pseudomonas aeruginosa. Chemical Biology and Drug Design, 80, 500- 515. http://dx.doi.org/10.1111/j.1747-0285.2012.01440.x
- McGeary, R.P., Schenk, G. and Guddat, L.W. (2014) The Applications of Binuclear Metallohydrolases in Medicine: Recent Advances in the Design and Development of Novel Drug Leads for Purple Acid Phosphatases, Metallo-β-Lac- tamases and Arginases. European Journal of Medicinal Chemistry, 76, 132-144. http://dx.doi.org/10.1016/j.ejmech.2014.02.008
- Murphy, T.A., Catto, L.E., Halford, S.E., Hadfield, A.T., Minor, W., Walsh, T.R. and Spencer, J. (2006) Crystal Structure of Pseudomonas aeruginosa SPM-1 Provides Insights into Variable Zinc Affinity of Metallo-β-Lactamases. Journal of Molecular Biology, 357, 890-903. http://dx.doi.org/10.1016/j.jmb.2006.01.003
- Jacquin, O., Balbeur, D., Damblon, C., Marchot, P., De Pauw, E., Roberts, G.C., Frere, J.M. and Matagne, A. (2009) Positively Cooperative Binding of Zinc Ions to Bacillus cereus 569/H/9 β-Lactamase II Suggests that the Binuclear Enzyme Is the Only Relevant Form for Catalysis. Journal of Molecular Biology, 392, 1278-1291. http://dx.doi.org/10.1016/j.jmb.2009.07.092
- Yang, Y., Rasmussen, B.A. and Bush, K. (1992) Biochemical Characterization of the Metallo-β-Lactamase CcrA from Bacteroides fragilis TAL3636. Antimicrobial Agents and Chemotherapy, 36, 1155-1157. http://dx.doi.org/10.1128/AAC.36.5.1155
- Crowder, M.W., Wang, Z., Franklin, S., Zovinka, E.P. and Benkovic, S.J. (1996) Characterization of the Metal-Binding Sites of the β-Lactamase from Bacteroides fragilis. Biochemistry, 35, 12126-12132. http://dx.doi.org/10.1021/bi960976h
- Yanchak, M.P., Taylor, R.A. and Crowder, M.W. (2000) Mutational Analysis of Metallo-β-Lactamase CcrA from Bacteroides fragilis. Biochemistry, 39, 11330-11339. http://dx.doi.org/10.1021/bi0010524
- Park, H., Brothers, E.N. and Merz, K.M. (2005) Hybrid QM/MM and DFT Investigations of the Catalytic Mechanism and Inhibition of the Dinuclear Zinc Metallo-β-Lactamase CcrA from Bacteroides fragilis. Journal of the American Chemical Society, 127, 4232-4241. http://dx.doi.org/10.1021/ja042607b
- Hawk, M.J., Breece, R.M., Hajdin, C.E., Bender, K.M., Hu, Z.X., Costello, A.L., Bennett, B., Tierney, D.L. and Crowder, M.W. (2009) Differential Binding of Co(II) and Zn(II) to Metallo-β-Lactamase Bla2 from Bacillus anthracis. Journal of the American Chemical Society, 131, 10753-10762. http://dx.doi.org/10.1021/ja900296u
- Chen, R.F. (1967) Fluorescence Quantum Yields of Tryptophan and Tyrosine. Analytical Letters, 1, 35-42. http://dx.doi.org/10.1080/00032716708051097
- Hunt, J.B., Neece, S.H. and Ginsburg, A. (1985) The Use of 4-(2-pyridylazo) Resorcinol in Studies of Zinc Release from Escherichia coli Aspartate Transcarbamoylase. Analytical Biochemistry, 146, 150-157. http://dx.doi.org/10.1016/0003-2697(85)90409-9
- Eftink, M.R. (1991) Fluorescence Techniques for Studying Protein Structure. Methods of Biochemical Analysis, 35, 127-205. http://dx.doi.org/10.1002/9780470110560.ch3
- Simons, T.J.B. (1993) Measurement of Free Zn2+ Ion Concentration with the Fluorescent Probe Mag-fura-2 (furaptra). Journal of Biochemical and Biophysical Methods, 27, 25-37. http://dx.doi.org/10.1016/0165-022X(93)90065-V
- McCall, K.A. and Fierke, C.A. (2000) Colorimetric and Fluorimetric Assays to Quantitate Micromolar Concentrations of Transition Metals. Analytical Biochemistry, 284, 307-315. http://dx.doi.org/10.1006/abio.2000.4706
- Valladares, M.H., Felici, A., Weber, G., Adolph, H.W., Zeppezauer, M., Rossolini, G.M., Amicosante, G., Frère, J.M. and Galleni, M. (1997) Zn(II) Dependence of the Aeromonas hydrophila AE036 Metallo-β-Lactamase Activity and Stability. Biochemistry, 36, 11534-11541. http://dx.doi.org/10.1021/bi971056h
- Wommer, S., Rival, S., Heinz, U., Galleni, M., Frere, J.M., Franceschini, N., Amicosante, G., Rasmussen, B., Bauer, R. and Adolph, H.W. (2002) Substrate-Activated Zinc Binding of Metallo-β-Lactamases: Physiological Importance of the Mononuclear Enzymes. Journal of Biological Chemistry, 277, 24142-24147. http://dx.doi.org/10.1074/jbc.M202467200
- Gonzàlez, J.M., Martìn, F.J., Costello, A.L., Tierney, D.L. and Vila, A.J. (2007) The Zn2 Position in Metallo-β-Lac- tamases Is Critical for Activity: A Study on Chimeric Metal Sites on a Conserved Protein Scaffold. Journal of Molecular Biology, 373, 1141-1156. http://dx.doi.org/10.1016/j.jmb.2007.08.031
- Selevsek, N., Rival, S., Tholey, A., Heinzle, E., Heinz, U., Hemmingsen, L. and Adolph, H.W. (2009) Zinc Ion-In- duced Domain Organization in Metallo-β-Lactamases: A Flexible “Zinc Arm” for Rapid Metal Ion Transfer? Journal of Biological Chemistry, 284, 16419-16431. http://dx.doi.org/10.1074/jbc.M109.001305
- Cricco, J.A., Orellano, E.G., Rasia, R.M., Ceccarelli, E.A. and Vila, A.J. (1999) Metallo-β-Lactamases: Does It Take Two to Tango? Coordination Chemistry Review, 190-192, 519-535. http://dx.doi.org/10.1016/S0010-8545(99)00113-7
- Badarau, A. and Page, M.I. (2008) Loss of Enzyme Activity during Turnover of the Bacillus cereus β-Lactamase Cata- lysed Hydrolysis of β-Lactams Due to Loss of Zinc Ion. Journal of Biological Inorganic Chemistry, 13, 919-928. http://dx.doi.org/10.1007/s00775-008-0379-2
- Moran-Barrio, J., Gonzalez, J.M., Lisa, M.N., Costello, A.L., Peraro, M.D., Carloni, P., Bennett, B., Tierney, D.L., Limansky, A.S., Viale, A.M. and Vila, A.J. (2007) The Metallo-β-Lactamase GOB Is a Mono-Zn(II) Enzyme with a Novel Active Site. Journal of Biological Chemistry, 282, 18286-18293. http://dx.doi.org/10.1074/jbc.M700467200
- Horsfall, L.E., Izougarhane, Y., Lassaux, P., Selevsek, N., Lienard, B.M.R., Poirel, L., Kupper, M.B., Hoffmann, K.M., Frere, J.-M., Galleni, M. and Bebrone, C. (2011) Broad Antibiotic Resistance Profile of the Subclass B3 Metallo-b- Lactamase GOB-1, a Di-Zinc Enzyme. FEBS Journal, 278, 1252-1263.
NOTES
*These authors contributed equally.
#Corresponding author.