Natural Science
Vol.4 No.11(2012), Article ID:24855,12 pages DOI:10.4236/ns.2012.411115
A two-dimensional electrophoresis reference map of the earthworm Eisenia fetida
![]()
1College of Resources and Environmental Sciences, China Agricultural University, Beijing, China; *Corresponding Author: Hsunzj108@sina.com
2School of Life Sciences, Huaibei Normal University, Huaibei, China
Received 29 September 2012; revised 30 October 2012; accepted 14 November 2012
Keywords: 2-DE; Invertebrates; Animal Proteomics; Eisenia fetida; MALDI-TOF/TOF-MS
ABSTRACT
The molecular mechanisms underlying innate immunity in the earthworm E. fetida remain unclear. For the recognition of innate immunity in the earthworm E. fetida, a detailed knowledge of this proteome is a prerequisite. The absence of a high-resolution E. fetida proteome map prompted us to determine E. fetida protein spots that can be visualised on 2-D protein gels. In this study, we present a preliminary description of the whole earthworm E. fetida proteome. A highly detailed two-dimensional gel electrophoresis (2-DE) map of the E. fetida proteome was established and approximately 1500 protein spots were detected from the earthworm sample when applying a 500 µg protein 2-DE in the pH range 3.0 - 10.0. We present a 2-DE proteome map of E. fetida, identifying 76 different proteins by matrix-assisted laser desorption/ionization-tandem time of flight mass spectrometry (MALDI-TOF/TOFMS) analysis. These identified proteins, including heat shock protein 90 (Hsp90), chaperonine protein HSP60, caspase-8, fibrinolytic protease 0, gelsolin-like protein, lombricine kinase, coelomic cytolytic factor1 (CCF 1), manganous superoxide dismutase (MnSOD), triosephosphate isomerase, extracellular globin-4, lysenin, and intermediate filament protein, glyceraldehyde-3- phosphate dehydrogenase, et al., are involved in several processes, including transcripttion, translation, the tricarboxylic acid cycle, the cellular amino acid metabolic process, protein amino acid phosphorylation, glycolysis, and the glucose metabolic process. These 2-DE data will enhance future comparisons of immunity, toxicology, biotic processes and other challenges, thereby allowing for further study of the molecular mechanisms in response to environmental stressors, and it will be useful to investigate environmental proteomics in invertebrate earthworms.
1. INTRODUCTION
As a key representative of soil fauna, earthworms are essential in maintaining soil fertility through their burrowing, ingestion, and excretion [1]. Earthworms are increasingly recognized as indicators of agroecosystem health and an ecotoxicological sentinel species because they are constantly exposed to soil contaminants [2]. Working with earthworms presents some common challenge: none has a fully sequenced genome; they are often so small as to preclude ready dissection of internal organs or tissues [3]. Modern genomic approaches offer a potential opportunity to circumvent some of these drawbacks [4-9]. This study aims at creating a proteome reference map for to be used for the study of its ecotoxicoproteomics.
Proteomics is a large-scale study of the proteins that gene transcription and translation, which ultimately provides direct measurement of protein expression levels and insight into the activity state of all relevant proteins [10]. Like in many other ecologically important species, proteomics research in earthworms lags far behind other model species, such as Mus musculus and Caenorhabditis elegans. There are only five research papers about proteomics of earthworm until now [11-15]. The key technologies of the proteome analysis are protein separation by 2-DE and mass spectrometric protein identification, which result in the efficient analysis of hundreds to thousands of proteins expressed in cells at one time [16].
Protein identification of earthworm is difficulty for most of proteins in earthworm have unknown function. The proteomics map of the earthworm E. fetida will provide information on earthworm proteomics that will be useful in our knowledge of the innate immunity, ecotoxicology, environmental science, et al.
2. MATERIALS AND METHODS
2.1. The Experimental Organism and Preparation of an Artificial Soil
Adult earthworm species E. fetida (weighing between 300 and 400 mg) reared in composted cattle feces were purchased from an earthworm farm of the China Agricultural University and kept in test substrates for a week before the experiment. Artificial substrates that used in laboratory and could maintain the same conditions were prepared as described in the OECD (1984) guidelines [17]. Briefly, the substrate was composed (percentages refer to dry weight) of 10% sphagnum, 20% kaolinite clay, and 70% quartz sand. The pH was adjusted to 6.0 ± 0.5 with CaCO3. The artificial climate chamber was maintained at 23˚C ± 1˚C and a lighting of about 1000 lx with a 12/12-h photo-period; water was supplemented to ensure appropriate water content of the substrate by weighing the containers. Two days before the experiments, the earthworms were transferred onto filter paper soaked with isotonic Lumbricus balanced salt solution12 at 22˚C ± 1˚C to void the gut load. Whole E. fetida were stored at –80˚C for protein extraction. The treatment was repeated three times for each sample (three biological replicates).
2.2. Protein Extraction
Proteins were extracted according to the TCA-A procedure described previously [13]. Briefly, fresh whole E. fetida samples (2 g) collected from three biologically replications, three protein samples, and each sample made from three worms were pulverized to a fine powder with liquid nitrogen and a mortar and pestle. The powder was resuspended and the homogenate was precipitated overnight with 10 ml protein extraction buffer (acetone/20% TCA/0.1%DTT) (–20˚C). The protein pellets were washed with ice-cold methanol followed by multiple ice-cold acetone washes, dried in a vacuum freeze-drying machine, and redissolved in IEF buffer (7 M urea, 2 M thiourea, 2% CHAPS, 65 mM DTT, and 0.5% w/v carrier ampholytes (nonlinear pH 3 - 10; Amersham Biosciences, Piscataway, NJ, USA). Protein concentrations were quantified using the 2D-Quant kit protein assay (GE Healthcare Biosciences, Milwaukee, WI, USA). The supernatants were stored in aliquots at –80˚C or directly loaded for isoelectric focusing.
2.3. IEF and SDS-PAGE
IPG strips (24 cm, nonlinear pH 3 - 10, GE Healthcare Biosciences Immobiline DryStrips) were rehydrated overnight with 450 µl IEF buffer containing an estimated 500 µg protein and focused using a Ettan IPGphor Multiphor III (GE Healthcare Biosciences) at 20˚C, applying the following program: 30 V for 6 h, 60 V for 6 h, 100 V for 1 h, 200 V for 1 h, 500 V gradient for 1 h, 1,000 V gradient for 30 min, 5,000 V gradient for 2 h, 10,000 V for 1 h, and 10,000 V for 60,000 V·h. After focusing, the proteins reduction were in equilibration buffer (6 M urea, 29.3% w/v glycerol, 2% SDS, and 1.5 M Tris-HCl, pH 8.8) + 1% w/v DTT for 15 min and alkylation followed this in a separate incubation for an additional 15 min in equilibration buffer + 2.5% w/v IAA. The strips were then transferred to 12.5% SDS-PAGE gels for 2-DE using Amersham’s Ettan DALTsix gel system with SDS electrophoresis buffer [250 mM Tris (pH 8.3), 1.92 M glycine, and 1% w/v SDS)] and a 2 W/gel for 20 min and a 16.7 W/gel for 5 h. At least three gels were run for each sample.
2.4. Image Analysis
Protein spots were detected by Coomassie Blue G-250 staining [18]. After the 2-DE run, gel cassettes were opened and the gels carefully removed and placed in fixing solution (10% v/v acetic acid, 40% ethanol) for 1.5 h. Gels were then stained using Coomassie solution (0.1% w/v CBB G-250, 1.6% w/v o-phosphoric acid, 10% (NH4)2SO4, and 20% ethanol) overnight. The gels were then washed with deionized water until the gel background was clear. Stained gels were scanned and calibrated using LabScan 5 software (Amersham Biosciences). Image analysis was performed using ImageMasterTM 2D Platinum 7 (GE Healthcare Amersham Biosciences). Detection and matching of the protein spots was facilitated by the use of software and re-evaluated by visual inspection. Three well separated gels of each sample were used to create “replicate groups”.
2.5. Protein Identification and Database Search
Protein spots of interest were manually excised from the 2-D gels. The gel plugs were washed three times with ultrapure water, destained twice with 100 mM NH4HCO3 in 50% acetonitrile, reduced with 10 mM DTT in 100 mM NH4HCO3 for 1 h at 56˚C and incubated with 40 mM IAA in 100 mM NH4HCO3 for 45 min., dried twice with 100% acetonitrile, and digested overnight at 37˚C with 0.1 mg/ml trypsin (Promega, Madison, WI, USA) for 1:40 (w/w) (trypsin:protein) according to manufacturer’s instructions. The resulting peptides were subjected to sequential extraction (3 times at 37˚C) with 8 µl each of 5% trifluoroacetic acid (TFA) for 1 h, 2.5% TFA in 50% ACN for 1 h, and 100% ACN for 1 h. Extracted peptides were dried in a vacuum centrifugation and stored at –80˚C. The digests were mixed with a saturation solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile/0.1% trifluoroacetic acid and spotted on a MALDI target plate.
Peptide masses were measured using a MALDI-TOF/ TOF ultraflex mass spectrometer (Bruker Daltonics, Billerica, MA, USA). Mass spectra were recorded in positive reflector mode at a maximum accelerating potential of 25 kV in both manual and automatic modes. Internal calibration of the standard spectra was performed after every 10 consecutive spectra using Pepmix peptide calibration standards (Bruker Daltonics). The spectra were analyzed and a peak list was generated using XTOF/ XMASS spectrum analysis software, prior to database searching using MASCOT software (Matrix Science, London, UK). The peptide mass data were analyzed for corresponding protein matching in the NCBInr database with settings of 100 ppm mass accuracy and 0 to 2 peptide cleavage sites in the MASCOT search engine. Positive identification was considered only with a high MASCOT score, maximum peptide coverage, and additional experimental confirmation of the Mr and pI of the protein based on the 2-DE studies. The database was set to NCBInr (released on September 20, 2011; 9363125 sequences; 3207829265 residues). Search parameters included: 1) MS/MS Ion Search, as the type of search; 2) Trypsin, as the enzyme of protein digestion; 3) Carbamidomethyl (C) and Oxidation (M), as Variable modifications; 4) MONOISOTOPIC, as Mass value; 5) Unrestricted, as Protein Mass; 6) ±50 ppm, as Peptide Mass Tolerance; 7) 0.25 Da, as Fragment Mass Tolerance; 8) 1, as Max Missed Cleavages; and 9) MALDI-TOF-TOF, as Instrument type. The identification was considered with MASCOT score, total ion C.I.%. The minimum MASCOT score was 45, the minimum total ion C.I.% for positive identification was 95%, the minimum number of unique peptides was 1. The experimental Mr of the protein spots was calculated using ladder. The experimental pI of the protein spots was calculated from their position on the IEF strips as mentioned by the manufacturer’s specification (GE Healthcare).
Functional characterization of the identified proteins was determined by gene ontology tool available at Hhttp://www.uniprot.orgH. Due probably to the lack of resolution sometimes associated with 2-DE, the MASCOT searches for some protein spots occasionally gave several confident identifications [19]. Only the top scoring proteins identified within each sample are reported in the Results section.
3. RESULTS
The proteome of the whole earthworms E. fetida was profiled using 2-DE. Multiple 2-DE gels were acquired, and we present a 2-DE map of the earthworms E. fetida (Figure 1). Analytical and biological variances were calculated for triplicate 2-DE gels obtained for similar but independent culture conditions. MALDI-TOF/TOFMS allowed the identification of the proteins from all indicated spots, corresponding to 76 different proteins. The identified proteins are listed in Table 1 according to their functional categories. These data have allowed us to establish a earthworm E. fetida database, available worldwide, that contains information on protein species identified in the gels.
The resulting master image, representing all reproduceble features, contains 1500 protein spots in the range pH 3 - 10. We excised 230 spots from gels and a total of 76 spots were identified after tryptic digestion by MALDITOF/TOF-MS. Representative MALDI-TOF/TOF-MS data in Figure 2, and a database search result are shown in Figure 3. However, it cannot be ruled out that the multiple protein identifications due to streaky protein spots or due to diffusion of protein spots in close proximity. Some proteins were present as multiple spots, including lysenin, lombricine kinase, extracellular globin-4, intermediate filament protein, etc. According to the database, the identified proteins were classified by Gene Ontology Tool (Hhttp://www.uniprot.org)H.
Spot 1560 (Predicted protein XP_001503943) corresponded to an MS/MS sequences (i.e. K.AAESPPHPLEESEVLVEK.S) indicating similarity to the cardiomyopathy associated 5, which is expressed in the Equus caballus. Spot 1553 corresponded to an MS/MS sequence of R.NVSNVNYVSPVR.N. Homology searches revealed that this sequence corresponds to hypothetical protein TRIADDRAFT_28083, Trichoplax adhaerens. Thus, we have determined the most inclusive 2-DE map of the earthworms E. fetida proteins published to date.
The individual protein spots, the putative names of proteins, the organism from which the protein was identified, the MASCOT score together with the sequence coverage, the values for experimental and theoretical pI, and the molecular mass. The identified proteins could be classified into five major groups according to their biological function as shown in Figure 3: 1) Metabolism, 32 spots: seven are associated with transporter activity/ATP synthesis coupled proton transport/transferase/microtubule-based movement/synaptic transmission /regulation of GTPase activity, three other spots are involved in catalytic activity, nine spots are associated with zinc ion-binding/calcium ion-binding/ATP-binding/sugar binding/actin binding, three spots are associated with proteolysis, nine spots are associated with the tricarboxylic acid cycle/cellular amino acid metabolic process/protein amino acid phosphorylation/nucleobase, nucleoside, nucleotide and nucleic acid metabolic process/glycolysis/ glucose metabolic process, and one spot is associated with oxidation reduction; 2) Stress response related proteins, 12 spots in total: one spot is associated with electron transport, two spots are associated with oxidation
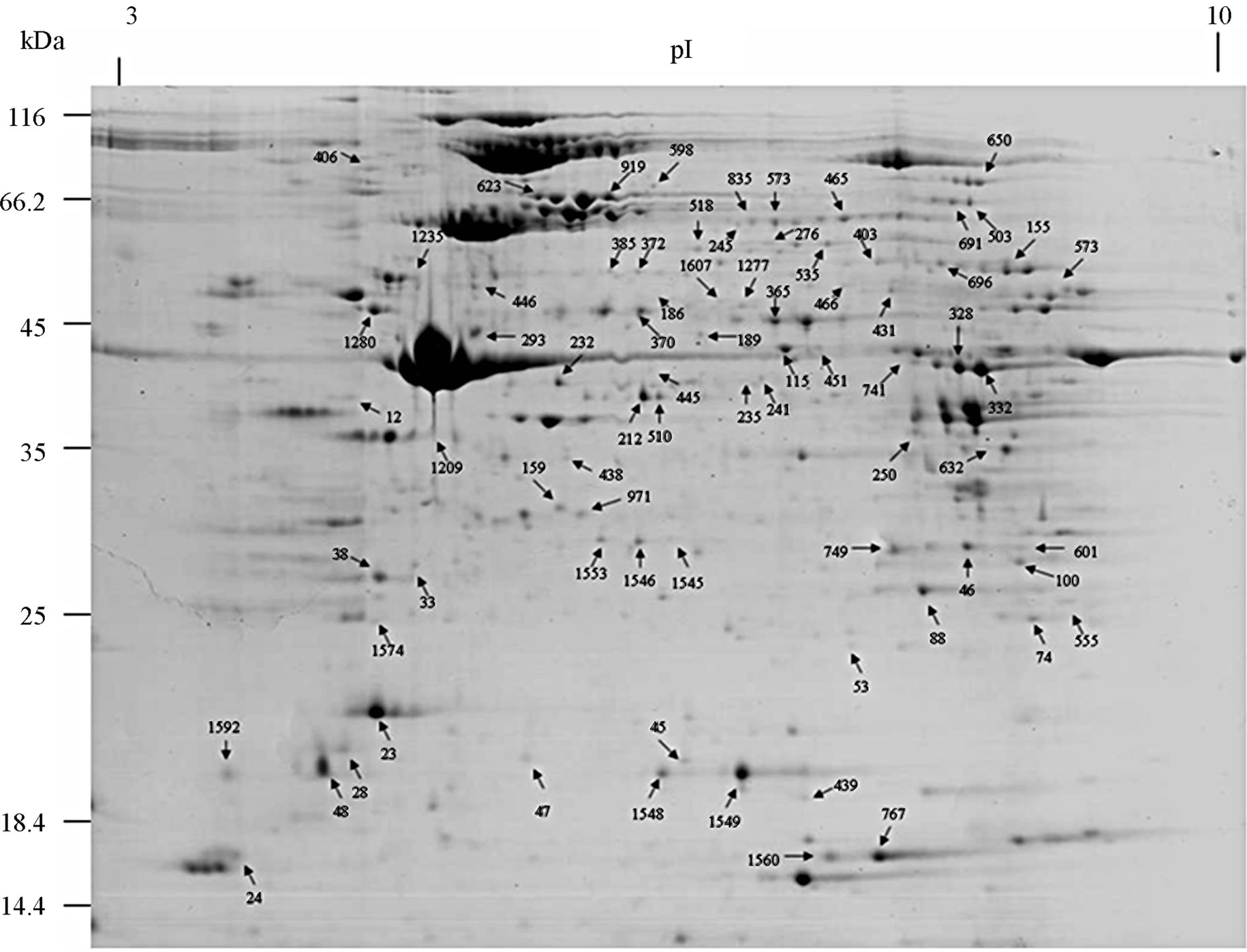
Figure 1. A representative 2-DE map of the earthworms E. fetida proteins. 2-DE was performed using 500 µg of protein, IPG strips [24 cm, nonlinear pH 3 - 10], and a 12.5% [w/v] SDS-PAGE gels for second dimension.
reduction, one spot is associated with positive regulation of apoptosis, one spot is associated with protein folding, two spots are associated with oxygen transport, two spots are associated with response to stress and oxidative stress, one spot is associated with kinase activity, and two spots are associated with actin-binding; 3) Defense-related, 4 spots: two spots are associated with lysenin, whose function is a defense response to bacteria, one spot is associated with antimicrobial peptide lumbricin 1, and one spot is associated with coelomic cytolytic factor 1, whose function is a defense response to microbial stimulation; 4) Transcription, two spots are associated with the regulation of transcription, and one spot is associated with transcription; 5) Translation, one spot is associated with the regulation of translation; 6) Signal transduction, one spot is associated with signal transduction.
4. DISCUSSION
The pattern of protein expression in the earthworms E. fetida was studied by 2-DE and Coomassie staining method. We were able to visualize and quantify around 1500 protein spots. 76 different proteins were identified by MALDI-TOF/TOF-MS analysis. We found that these spots could be classified into eight major categories according to their putative biological role (Figure 4): metabolism, stress, defense, transcription, translation, signal transduction, predicted and hypothetical, and other. Several proteins produced multiple spots, which was most likely due to posttranslational modifications (PTMs). In most cases, the shift in position is horizontal, suggesting that the modification affects only the pI without changes in molecular mass [20]. Their functional significance is discussed below.
Of the identified proteins, the most interesting group plays a role in invertebrate immunity function. Members of this group include caspase-8, manganese superoxide dismutase (MnSOD), chaperonine protein HSP60, Extracellular globin-4, gelsolin-like proteins, lysenin, coelomic cytolytic factor 1 (CCF 1). HSP60 is known to be up-regulated in the early period after Cd exposure [21]. A study reported that the potential functions of hemoglobin may include anti-oxidative defense [22]. Gelsolins all likely play an important role in altering the contractile function and motility of vascular smooth muscle [23]. Lysenin, which has been shown to cause
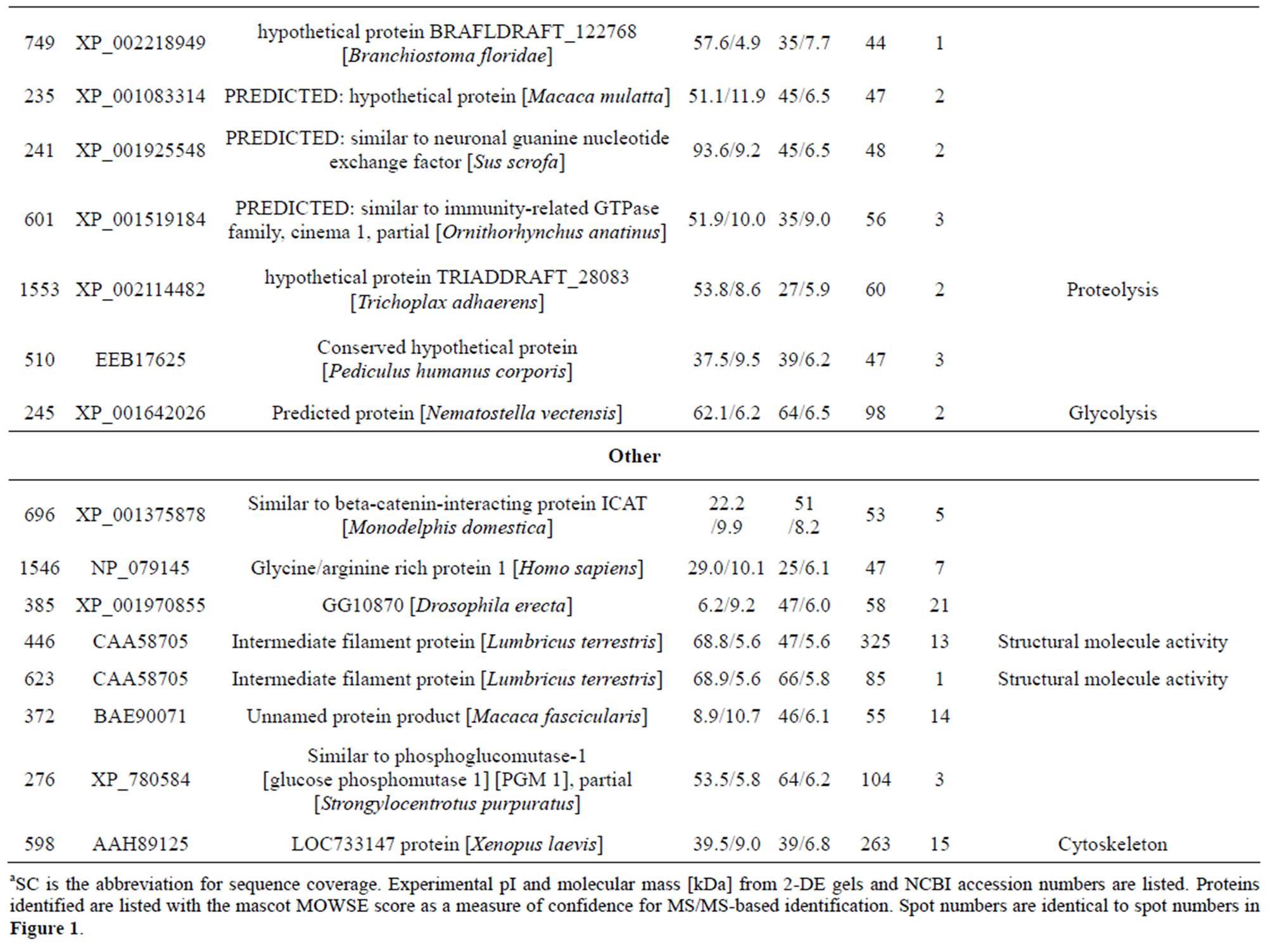
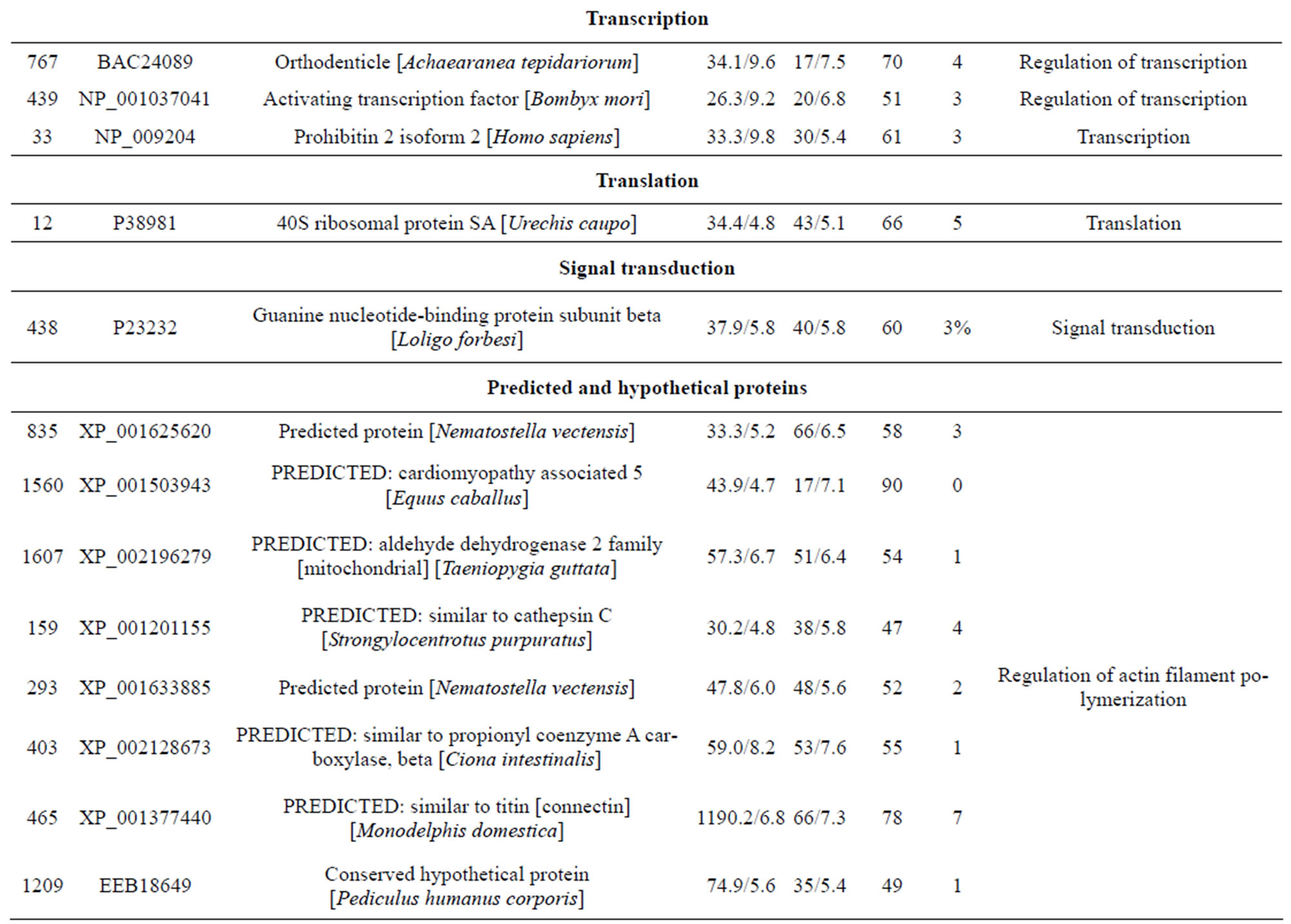
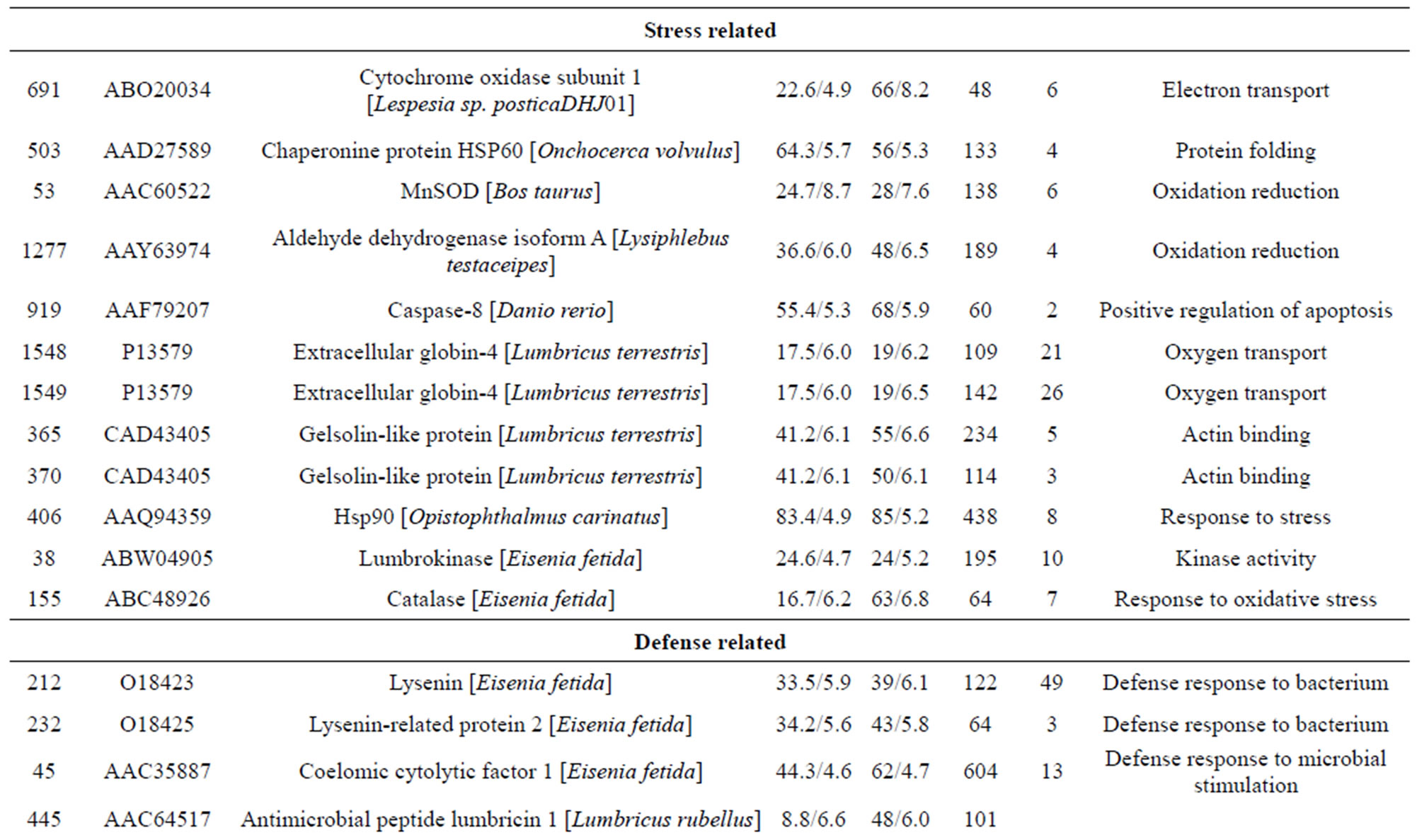
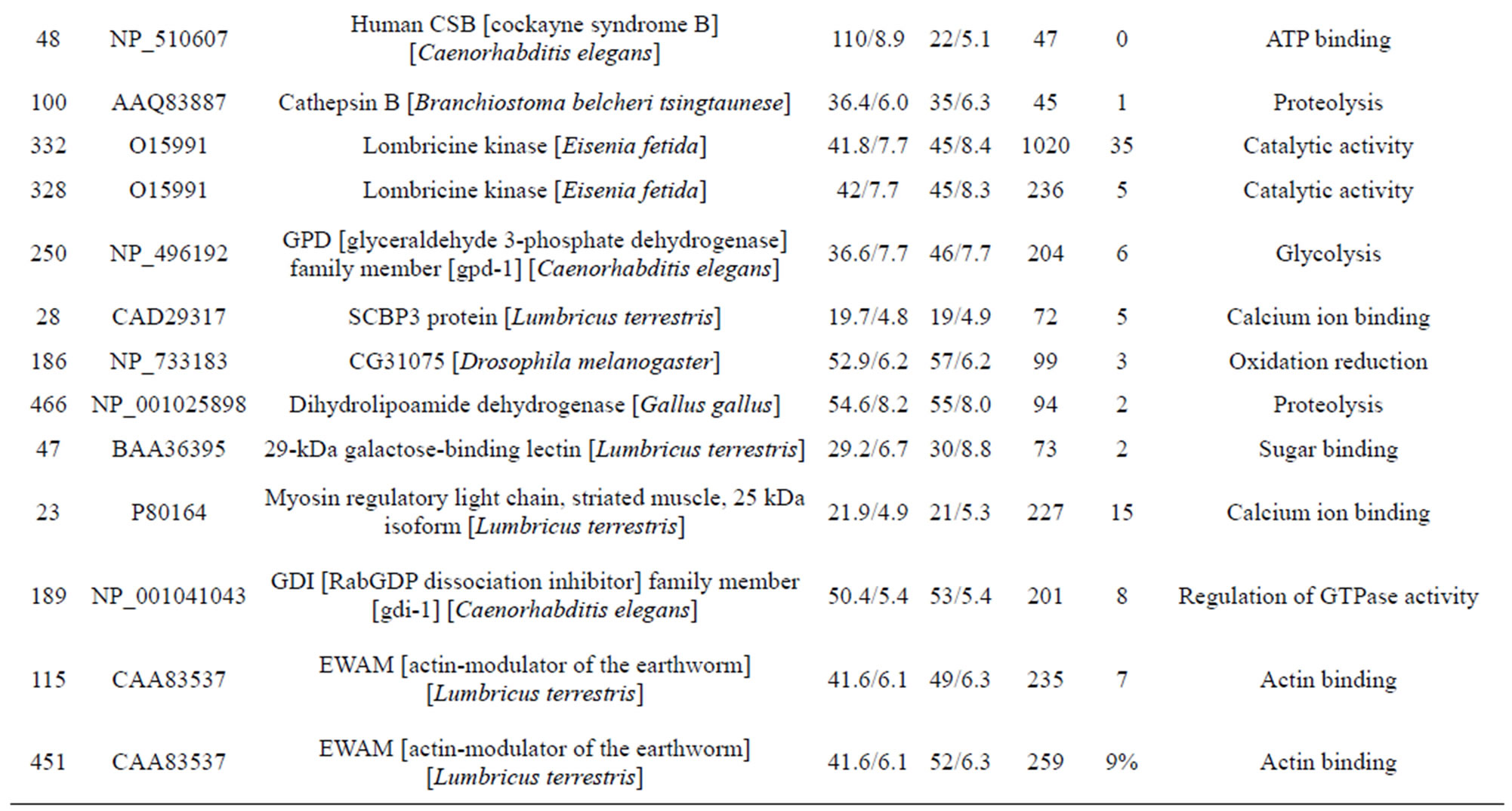
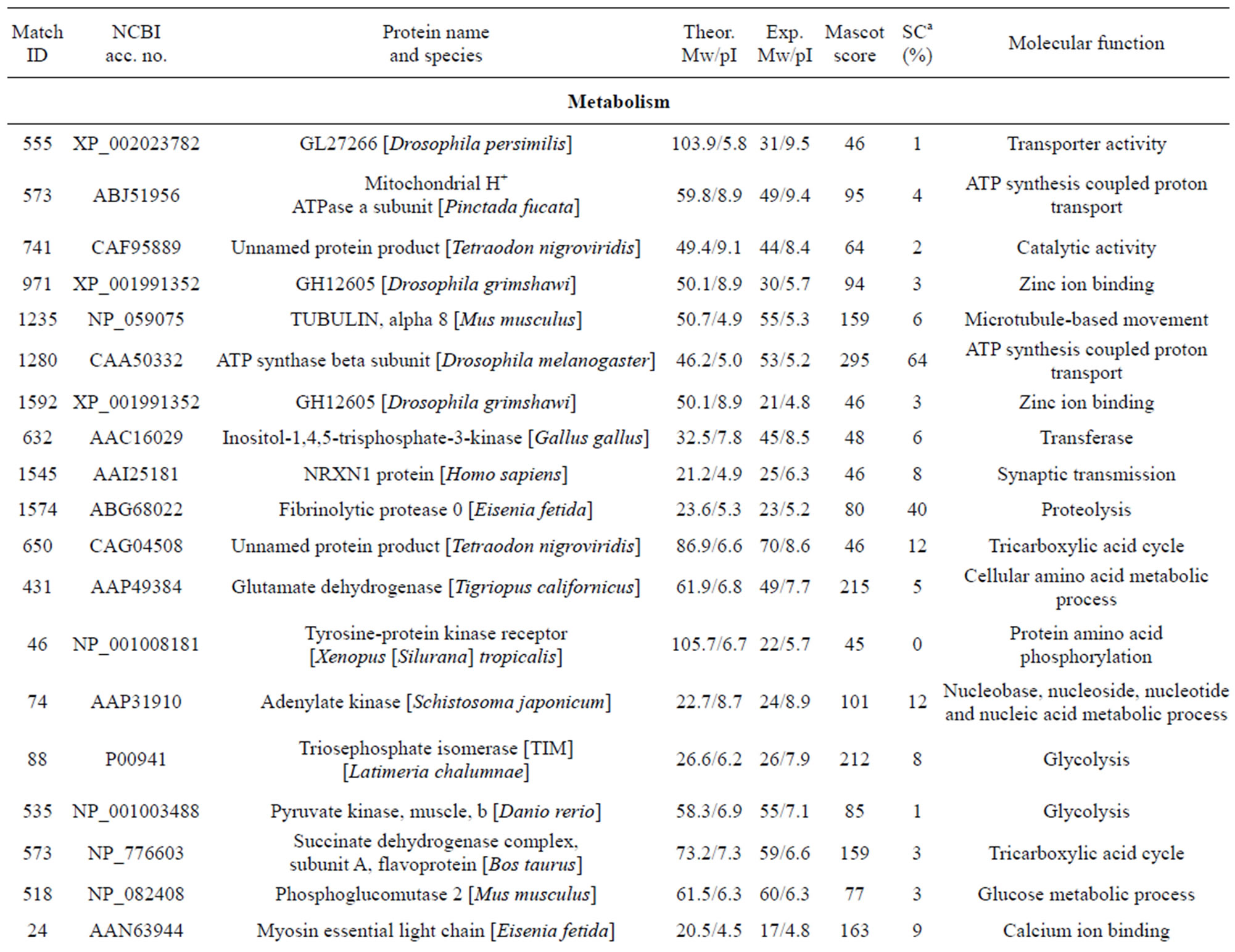
Table 1. List of identified proteins grouped in functional categories.
contraction of rat vascular smooth muscles [24]. The cytotoxic effects of lysenin seem to be due to membrane damage that is caused by the binding of lysenin to SM in the plasma membrane [25]. Lysenin produced two spots (232 and 212) most likely due to PTMs. In annelids, CCF is a defense molecule with functional analogies to mammalian tumor necrosis factor (TNF), which plays an important role in regulating immune responses [26,27]. In invertebrates, the orthodenticle gene is involved in regulating morphogens by activating transcription [28].
One spot, corresponding to a predicted mitochondrial aldehyde dehydrogenase 2 family protein. Moreover, aldehyde dehydrogenase was reported as inducible in response to various environmental stress conditions in mice [29]. McKim et al. found that apo-MT was degraded by cathepsins. Another predicted protein spot, identified as cinema1 belonging to the immunity-related GTPase family [30]. The powerful biological activity of the immunity-related GTPases (IRG) against a range of pathogens has been extensively reviewed [31]. Djabali et al. found that the intracellular state of alphaB-crystallin seemed to correlate directly with the remodeling of the intermediate filament network in response to stress [32]. One identified protein lombricine kinase which plays key roles in the coupling of energy production and utilization in animals [33]. Fibrinolytic proteases dissolve blood fibrin clots, and are important chemotherapeutic agents [34]. One spot (spot 919), identified as caspase-8, correspond to enzymes that has been demonstrated to play a key role in mediating Fas-induced apoptosis [35]. Apoptosis is executed by caspases, a family of proteases that disassemble a cell [36]. Caspase-8 is an initiator of the downstream apoptotic process that includes the activetion of Casp3, -6, and -7 and mitochondrial damage [37]. One spot (spot 155) corresponding to catalase which is important for peroxide and radical detoxification [38]. Catalase activity was studied as possible biomarkers of toxic stress in earthworms [39]. One spot (spot 406), identified as HSP90, Hsp90 is known to be involved in the inhibition of stress-induced apoptotic cell death and is a ubiquitous molecular chaperone found in eubacteria and all branches of Eukarya [40]. It plays a central role in cellular signaling because it is essential for maintaining the activity of key signaling factors, including steroid
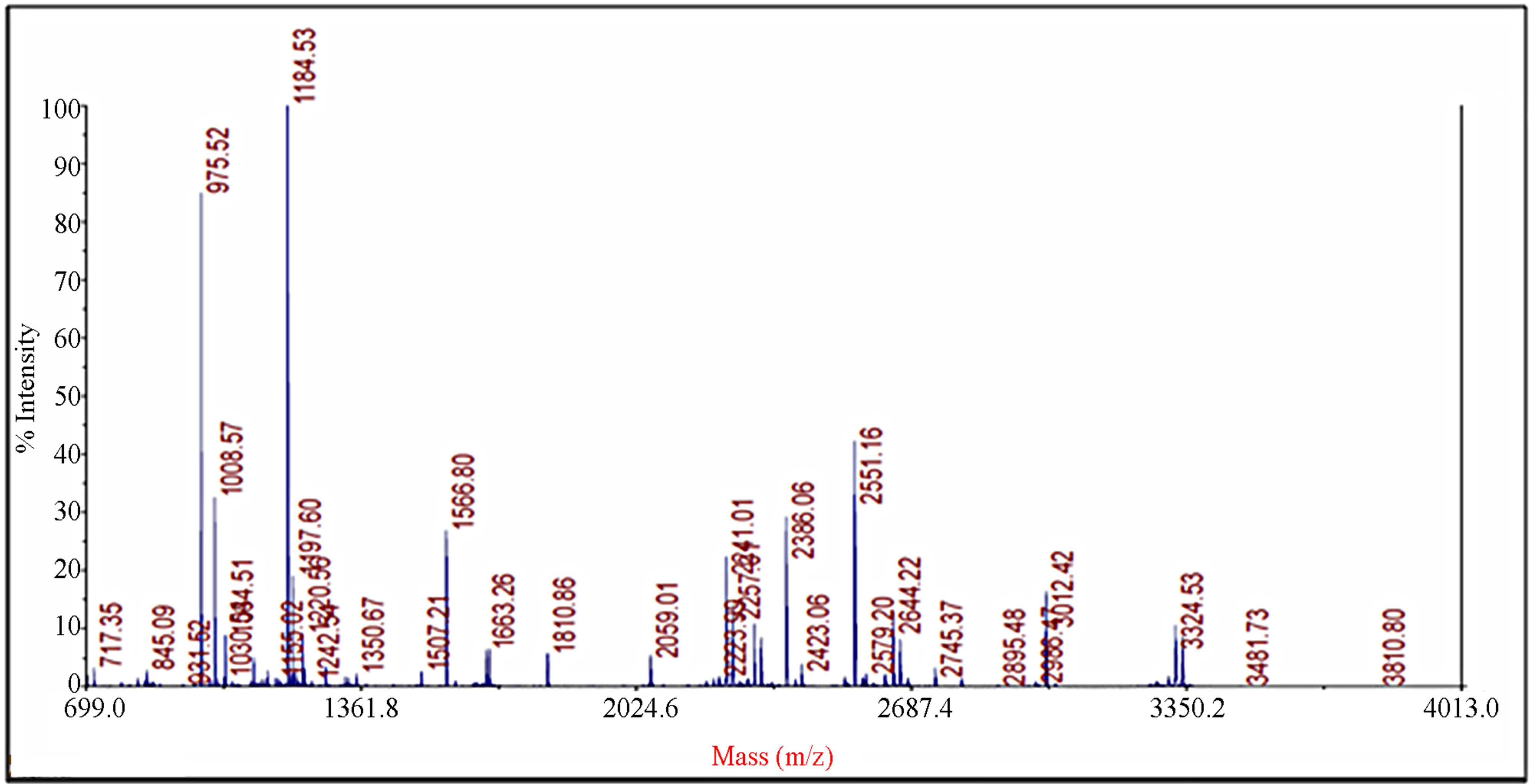
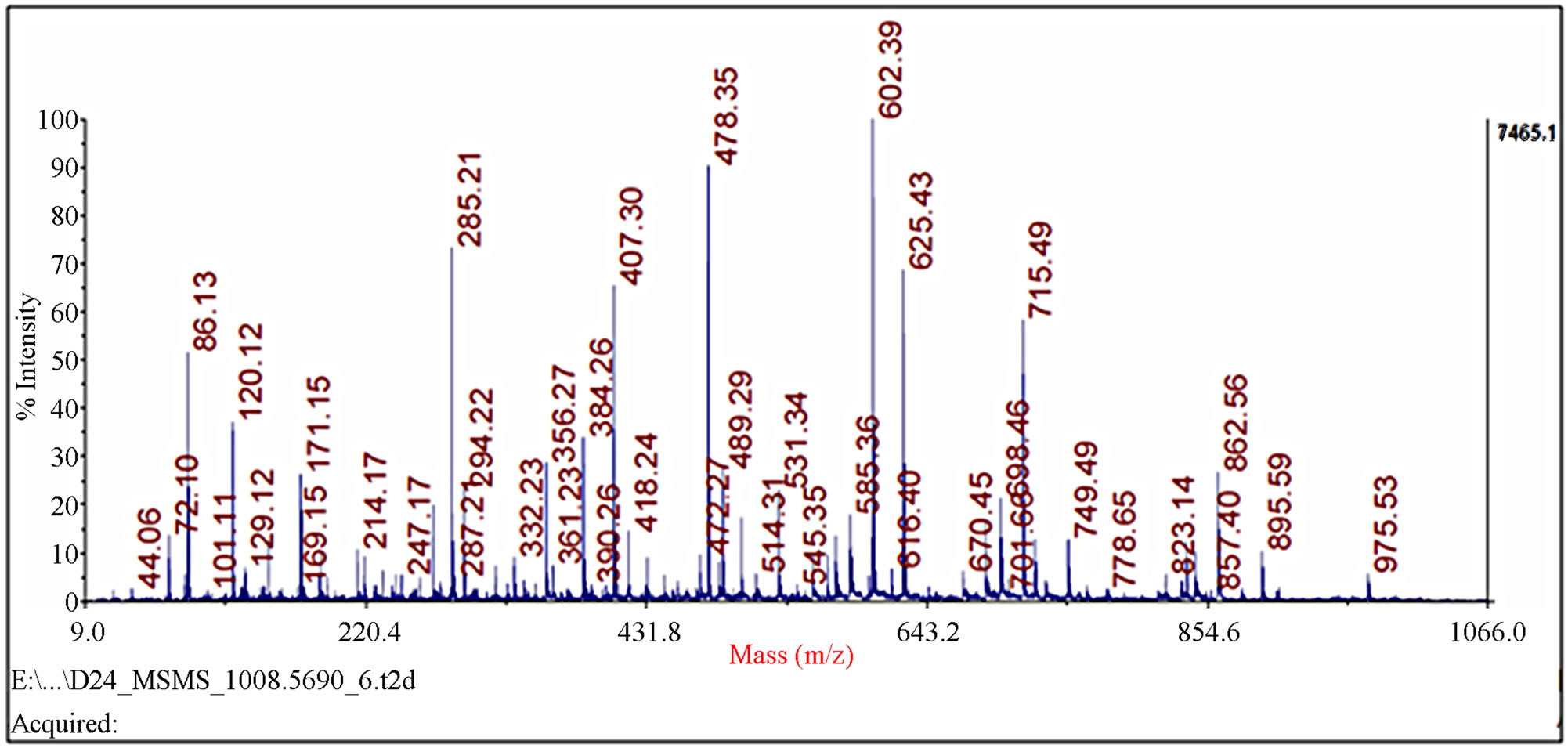
Figure 2. Identification of spot 23 via MALDI-TOF/TOF-MS. The protein excised from the gels was digested with trypsin and the resulting peptides were analyzed by a 4800 MALDI-TOF/TOF Proteomics Analyzer [Applied Biosystems, Foster City, CA]. [A] The MS spectra. The 975.52 ion marked with an asterisk was analyzed by MS/MS. [B] MS/MS spectra of ion 975.52.
hormone receptors and protein kinases [41]. One spot (spot 155) corresponding to antimicrobial peptide lumbricin 1. Antimicrobial peptides have been considered to be a universal host defense tool used by earthworms against microbial infection. Lumbricin 1 has been isolated and characterized from the earthworm L. rubellus, which showed antimicrobial activity in vitro against a broad spectrum of microorganisms without hemolytic activity [42].
Analyzing gene expression at the protein level is potentially more informative than mRNA analysis. In fact, proteins have a number of important features that reveal their actual, rather than potential, function. Consequently, a series of proteome database projects have been undertaken to identify all proteins in an organized cell and to explain the role of each protein using published biological data. Furthermore, in contrast to the predominantly static genome, a proteome is exceedingly dynamic: Mechanisms such as cell activation, disease, infection, and differentiation can considerably transform the proteome [43].
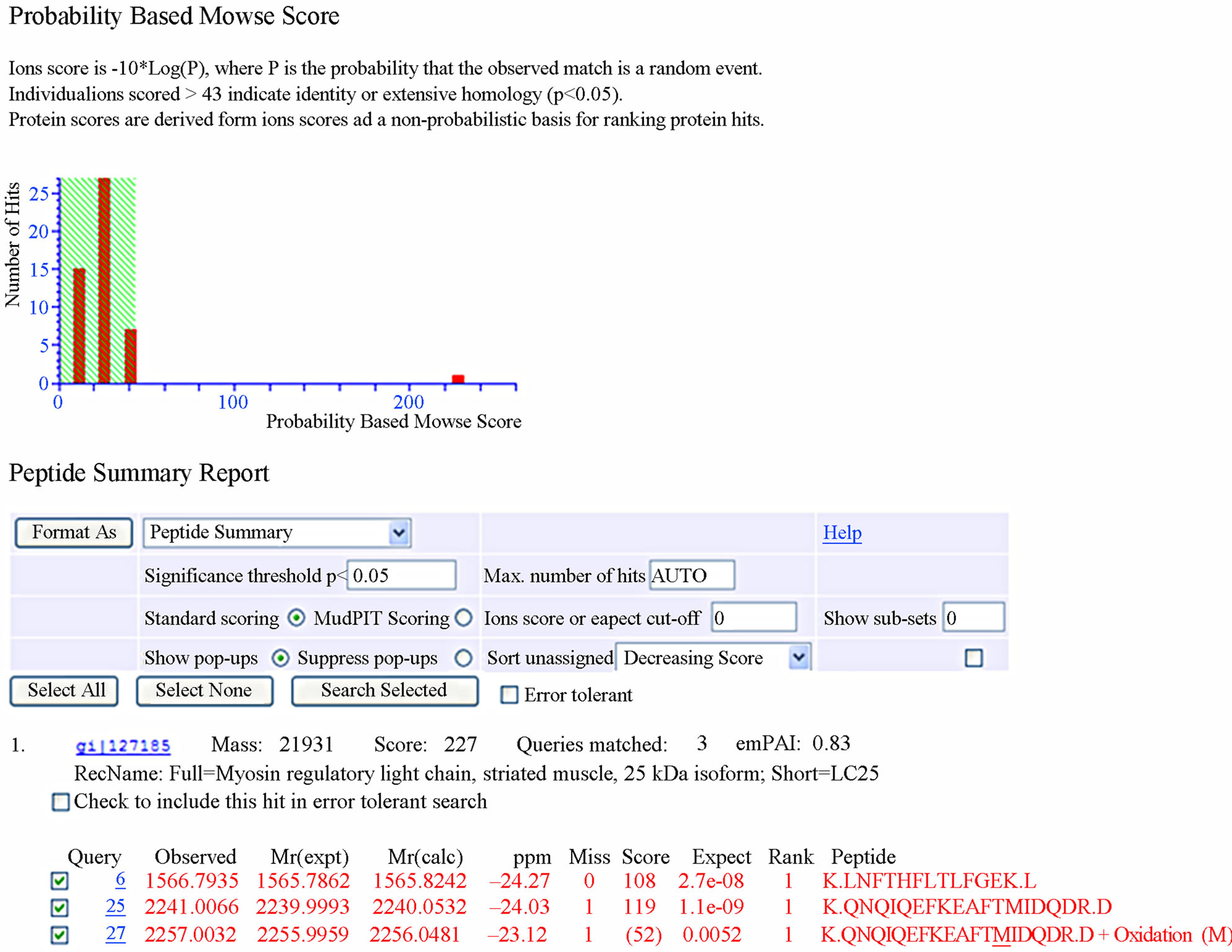
Figure 3. Database search result of spot 23.
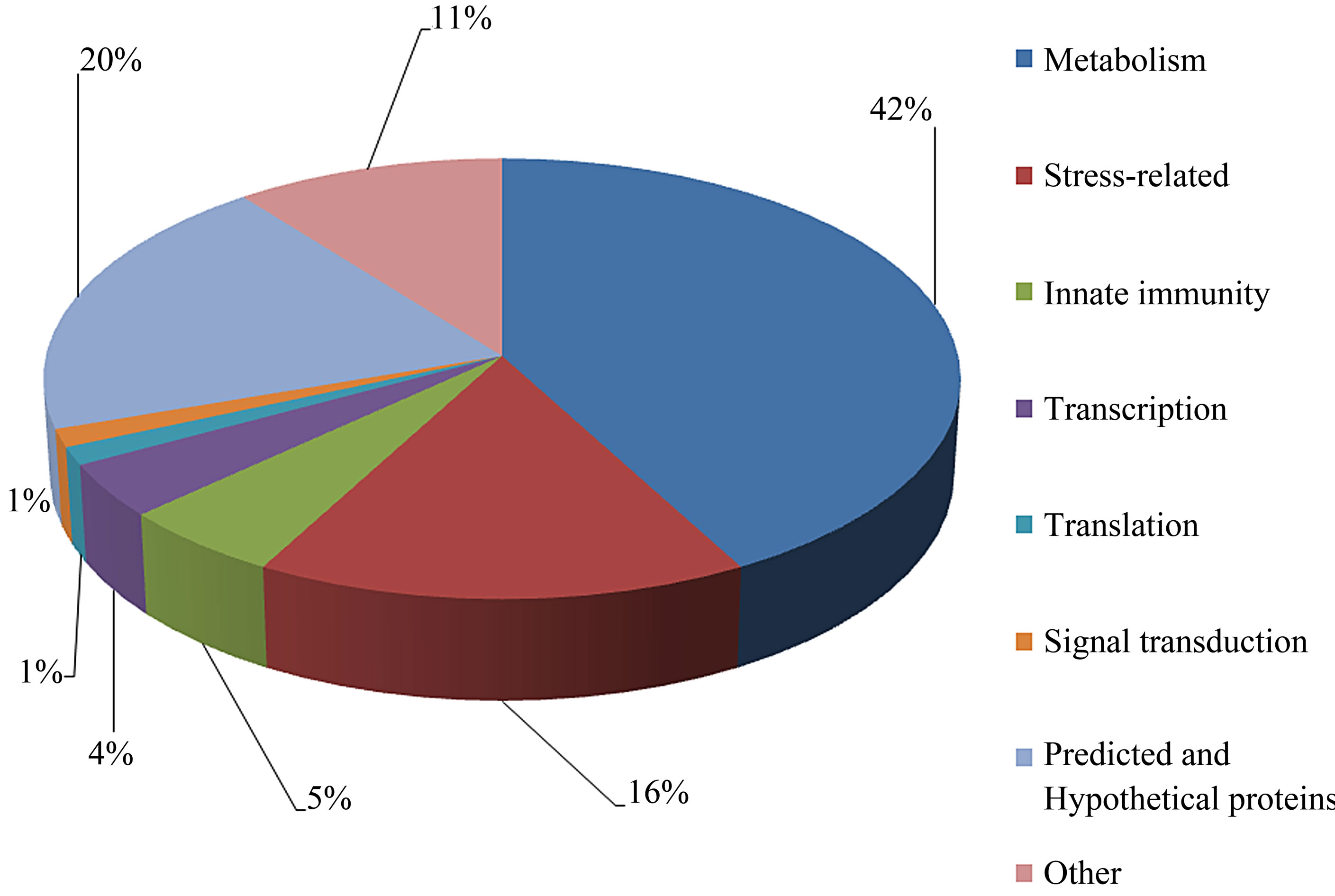
Figure 4. Summary of the classification of the biological functions with gene ontology analysis of identified proteins.
Our data support a resource for the proteomics profile of the whole earthworm E. fetida. We used a gel image to identify and localize 76 distinct proteins. To our knowledge, this is the first description of a 2-DE reference map of the whole earthworm E. fetida proteins. These 2-DE data will enhance future comparisons of immunity, toxicology, biotic processes and other challenges, thereby allowing for further study of the molecular mechanisms of innate immunity.
5. ACKNOWLEDGEMENTS
This work was financially supported by the National Natural Science Foundation of China (Nos. 31172091 and 30770275), the National Key Technology R & D Program in the 11th Five-year Plan of China (2008BADA7B04), the Key Beijing Discipline of Ecology (XK10019440), Innovation Fund for Graduate Student of China Agricultural University (kycx09068), Chinese University Scientific Fund (2009-02-15), and Academic Newcomer Award for Ph.D. of Ministry of Education of the People’s of China.
REFERENCES
- Edwards, C.A. (2004) Earthworm ecology. 2nd Edition, CRC Press, Boca Raton.
- Pirooznia, M., Gong, P., Guan, X., Inouye, L.S., Yang, K., Perkins, E.J. and Deng, Y.P. (2009) Cloning, analysis and functional annotation of expressed sequence tags from the earthworm Eisenia fetida. BMC Bioinformatics, 8, 1-16. Hdoi:10.1186/1471-2105-8-S7-S7
- Bundy, J.G., Sidhu, J.K., Rana, F., Spurgeon, D.J., Svendsen, C.S., Wren, J.F. Stürzenbaum, S.R., Morgan, A. J. and Kille, P. (2008) “Systems toxicology” approach identifies coordinated metabolic responses to copper in a terrestrial non-model invertebrate, the earthworm Lumbricus rubellus. BMC Biology, 6, 25. Hdoi:10.1186/1741-7007-6-25
- Amelina, H., Apraiz, I., Sun, W. and Cristobal, S. (2007) Proteomics-based method for the assessment of marine pollution using liquid chromatography coupled with twodimensional electrophoresis. Journal of Proteome Research, 6, 2094-2104. Hdoi:10.1021/pr060689s
- Dowling, V.A. and Sheehan, D. (2006) Proteomics as a route to identification of toxicity targets in environmental toxicology. Proteomics, 6, 5597-5604. Hdoi:10.1002/pmic.200600274
- Lee, S.E., Yoo, D.H., Son, J. and Cho, K. (2006) Proteomic evaluation of cadmium toxicity on the midge Chironomus riparius Meigen larvae. Proteomics, 6, 945- 957. Hdoi:10.1002/pmic.200401349
- Knigge, T., Monsinjon, T. and Andersen, O.K. (2004) Surface-enhanced laser desorption/ionization-time of flight-mass spectrometry approach to biomarker discovery in blue mussels [Mytilus edulis] exposed to polyaromatic hydrocarbons and heavy metals under field conditions. Proteomics, 4, 2722-2727. Hdoi:10.1002/pmic.200300828
- Owen, J., Hedley, B.A., Svendsen, C., Wren, J., Jonker, M. J., Hankard, P.K., Lister, L.J., Stürzenbaum, S.R., Morgan, A.J., Spurgeon, D.J., Blaxter, M.L. and Kille, P. (2008) Transcriptome profiling of developmental and xenobiotic responses in a keystone soil animal, the oligochaete annelid Lumbricus rubellus. BMC Genomics, 9, 226. Hdoi:10.1186/1471-2164-9-266
- Gong, P., Guan, X., Inouye, L.S., Deng, Y., Pirooznia, M. and Perkins, E.J. (2008) Transcriptomic analysis of RDX and TNT interactive sublethal effects in the earthworm Eisenia fetida. BMC Genomics, 9, S15. Hdoi:10.1186/1471-2164-9-S1-S15
- Nabby-Hansen, S., Waterfield, M.D. and Cramer, R. (2001) Proteomics-post-genomic cartography to understand gene function. Trends in Pharmacological Sciences, 22, 376- 384. Hdoi:10.1016/S0165-6147(00)01663-1
- Kuperman, R.G., Checkai, R.T., Ruth, L.M., Henry, T., Simini, M., Kimmel, D.G., Phillips, C.T. and Bradley, B.P. (2003) A proteome-based assessment of the earthworm Eisenia fetida: Response to chemical warfare agents in a sandy loam soil. Pedobiologia, 47, 617-621. Hdoi:10.1016/S0031-4056(04)70245-0
- LaCourse, E.J., Hernandez-Viadel, M., Jefferies, J.R., Svendsen, C., Spurgeon, D.J., Barrett, J., Morgan, A.J., Kille, P. and Brophy, P.M. (2009) Glutathione transferase [GST] as a candidate molecular-based biomarker for soil toxin exposure in the earthworm Lumbricus rubellus. Environmental Pollution, 157, 2459-2469. Hdoi:10.1016/j.envpol.2009.03.015H
- Wang, X., Chang, L., Wang, G.C., Sun, Z.J., Ma, H.B., Sun, Q. and Li, J. (2010) Protein extraction from the earthworm Eisenia fetida for 2-DE. Proteomics, 10, 1095- 1099. Hdoi:10.1002/pmic.200900488
- Wang, X., Chang, L., Sun, Z.J., Zhang, Y.F. and Yao, L. (2010) Analysis of earthworm Eisenia fetida proteomes during cadmium exposure: An ecotoxicoproteomics approach. Proteomics, 10, 4476-4490. Hdoi:10.1002/pmic.201000209
- Wang, X., Chang, L., Sun, Z.J. and Zhang, Y.F. (2010) Comparative proteomic analysis of differentially expressed proteins in the earthworm Eisenia fetida during Escherichia coli O157:H7 stress. Journal of Proteome Research, 9, 6547-6560. Hdoi:10.1021/pr1007398
- Hernández, R., Nombela, C., Diez-Orejas, R. and Gil, C. (2004) Two-dimensional reference map of Candida albicans hyphal forms. Proteomics, 4, 374-382. Hdoi:10.1002/pmic.200300608
- OECD (2004) OECD (Organization for Economic Cooperation and Development) guideline for testing chemicals. Section 2: Effects on biotic systems. Earthworm, Acute Toxicity Tests, Paris.
- Carpentier, S.C., Witters, E., Laukens, K., Deckers, P., Swennen, R. and Panis, B. (2005) Preparation of protein extracts from recalcitrant plant tissues: An evaluation of different methods for two-dimensional gel electrophoresis analysis. Proteomics, 5, 2497-2507. Hdoi:10.1002/pmsic.200401222
- Lim, H., Eng, J., Yates, J.R., Tollaksen, S.L., Giometti, C. S., Holden, J.F., Adams, M.W.W., Reich, C.I., Olsen, G.J. and Hays, L.G. (2003) Identification of 2D-gel proteins: A comparison of MALDI/TOF peptide mass mapping to mu LC-ESI tandem mass spectrometry. Journal of the American Society for Mass Spectrometry, 14, 957-970. Hdoi:10.1016/S1044-0305(03)00144-2
- Huang, F., Fulda, S., Hagemann, M. and Norling, B. (2006) Proteomic screening of salt-stress-induced changes in plasma membranes of Synechocystis sp. strain PCC 6803. Proteomics, 6, 910-920. H doi:10.1002/pmic.200500114
- Sarry, J.E., Kuhn, L., Ducruix, C., Lafaye, A., Junot, C., Hugouvieux, V., Jourdain, A., Bastien, O., Fievet, J.B., Vailhen, D., Amekraz, B., Moulin, C., Ezan, E., Garin, J. and Bourguignon, J. (2006) The early responses of Arabidopsis thaliana cells to cadmium exposure explored by protein and metabolite profiling analyses. Proteomics, 6, 2180-2198. Hdoi:10.1002/pmic.200500543
- Nishi, H., Inagi, R., Kato, H., Tanemoto, M., Kojima, I., Son, D., Fujita, T. and Nangaku, M. (2008) Hemoglobin is expressed by mesangial cells and reduces oxidant stress. Journal of the American Society of Nephrology, 19, 1500- 1508. Hdoi:10.1681/ASN.2007101085H
- McGregor, E., Kempster, L., Wait, R., Gosling, M., Dunn, M.J. and Powell, J.T. (2004) F-actin capping [CapZ] and other contractile saphenous vein smooth muscle proteins are altered by hemodynamic stress. Molecular & Cellular Proteomics, 3, 115-124. Hdoi:10.1074/mcp.M300046-MCP2004
- Sekizawa, Y., Kubo, T. and Kobayashi, H. (1997) Molecular cloning of cDNA for lysenin, a novel protein in the earthworm Eisenia foetida that causes contraction of rat vascular smooth muscle. Gene, 191, 97-102. Hdoi:10.1016/S0378-1119(97)00047-4
- Yamaji, A., Sekizawa, Y., Emoto, K., Sakuraba, H., Inoue, K., Kobayashi, H. and Umeda, M. (1998) Lysenin, a novel sphingomyelin-specific binding protein. Journal of Biological Chemistry, 273, 5300-5306. Hdoi:10.1074/jbc.273.9.5300
- Köhlerová, P., Beschin, A., Šilerová, M., De Baetselier, P. and Bilej, M. (2004) Effect of experimental microbial challenge on the expression of defense molecules in Eisenia foetida earthworm. Developmental & Comparative Immunology, 28, 701-711. Hdoi:10.1016/j.dci.2004.01.001
- Zhang, X.M., Luan, W., Jin, S.J. and Xiang, J. (2008) A novel tumor necrosis factor ligand superfamily member [CsTL] from Ciona savignyi: Molecular identification and expression analysis. Developmental & Comparative Immunology, 32, 1362-1373. Hdoi:10.1016/j.dci.2008.05.009
- Lynch, J.A., Brent, A.E., Leaf, D.S., Pultz, M.A. and Desplan, C. (2006) Localized maternal orthodenticle patterns anterior and posterior in the long germ wasp Nasonia. Nature, 39, 728-732. Hdoi:10.1038/nature04445
- Törrönen, R., Korkalainen, M. and Kärenlampi, S.O. (1992) Induction of class 3 aldehyde dehydrogenase in the mouse hepatoma cell line Hepa-1 by various chemicals. Chemico-Biological Interactions, 83, 107-119. Hdoi:10.1016/0009-2797(92)90040-R
- McKim, J.M., Choudhuri, S. and Klaassen, C.D. (1992) In vitro degradation of apo-, zinc-, and cadmium-metallothionein by cathepsins B, C, and D. Toxicology and Applied Pharmacology, 116, 117-124. Hdoi:10.1016/0041-008X(92)90152-I
- Martens, S. and Howard, J. (2006) The interferon-inducible GTPases. Annual Review of Cell and Developmental Biology, 22, 559-589. Hdoi:10.1146/annurev.cellbio.22.010305.104619
- Djabali, K., de Nechaud, B., Landon, F. and Portier, M.M. (1997) AlphaB-crystallin interacts with intermediate filaments in response to stress. Journal of Cell Science, 110, 2759-2769.
- Ellington, W.R. (2001) Evolution and physiological roles of phosphagen systems. Annual Review of Physiology, 63, 289-325.Hdoi:10.1146/annurev.physiol.63.1.289H
- Park, Y., Ryu, E., Kim, H., Jeong, J., Kim, J., Shim, J., Jeon, S., Jo, Y., Kim, W. and Min, B. (1999) Characterization of antithrombotic activity of lumbrokinase-immobilized polyurethane valves in the total artificial heart. Artificial Organs, 23, 210-214. Hdoi:10.1046/j.1525-1594.1999.06013.x
- Boldin, M.P., Goncharov, T.M., Goltsev, Y.V. and Wallach, D. (1996) Involvement of MACH, a novel MORT1/FADDinteracting protease, in Fas/APO-1- and TNF receptorinduced cell death. Cell, 85, 803-815. Hdoi:10.1016/S0092-8674(00)81265-9H
- Hengartner, M.O. (2000) The biochemistry of apoptosis. Nature, 407, 770-776. Hdoi:10.1038/35037710
- Salvesen, G.S. and Dixit, V.M. (1997) Caspases: Intracellular signaling by proteolysis. Cell, 91, 443-446. Hdoi:10.1016/S0092-8674(00)80430-4
- Ram, R.J., VerBerkmoes, N.C., Thelen, M.P., Tyson, G. W., Baker, B.J., Blake II, R.C., Shah, M., Hettich, R.L. and Banfield, J.F. (2005) Community proteomics of a natural microbial biofilm. Science, 308, 1915-1920. Hdoi:10.1126/science. 1109070
- Brown, P.J., Long, S.M., Spurgeon, D.J., Svendsen, C. and Hankard, P.K. (2004) Toxicological and biochemical responses of the earthworm Lumbricus rubellus to pyrene, a non-carcinogenic polycyclic aromatic hydrocarbon. Chemosphere, 57, 1675-1681.
- Pandey, P., Saleh, A., Nakazawa, A., Kumar, S., Srinivasula, S.M., Kumar, V., Weichselbaum, R., Nalin, C., Alnemri, E.S., Donald, K. and Kharbanda, S. (2000) Negative regulation of cytochrome c-mediated oligomerization of Apaf-1 and activation of procaspase-9 by heat shock protein 90. The EMBO Journal, 19, 4310-4322. Hdoi:10.1093/emboj/19.16.4310
- Zhao, R., Davey, M., Hsu, Y.C., Kaplanek, P., Tong, A., Parsons, A.B., Krogan, N., Cagney, G., Mai, D., Greenblatt, J., Boone, C., Emili, A. and Houry, W.A. (2005) Navigating the chaperone network: An integrative map of physical and genetic interactions mediated by the Hsp90 chaperone. Cell, 120, 715-727. Hdoi:10.1016/j.cell.2004.12.024
- Cho, J.H., Park, C.B., Yoon, Y.G. and Kim, S.C. (1998) Lumbricin I, a novel proline-rich antimicrobial peptide from the earthworm: Purification, cDNA cloning and molecular characterization. Biochimica et Biophysica Acta, 1408, 67-76. Hdoi:10.1016/S0925-4439(98)00058-1
- Kim, H.R., Han, R.X., Yoon, J.T., Park, C.S. and Jin, D.I. (2010) A two-dimensional electrophoresis reference map for the bovine placenta during late pregnancy. Proteomics, 10, 564-573. Hdoi:10.1002/pmic.200900508

