Advances in Bioscience and Biotechnology
Vol. 4 No. 12 (2013) , Article ID: 41283 , 6 pages DOI:10.4236/abb.2013.412143
A ConA-like lectin isolated from Canavalia maritima seeds alters the expression of genes related to virulence and biofilm formation in Streptococcus mutans
![]()
1Faculty of Dentistry, Institute of Applied Theology (INTA), Sobral, Brazil
2Integrate Biomolecules Laboratory (LIBS), Faculty of Medicine of Sobral, Federal University of Ceará, Sobral, Brazil
3Biologically Active Molecules Laboratory (Biomol-Lab), Department of Biochemistry and Molecular Biology, Federal University of Ceará, Fortaleza, Brazil
4Integrate Biomolecules Laboratory (LIBS), Department of Pathology and Legal Medicine, Federal University of Ceará, Fortaleza, Brazil
5Center of the Exact Sciences and Technology, Acaraú Valley State University, Sobral, Brazil
6Center of Agricultural Science and Biological, Acaraú Valley State University, Sobral, Brazil
Email: *edsonlec@gmail.com
Copyright © 2013 Theodora Thays Arruda Cavalcante et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 24 September 2013; revised 28 October 2013; accepted 1 December 2013
Keywords: Streptococcus mutants; Lectin; Canavalia maritima; Biofilm; Expression
ABSTRACT
Bacteria form biofilms as an adaptive mechanism in response to environmental changes. Streptococcus mutans is the biofilm-forming bacterium that is primarily associated with dental caries. The expression of important genes by bacteria in biofilms is different from that of planktonic cells. Lectins are proteins that bind specifically to carbohydrates and may have important biological activities on bacterial cells, acting as antibacterial and anti-biofilm agents. ConM (Canavalia maritima lectin) is a protein that is able to inhibit the planktonic growth and biofilm formation of S. mutans. In this context, this study aimed to evaluate the effects of ConM and concanavalin A (ConA) on the expression of genes related to virulence and biofilm formation in S. mutans. The results showed that ConM significantly reduced the expression of genes encoding enzymes related to adhesion, formation and regulation of biofilms. On the contrary, ConA did not alter the expression of the genes studied. Because the two lectins have a high degree of similarity, the differences in the actions of ConM and ConA may be explained by the small structural differences in the carbohydrate recognition domain of the lectins.
1. INTRODUCTION
Most microorganisms have a natural tendency to develop cell aggregates surrounded by a self-produced polysaccharide matrix called a biofilm [1]. Biofilms formed on tooth surfaces (dental plaque) are the main etiologic factor for the majority of dental disorders such as caries, gingivitis and periodontitis [2,3]. Furthermore, oral biofilms are the main cause of the dental implant failure [4]. Concerning the microorganisms involved in oral biofilms, Streptococcus mutans is commonly found in the human oral cavity. This species is a potent initiator of caries due to a wide range of factors, which plays an important role in caries formation [5].
When such microorganisms adhere to a surface, they develop biofilms and express an altered phenotype through the repression or activation of specific genes [6, 7]. In fact, biofilm formation by S. mutans is followed by differential expression of several genes compared to planktonic cells. This distinct pattern of expression is frequently associated with biofilm regulation, formation, resistance and bacterial physiology [8,9].
According to Wen and Burne [10], the expression of genes associated with biofilm development, such as spaP and gbpB, regulation (brpA), and those involved in the formation of extracellular glucans (gtfB), plays an important role in the regulation of biofilm development by S. mutans.
Lectins are a group of proteins of non-immune origin that have the ability to bind specifically and reversibly to carbohydrate epitopes without modifying them [11]. These proteins work as mediators of information in various biological systems, acting through interactions with glycoproteins, glycolipids and oligosaccharides [12, 13]. The ability of lectins to detect small differences in the configuration of carbohydrates [14] makes these proteins important in several studies involving microbial biofilms. Among the biological applications of lectins, the antibacterial activity deserves special attention [15], as lectins have been shown to inhibit biofilm formation in some studies [16,17]. Thus, to verify the different effects of lectins on the expression of virulence genes in S. mutans, the bacterium was grown in the presence of lectins, and the relative expression of spaP, gtfB, gbpB, brpAI and ldh was analyzed using burk16S as a normalizing reference gene.
2. MATERIALS AND METHODS
2.1. Microorganism
Streptococcus mutans UA159 was maintained in BHI (Brain Heart Infusion) broth with 20% glycerol at −80˚C until use. For experiments, an aliquot of 100 ml of the stock was inoculated into 10 ml of fresh BHI broth and incubated at 37˚C with 10% CO2. After growth, the cells were centrifuged at 1500 × g for 20 min at 4˚C and washed twice with 0.9% NaCl.
2.2. Lectins
Seeds of Canavalia maritima and C. ensiformis were separately ground into a fine powder using a coffee mill. The powder was homogenized with 0.15 M NaCl (1:10 w/v) at room temperature overnight. The extract was then centrifuged at 20,000 × g for 20 min at 4˚C, and the supernatant was applied to a Sephadex G-50 column (Amersham-Bio-sciences, USA) that had been preequilibrated with 0.15 M NaCl containing 5 mM CaCl2 and MnCl2. After removal of the unbound material, each lectin was eluted with 0.1 M of glucose in 0.15 M NaCl and submitted to a 1 h dialysis against 0.1 M acetic acid, which was succeeded by an extensive dialysis against distilled water. Finally, the lectins were freeze-dried and stored at 4 °C for later use. The purity of the lectins was assessed by SDS-PAGE as described by Laemmili [18].
2.3. Experimental Groups
Prior to RNA extraction, the bacterial suspension was washed three times by centrifugation at 5000 × g for 5 min at 4˚C with 0.9% NaCl. The cells were adjusted to 2 × 108 CFU/ml in BHI broth and transferred to a 96 well polystyrene plate. The lectins ConM and ConA (250 µg/ml) were then added to wells containing bacterial suspensions. The negative control was carried out with 0.9% NaCl. The plate was then incubated for 1 h at 37˚C with 10% CO2. After 1 h, 300 ml of the bacterial suspension from each condition was inoculated into 30 ml of fresh BHI broth and incubated at 37˚C with 10% CO2. The RNA extraction was performed when bacterial growth reached an absorbance of 0.4 to 0.5 absorbance units (AU), corresponding to the beginning of the logarithmic phase.
2.4. Extraction of total RNA and cDNA synthesis
Extraction steps were performed using RNase-free water, reagents and plastics. Briefly, as soon as bacterial growth reached 0.4 to 0.5 AU, the cells were centrifuged at 6000 × g for 10 min at 4˚C, and the pellet was resuspended in 1 ml of Tris-EDTA (100 mM Tris, 2 mM EDTA, pH 8.0) containing 15 mg/ml lysozyme and incubated for 30 min in a water bath at 37˚C. The pellet was then centrifuged as previously described, and the supernatant was discarded. The pellet was resuspended with 1 ml of TRIZOL® reagent and homogenized through up-down movements in a pipette. To maximize the action of the reagent and prevent degradation of RNA, the samples were kept on ice during all intervals between procedures. Following this step, 200 ml of cooled chloroform was added, and the tubes were vigorously mixed by inversion for 15 s. The resulting solution was centrifuged at 12,000 × g for 15 min at 4˚C to obtain a three-phase system, in which the upper phase, which is aqueous and colorless, corresponds to the extracted RNA. The RNA was then transferred to a new tube containing 500 ml of isopropanol and kept on ice for 10 min to allow its precipitation and DNA dehydration. The RNA was then centrifuged at 15,000 × g for 10 min at 4˚C, and the pellet was resuspended with 1 ml of cold 75% ethanol (DNase/RNasefree DEPC water). This solution was centrifuged at 7000 × g for 5 min at 4˚C, and then the tube was immersed on ice and kept open for 5 min inside a flow chamber to prevent contamination. Finally, the pellet was resuspended in 30 ml of DEPC water and assessed for purity and concentration using a spectrophotometer (GENquant, Amersham Biosciences) [19]. The integrity and quality of the total RNA samples were determined by electrophoresis on a 1.2% agarose gel [20].
To avoid contamination with DNA, samples were treated with DNase (Invitrogen) according to the manufacturer’s recommendations [19]. The synthesis of complementary DNA (cDNA) was performed using SuperScript Reverse Transcriptase® (Invitrogen) and Random Hexamers® primers (Qiagen). All procedures were performed in triplicate.
2.5. Real-Time Reverse Transcriptase PCR (qRT-PCR) Analysis
The quantitative analysis of gene expression was carried out by qRT-PCR. For this purpose, 300 ng of cDNA from each sample was used in each reaction. In addition, primers for specific genes, which were previously optimized, as well as 10 ml of 2x Power SYBR® Green Master Mix (Applied Biosystems) were added to a final volume of 20 ml. The amplification reaction was performed with 40 cycles at 95˚C for 5 s, 55˚C for 5 s and 68˚C for 20 s. The initial denaturation was performed at 95˚C for 5 min.
Primer selection was based on the study by Wen and colleagues [21] (Table 1). To evaluate the effect of lectins on selected genes, the expression of an unaffected gene (Burkholderia 16S ribosomal RNA) was used as the endogenous control and compared to the expression of target genes. The assays were carried out with a RealPlex 4S thermocycler® (Eppendorf) using the 2x Power SYBR® Green Master Mix Kit (Applied Biosystems). All reactions were performed in quadruplicate. The cycle threshold (Cts) used in the analysis corresponded to the arithmetic mean between the quadruplicate of the target and the endogenous control genes. To achieve the relative expression of the target genes, the 2ΔΔCt method was used as previously described by Livak and Schmittgen [22].
2.6. Statistical Analysis
All experiments were performed in triplicate and repeated three times independently. Differences in relative expression between samples were analyzed by one-way ANOVA with Tukey’s posttest by the GraphPad Prism® software. Values were considered statistically significant when p < 0.05.
3. RESULTS AND DISCUSSION
To verify the purity of the lectins, the proteins were submitted to SDS-PAGE. Both lectins showed an electrophoretic pattern that was characteristic of the Diocleinae subtribe as three different bands were visualized: a chain of 25 kDa (α-chain) and two fragments of 14 and 12 kDa (β and γ chains, respectively). Moreover, the lectins showed hemagglutinating activity on rabbit erythrocytes and were inhibited by 0.1 M D-glucose and D-mannose (data not shown). Concerning the activity of Diocleinae lectins on S. mutans growth, Cavalcante and colleagues [17] showed that ConM was able to inhibit the planktonic growth and biofilm formation of S. mutans, whereas ConA, although similar to ConM, had no effect
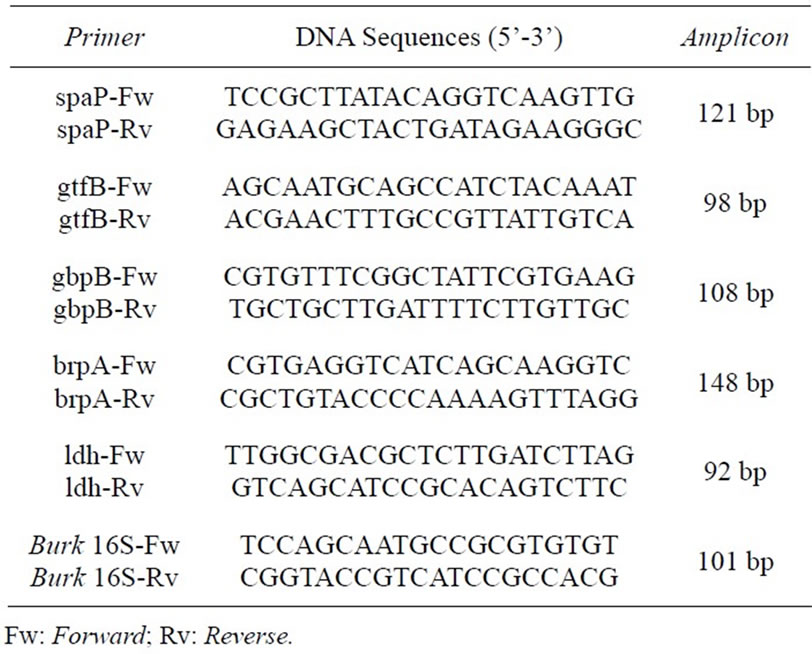
Table 1. List of primers used for qRT-PCR, their DNA sequences and amplicon length (adapted from Wen and colleagues [21]).
on planktonic growth and showed only a weak inhibition on biofilm formation after 18 hr.
The integrity of the RNA extraction from cells treated with ConM and ConA was analyzed by agarose gel electrophoresis. The results showed that all steps involved in the extraction were successful (Figure 1). cDNA synthesis and relative expression analysis by qRT-PCR showed that ConM significantly reduced the expression of all tested genes (Figure 2). For example, the expression of gtfB decreased approximately 7-fold in cells treated with ConM as compared to the negative control. gtfB encodes the enzyme glucosyltransferase B (GtfB), which synthesizes extracellular insoluble glucan polymers from sucrose. In addition to being a critical virulence factor involved in the pathogenesis of dental caries, GtfB plays an important role in biofilm formation [23]. In addition to gtfB, ConM also reduced the expression of spaP and gbpB by 4.2 and 7.7-fold, respectively. These genes encode important adhesins, which bind to surface receptors and are important for the successful establishment of S. mutans biofilms [24,25]. In addition, ConM reduced the expression of brpA by 9.5-fold. This gene encodes a regulatory protein that plays a critical role in environmental stress responses and biofilm formation in S. mutans [10,26]. Interestingly, expression of the gene that encodes the enzyme lactate dehydrogenase (ldh) is also reduced by 8.5-fold. The ldh gene is constitutively expressed in S. mutans and is responsible for the synthesis of lactic acid from pyruvate, in addition to being an important virulence factor in the bacterium [27,28]. Hillman and colleagues [29] showed that the deficiency in LdH production might be lethal to S. mutans due to its importance in the generation of energy for the cell.
As previously demonstrated by Teixeira and colleagues [16], lectins have the ability to block receptor
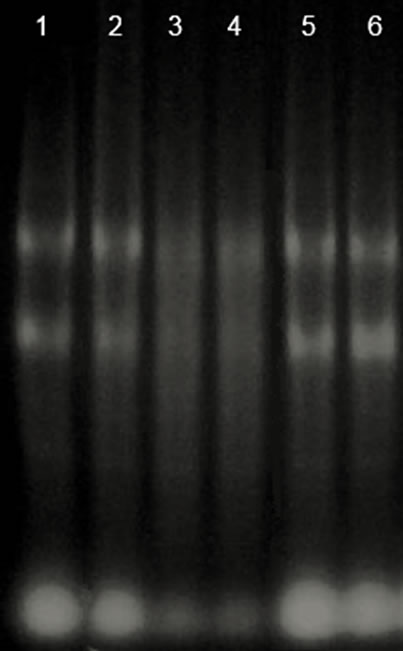
Figure 1. Integrity of the RNA extracted in the presence of lectins by agarose gel electrophoresis. Lane 1 and 2: Control; lane 3 and 4: ConA; lane 5 and 6: ConM.
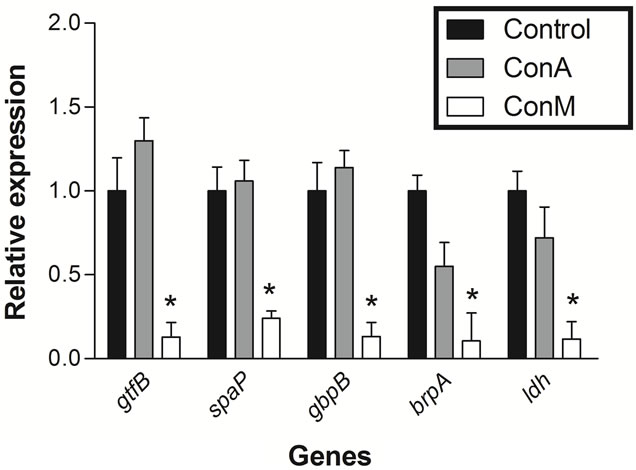
Figure 2. Analysis of the relative expression of genes related to biofilm formation of S. mutans in presence of ConM or ConA. Control (black bars), ConA (gray bars) and ConM (white bars). *p < 0.01 Relative to control.
molecules present in the acquired enamel pellicle in vitro, thus interfering with early adhesion of colonizers to the tooth surface and preventing the growth of cariogenic biofilms. Furthermore, Canavalia lectins have been demonstrated to be proteins with antimicrobial activity against Streptococcus mutans [17]. The antibacterial effect of ConM on planktonic growth and biofilm formation in S. mutans as shown by Cavalcante and colleagues [17] can be attributed not only to a direct action on cells but also to its ability to inhibit some genes related to biofilm formation and virulence.
Despite the high degree of similarity between ConA and ConM, treatment with ConA did not alter the expression of any gene investigated in this work (Figure 2). However, this is not surprising because several studies have shown that lectins from Diocleinae subtribe, although sharing a high degree of similarity, differ in their
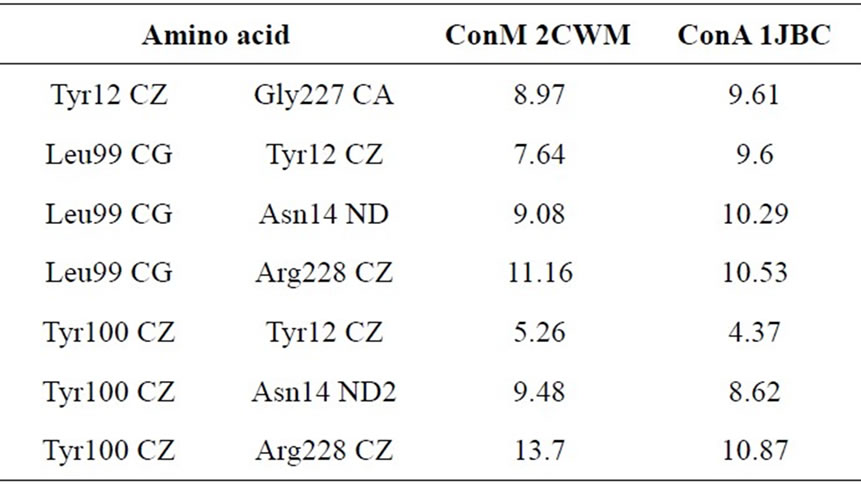
Table 2. Distances (Å3) between amino acid residues of the CDR of ConM and ConA that are involved in interactions with carbohydrates (adapted from Bezerra and colleagues [34]).
biological activities [30-33]. According to Cavada and colleagues [34], small changes in the configuration of important amino acids from the carbohydrate recognition domain (CRD), as well as variations in pH-dependent oligomerization, may explain the differences in biological activities. Moreover, Bezerra and colleagues [35] showed that not only are the configuration and distances between residues in the CRD crucial to explain the difference in activity between Diocleinae lectins, but the volume of the CRD also plays an important role in the lectin activity. The distances between the amino acid residues involved in interactions with carbohydrates are different in ConM and ConA (Table 2), and the CRD of ConA shows a lower volume when compared with ConM. The volumes for ConA PDB 1JBC and ConM PDB 2CWM are 151 and 135 Å3 respectively, suggesting a plausible explanation for the differences in the antimicrobial actions of such lectins [35].
Although the mechanism of action of these lectins requires a better understanding, the results reported in the present article suggest that ConM acts by starting or interrupting intracellular signaling pathways that culminate with the lowest expression of genes associated with virulence and biofilm formation in S. mutans. Future experiments will be performed to investigate the mechanisms that lead to changes in expression of the selected genes and the impact of such changes on biofilm formation.
4. ACKNOWLEDGEMENTS
This work was financed by Fundação Cearense de Apoio ao Desenvolvimento Científico e Tecnológico (FUNCAP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). We acknowledge American Journal Experts (AJE) for English editing services. BSC and EHT are senior researches of CNPq.
REFERENCES
- McDougald, D., Rice, S.A., Barraud, N., Steinberg, P.D. and Kjelleberg, S. (2012) Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nature Reviews Microbiology, 10, 39-50. http://dx.doi.org/10.1038/nrmicro2695
- Paquette, D.W., Brodala, N. and Williams, R.C. (2006) Risk factors for endosseous dental implant failure. Dental Clinics of North America, 50, 361-374. http://dx.doi.org/10.1016/j.cden.2006.05.002
- Dhir, S. (2013) Biofilm and dental implant: The microbial link. Journal of Indian Society of Periodontology, 17, 5-11. http://dx.doi.org/10.4103/0972-124X.107466
- Buser, D. and Merickse-Stern, R. (1997) Long term evaluation of nonsubmerged ITI implants. Part 1: 8 year life table analysis of a prospective multicenter study with 2359 implants. Clinical Oral Implants Research, 8, 161- 172. http://dx.doi.org/10.4103/0972-124X.107466
- Marsh, P.D. (2005) Dental plaque: biological significance of a biofilm and community life-style. Journal of Clinical Periodontology, 32,7-15. http://dx.doi.org/10.1034/j.1600-0501.1997.080302.x
- Brooun, A., Liu, S. and Lewis, K.A. (2000) A Dose-Response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrobial Agents and Chemotherapy, 44, 640-646. http://dx.doi.org/10.1111/j.1600-051X.2005.00790.x
- Sauer, K. and Camper, A.K. (2001) Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. Journal of Bacteriology, 183, 6579-6589. http://dx.doi.org/10.1128/AAC.44.3.640-646.2000
- Shemesh, M., Tam, A. and Steinberg, D. (2007) Expression of biofilm-associated genes of Streptococcus mutans in response to glucose and sucrose. Journal of Medical Microbiology, 56, 1528-1535. http://dx.doi.org/10.1128/JB.183.22.6579-6589.2001
- Shemesh, M., Tam, A., Aharoni, R. and Steinberg, D. (2010) Genetic adaptation of Streptococcus mutans during biofilm formation on different types of surfaces. BMC Microbiology, 10, 51. http://dx.doi.org/10.1099/jmm.0.47146-0
- Wen, Z.T. and Burne, R.A. (2002) Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Applied and Environmental Microbiology, 68, 1196-1203. http://dx.doi.org/10.1186/1471-2180-10-51
- Bies, C., Lehr, C.M. and Woodley, J.F. (2004) Lectinmediated drug targeting: history and applications. Advanced Drug Delivery Reviews, 56, 425-435. http://dx.doi.org/10.1128/AEM.69.1.722.2003
- Rakhshandehroo, M., Stienstra, R., De Wit, N.J., Bragt, M.C., Haluzik, M., Mensink, R.P., Müller, M. and Kersten, S. (2012) Plasma mannose-binding lectin is stimulated by PPARα in humans. American Journal of Physiology-Endocrinology and Metabolism, 302, 595-602. http://dx.doi.org/10.1016/j.addr.2003.10.030
- Zappelli, C., Van Der Zwaan, C., Thijssen-Timmer, D.C., Mertens, K. and Meijer, A.B. (2012) Novel role for galectin-8 protein as mediator of coagulation factor V endocytosis by megakaryocytes. The Journal of Biological Chemistry, 287, 8327-8335. http://dx.doi.org/10.1152/ajpendo.00299.2011
- Hirabayashi, J. (2008) Concept, strategy and realization of lectin-based glycan profiling. Journal of Biochemistry, 144, 139-147. http://dx.doi.org/10.1074/jbc.M111.305151
- Wong, J.H., Ng, T.B., Cheung, R.C., Ye, X.J., Wang, H.X., Lam, S.K., Lin, P., Chan, Y.S., Fang, E.F., Ngai, P.H., Xia, L.X., Ye, X.Y., Jiang, Y. and Liu, F. (2010) Proteins with antifungal properties and other medicinal applications from plants and mushrooms. Applied Microbiology and biotechnology, 87, 1221-1235. http://dx.doi.org/10.1093/jb/mvn043
- Teixeira, E.H., Napimoga, M.H., Carneiro, V.A., De Oliveira, T.M., Cunha, R.M., Havt, A., Martins, J.L., Pinto, V.P., Gonçalves, R.B. and Cavada, B.S. (2006) In vitro inhibition of streptococci binding to enamel acquired pellicle by plant lectins. Journal of Applied Microbiology, 101, 111-116. http://dx.doi.org/10.1007/s00253-010-2690-4
- Cavalcante, T.T., Da Rocha, B.A.M., Carneiro, V.A., Arruda, F.V.S., Do Nascimento, A.S.F., Sá, N.C., Nascimento, K.S., Cavada, B.S. and Teixeira, E.H. (2011) Effect of lectins from Diocleinae subtribe against oral Streptococci. Molecules, 16, 3530-3543. http://dx.doi.org/10.1111/j.1365-2672.2006.02910.x
- Laemmili, U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680-685. http://dx.doi.org/10.3390/molecules16053530
- Ritz, M., Garenaux, A., Berge, M. and Federighi, M. (2009) Determination of rpoA as the most suitable internal control to study stress response in C. jejuni by RTqPCR and application to oxidative stress. Journal of Microbiological Methods, 76, 196-200. http://dx.doi.org/10.1016/j.mimet.2008.10.014
- Sambrook, J., Fritsch, E.F. and Maniatis, T. (1989) Molecular cloning: A laboratory manual. 2nd Edition, Cold Spring Harbor, New York.
- Wen, Z.T., Yates, D., Ahn, S.J. and Burne, R.A. (2010) Biofilm formation and virulence expression by Streptococcus mutans are altered when grown in dual-species model. BMC Microbiology, 10, 111. http://dx.doi.org/10.1186/1471-2180-10-111
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using Real-Time Quantitative PCR and the 2–ΔΔCT Method. Methods, 25, 402-408. http://dx.doi.org/10.1006/meth.2001.1262
- Tsumori, H. and Kuramitsu, H. (1997) The role of the Streptococcus mutans glucosyltransferases in the sucrose-dependent attachment to smooth surfaces: Essential role of the GtfC enzyme. Oral Microbiology and Immunology, 12, 274-280. http://dx.doi.org/10.1111/j.1399-302X.1997.tb00391.x
- Banas, J.A. and Vickerman, M.M. (2003) Glucan-binding proteins of the oral streptococci. Critical Reviews in Oral Biology & Medicine, 14, 89-99. http://dx.doi.org/10.1177/154411130301400203
- Jakubovics, N.S., Strömberg, N., Van Dolleweerd, C.J., Kelly, C.G. and Jenkinson, H.F. (2005) Differential binding specificities of oral streptococcal antigen I/II family adhesins for human or bacterial ligands. Molecular Microbiology, 55, 1591-1605. http://dx.doi.org/10.1111/j.1365-2958.2005.04495.x
- Wen, Z.T., Baker, H.V. and Burne, R.A. (2006) Influence of BrpA on critical virulence attributes of Streptococcus mutans. Journal of Bacteriology, 188, 2983-2992. http://dx.doi.org/10.1128/JB.188.8.2983-2992.2006
- Merritt J., Kreth, J., Qi, F., Sullivan, R. and Shi, W. (2005) Non-disruptive, real-time analyses of the metabolic status and viability of Streptococcus mutans cells in response to antimicrobial treatments. Journal of Microbiology Methods, 61, 161-170. http://dx.doi.org/10.1016/j.mimet.2004.11.012
- Yang, D.Q., Liu, T.J., Zhou, X.D., He, K.F., Li, S. and Zhuang, H. (2005) Study on lactate dehydrogenase activity of Streptococcus mutans isolates derived from caries-active and caries-free individuals. Hua Xi Kou Qiang Yi Xue Za Zhi, 23, 116-118.
- Hillman, J.D., Chen, A., Duncan, M. and Lee, S.W. (1994) Evidence that L-(+)-lactate dehydrogenase deficiency is lethal in Streptococcus mutans. Infection and Immunity, 62, 60-64. http://iai.asm.org/content/62/1/60
- Bezerra, G.A., Oliveira, T.M., Moreno, F.B., de Souza, E.P., Rocha, B.A., Benevides, R.G., Delatorre, P., De Azevedo Jr., W.F. and Cavada, B.S. (2007) Structural analysis of Canavalia maritima and Canavalia gladiata lectins complexed with different dimannosides: New insights into the understanding of the structure biological activity relationship in legume lectins. Journal of Structural Biology, 160, 168-176. http://dx.doi.org/10.1016/j.jsb.2007.07.012
- Nóbrega, R.B., Rocha, B.M., Gadelha, C.A., SantiGadelha, T., Pires, A.F., Assreuy, A.M.S., Nascimento, K.S., Nagano, C.S., Sampaio, A.H., Cavada, B.S. and Delatorre, P. (2012) Structure of Dioclea virgata lectin: Relations between carbohydrate binding site and nitric oxide production. Biochimie, 94, 900-906. http://dx.doi.org/10.1016/j.biochi.2011.12.009
- de Vasconcelos, M.A., Cunha, C.O., Arruda, F.V.S., Carneiro, V.A., Mercante, F.M., do Nascimento Neto, L.G., de Sousa, G.S., Rocha, B.A.M., Teixeira, E.H., Cavada, B.S. and dos Santos, R.P. (2012) Lectin from Canavalia brasiliensis seeds (ConBr) is a valuable biotechnological tool to stimulate the growth of Rhizobium tropici in vitro. Molecules, 17, 5244-5254. http://dx.doi.org/10.3390/molecules17055244
- Bezerra, M.J.B., Rodrigues, N.V.F.C.,; Pires, A.F., Bezerra, G.A., Nobre, C.B., Alencar, K.L.L., Soares, P.M.G., Nascimento, K.S., Nagano, C.S., Martins, J.L., Gruber, K., Sampaio, A.H., Delatorre, P., Rocha, B.A.M., Assreuy, A.M.S. and Cavada, B.S. (2013) Crystal structure of Dioclea violacea lectin and a comparative study of vasorelaxant properties with Dioclea rostrata lectin. International Journal of Biochemistry & Cell Biology, 45, 807-815. http://dx.doi.org/10.1016/j.biocel.2013.01.012
- Cavada, B.S., Barbosa, T., Arruda, S., Grangeiro, T.B. and Barral-Netto, M. (2001) Revisiting proteus: Do minor changes in lectin structure matter in biological activity? Lessons from and potential biotechnological uses of the Diocleinae subtribe lectins. Current Protein & Peptide Science, 2, 123-135. http://dx.doi.org/10.2174/1389203013381152
- Bezerra, E.H., Rocha, B.A., Nagano, C.S., Bezerra, G.D., Moura, T.R., Bezerra, M.J., Benevides, R.G., Sampaio, A.H., Assreuy, A.M.S., Delatorre, P. and Cavada, B.S. (2011) Structural analysis of ConBr reveals molecular correlation between the carbohydrate recognition domain and nitric oxide release from endothelial cells. Biochemical and Biophysical Research Communications, 408, 566-570. http://dx.doi.org/10.1016/j.bbrc.2011.04.061
NOTES
*Corresponding author.

