Journal of Biophysical Chemistry
Vol.2 No.3(2011), Article ID:6737,14 pages DOI:10.4236/jbpc.2011.23030
Effect of disulfide bridges deletion on the conformation of the androctonin, polyphemusin-I, and thanatin antimicrobial peptides: molecular dynamics simulation studies
Grupo de Simulação Molecular, Faculdade de Filosofia Ciências e Letras de Ribeirão Preto, Universidade de São Paulo, Ribeirão Preto, Brazil; *Corresponding Author: jrcastro@usp.br
Received 6 March 2011; revised 27 April 2011; accepted 15 May 2011.
Keywords: Androctonin; Polyphemusin-I; Thanatin; Antimicrobial Peptides; Cysteine Rich Peptides; Molecular Simulation; Disulfide Bridges Deletion
ABSTRACT
In this work, the role of the disulfide bridges in the maintenance of the secondary structure of the antimicrobial peptides androctonin, polyphemusin-I, and thanatin is analyzed on the basis of their structural characteristics and of three of their respective mutants, andry4, poly4, and thany2, in which all the cysteine residues have been replaced with tyrosine residues. The absence of the disulfide bridges in andry4, poly4, and thany2 seems to be compensated by an overall enforcement of the original hydrogen bonds and by extra attractive interactions between the aromatic rings of the tyrosine residues. In spite of the mutations, the original β-hairpin structures are maintained in the three mutants, but the best conformational similarities are found for the androctonin/andry4 pair.
1. INTRODUCTION
Microbial resistance to antimicrobial tools is a serious problem nowadays since the number of antibiotic-resistant microorganisms has been increasing over the last years [1,2]. The impact caused by such challenging question has motivated various research groups to develop new drugs capable of dealing with the adaptation strategy (namely, the selection of organisms adapted to some specific medium) that these microorganisms develop along the years [3-6]. The reasons for the appearance of resistant bacterial strains lie on the excessive, and frequently inadequate, use of antibiotics [7]. However, nature itself has endowed some organisms with antimicrobial peptides, which are distinctive tools for survival [8]. In this context, the production of antimicrobial peptides is a powerful evolutionary ancient defensive weapon [9] that allows living organisms like plants, vertebrate and invertebrate animals, as well as fungi and bacteria to survive. Many organisms make use of antimicrobial peptides to defend themselves against harmful invader microorganisms. These peptides are part of the innate defense systems of the organisms [6]. An important characteristic of such peptides is their broad spectrum of action since they can be active against several kinds of bacteria, as well as against some fungi. The hemolytic effect and the potential toxicity exhibited by these peptides make their use against infectious diseases difficult. Heavy investments in research that could improve their selectivity and decrease their toxicity are necessary, so that these peptides can be made active against pathogenic microorganisms only.
The androctonin peptide is an antimicrobial agent constitutive of the cell-free hemolymph of the scorpion Androctonus australis. Its primary structure contains 25 aminoacid residues, Table 1, including eight positive charges spread all over the peptide chain and four cysteine residues that produce two disulfide bridges linking Cys4 to Cys20 and Cys10 to Cys16 [10]. Its molecular mass is 3076.7 Da. It is active against Gram-positive and Gram-negative bacteria as well as against a broad range of filamentous fungi. Androctonin concentrations up to 150 µM do not exhibit hemolytic activity in porcine and bovine erythrocytes when assayed under conventional conditions [10]. Its β-hairpin structure was determined by conventional two-dimensional nuclear magnetic resonance spectroscopy (2D-NMR) and by molecular modeling [11]. The 3D structure is deposited in the Protein Data Bank, PDB, (www.rcsb.org/pdb) under the code 1CZ6.
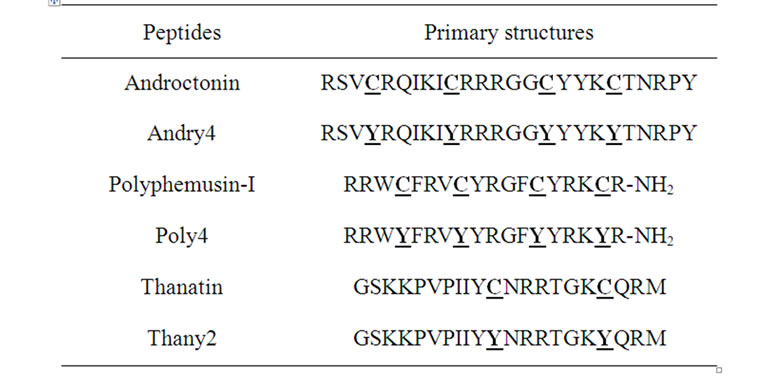
Table 1. Primary structures of the wild peptides and their respective mutants. The residues where the mutation was made are in bold letter.
Polyphemusin-I is an antimicrobial peptide encountered in the American horseshoe crab Limulus polyphemus. It has 18 aminoacid residues in length, Table 1, including seven positively charged residues and two disulfide bonds linking Cys4 to Cys17 and Cys8 to Cys13, as well as an amidated C-terminal arginine. It is active against Gram-positive and Gram-negative bacteria, and some fungi. Its solution structure determined using 1H-NMR spectroscopy is deposited in the PDB under the code 1RKK [12].
The thanatin peptide is an insect defense protein which has, at physiological concentrations, a broad spectrum of activity against Gram-positive and Gramnegative bacteria, and filamentous fungi. It presents a minimal inhibition concentration of approximately 1 µM for both Gram-positive and Gram-negative bacteria as well as fungi [13]. It appears in the haemolymph of the hemipteran insect Podisus maculiventris. Its primary structure contains 21 aminoacid residues, Table 1, including six positively charged residues, presenting a molecular mass of 2434.4 Da. Two cysteine residues, Cys11 to Cys18, forms one disulfide bridge. Its hemolytic activity has been tested in a conventional assay on porcine erythrocytes and it was concluded that it does not present hemolytic activity even at 40 µM [14]. Its three-dimensional (3D) structure in water was determined using two-dimensional (2D) 1H-NMR and molecular modeling methods which were deposited in the PDB under the code 8TFV [13].
In the present work, we will compare the structures of the abovementioned wild peptides with the structures of three mutants, namely andry4 (androctonin mutant), poly4 (polyphemusin-I mutant), and thany2 (thanatin mutant), whose primary structures are shown in Table 1. Our proposition is based on studies carried out with tachyplesin-I mutants, in which the four cysteine residues of the wild peptide were replaced with tyrosine residues [5,15]. In their studies with tachyplesin-I, these authors found a great similarity between the native and the mutant structures. They also reported that the deletion of all cysteines abolished the hemolytic effect while the antimicrobial activity remained unchanged. There is a great interest in the development of new antibiotic compounds, mainly ones free of cysteine residues, because these peptides are easier and cheaper to produce [5,15]. The detailed data provided in this work shall give support to a better understanding of the role of the disulfide bridges in the secondary structures of these peptides.
2. MODELS AND METHODS
The initial configurations of the three mutants (andry4, poly4, and thany2) were obtained using the PDB structures of the wild peptides and the mutations carried out with the DeepView/Swiss-PdbViewer 3.7 software [16]. The construction of the systems and the molecular dynamics simulations were carried out using the GROMACS 3.3.3 package [17] with the GROMOS96 force field [18], employed to model the peptides. The six systems were constructed by placing the center of mass (CM) of the peptides in the center of a cubic simulation box with edges equal to the peptide’s largest interatomic distance plus 4 nm. The box was fulfilled with water molecules using the SPC model [19]. Chloride and sodium ions were inserted, when necessary, into electrostatically favorable positions, in order to counterbalance the peptide charges, thus keeping the local electroneutrality of the systems. The covalent bonds of the peptides were constrained by the LINCS algorithm [20], while the SETTLE algorithm [21] was used to maintain the water SPC molecules structurally stable. The temperature (300 K) and the pressure (1 atm) of the systems (NpT ensemble) were regulated by the Berendsen’s algorithms [22], using correlation times of 0.1 ps and 0.5 ps for temperature and pressure coupling, respectively. A cut-off of 1.0 nm on the van der Waals interactions was used. The long range electrostatic interactions were calculated using the particle mesh Ewald summation method (PME) [23]. In order to eliminate bad contacts, the energy of the system was first energy minimized by the steepest descent algorithm. The simulation of each system was then carried out for 0.5 ns, under peptide atomic coordinate restrictions, with an integration time step of 0.5 fs. The next step of the simulations was carried out during an extra 0.5 ns period, still under peptide atomic coordinate restrictions, but an integration time step of 1.0 fs was employed. These two cycles of 0.5 ns each were performed in order to equilibrate the system. After that, the MD simulations were carried out without restrictions for 80 ns in the cases of androctonin and andry4; 85 ns for polyphemusin-I and poly4; 50 ns for thanatin and, finally, 70 ns for thany2, always using integration time steps of 2 fs. The Ramachandran angles in the wild peptides and their respective mutants were calculated, in order to compare their structures. The intramolecular hydrogen bonds (HB) were identified considering a donor-acceptor distance was smaller than 0.35 nm, and the angle between the hydrogen-donor and donor-acceptor vectors smaller than 30 degrees. Only HB occurrences greater than 20% of the total simulation time were considered. The radius of gyration (Rg) and the root mean square deviations (RMSD) were systematically used to analyze the structural and the evolutional behaviors of the peptides; the RMSD was obtained by least-square fitting of the peptide structures to the reference that was the initial structure of each peptide. The Rg provides global information about the packing of the molecules, while the RMSD is a measure of the similarity of a structure with its reference and also the temporal evolution of the structure. In these studies, the GROMACS analyzing tools package [24] was employed.
3. RESULTS
3.1. The Structures
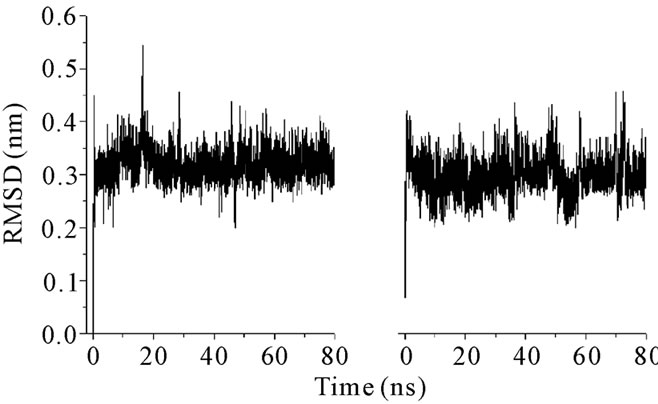
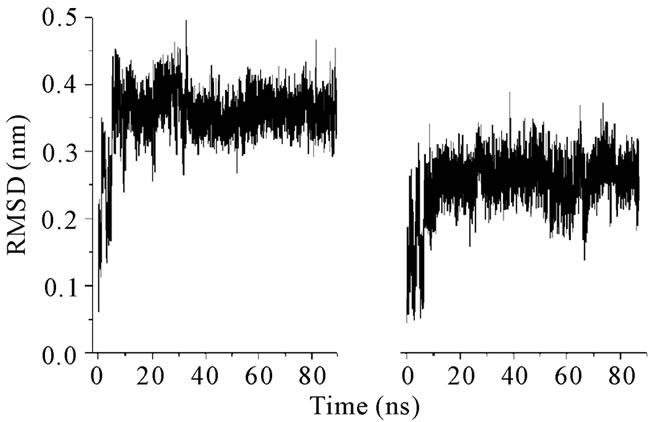
The structural characteristics of androctonin/andry4, polyphemusin-I/poly4, and thanatin/thany2 were first analyzed by comparison of their Cα RMSD evolution, Figures 1, 3, and 5, respectively; and RMSD per residue, Figures 2, 4, and 6, respectively.
The RMSDs for each pair of peptides (wild peptide and its mutant) can present, or not, very distinct behaveiors, indicating that the stability of the mutant is similar, or not, to that of the wild peptide. The RMSD time-averages and Rg for androctonin, polyphemusin-I, thanatin, and their respective mutants are listed in Table 2. The pair of peptides androctonin/andry4 presents very close mean RMSD values (0.33 and 0.30 nm, respect-tively, Table 2). These results are in good agreement with the mean RMSD values per residue (Figure 2), since the profiles of the two curves are very similar. The RMSD of the androctonin Arg1 residue is larger than that of the same andry4 residue, while the Val3 residue presents a smaller RMSD in androctonin compared with andry4. All the other residues of androctonin and andry4 present very close RMSDs, as shown in Figure 2. The Rg mean values, shown in Table 2, are very close to the experimental Rg values calculated from the PDB structures. The RMSD and Rg data, together with the RMSD per residue data, are a strong indication of the structural similarities between androctonin and its mutant andry4.
The distinct behaviors of the polyphemusin-I and poly4 RMSDs, presented in Figure 3, show that the mutant seems to assume configurations closer to that of reference structure than the ones assumed by the wild
 (a) (b)
(a) (b)
Figure 1. RMSD plots for (a) androctonin and (b) andry4.
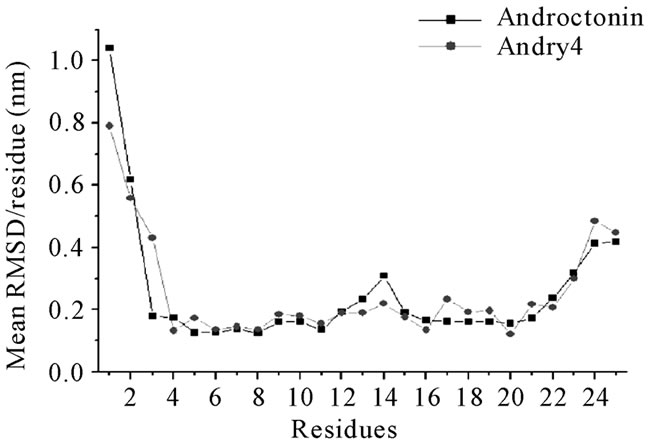
Figure 2. Androctonin and andry4 mean RMSD values per residue.
 (a) (b)
(a) (b)
Figure 3. RMSD plots for (a) polyphemusin-I and (b) poly4.
peptide. It is possible that the reference structure, which is an experimentally determined structure, is not a typical structure pertaining to a high density region of the phase space. The mean RMSD values (0.36 and 0.25 for polyphemusin-I and poly4, respectively) and their profiles show that the structure of the mutant is somehow different from that of the wild peptide. Figure 4 presents the RMSD per residue and shows that the motion of the residues in polyphemusin-I is greater compared with poly4, where they seem to be more locked. The close
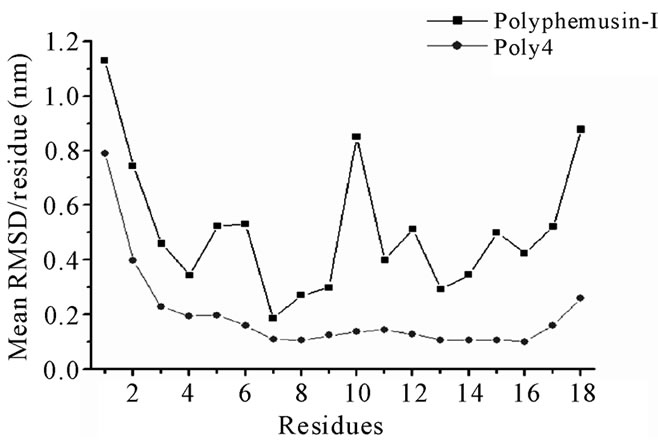
Figure 4. Polyphemusin-I and poly4 mean RMSD values per residue.
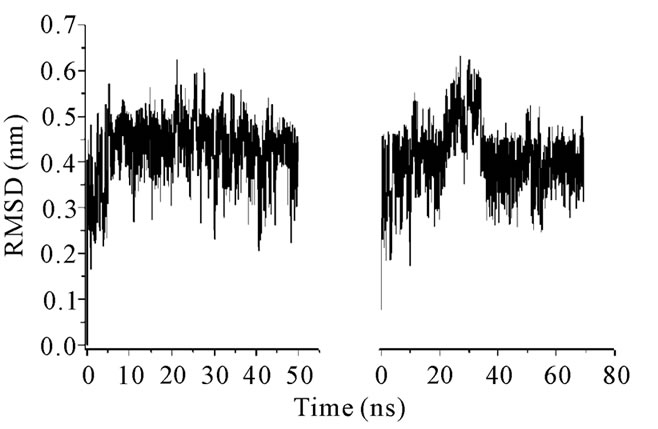
Figure 5. RMSD plots for (a) thanatin and (b) thany2.
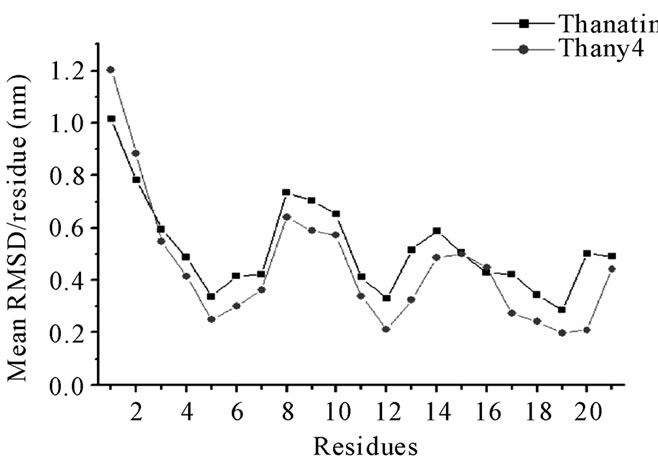
Figure 6. Thanatin and thany2 mean RMSD values per residue.
values of Rg (Table 2) together with the RMSDs for polyphemusin-I and poly4 suggest that the mutant poly4 presents structural similarities with the wild peptide polyphemusin-I, keeping the β-hairpin structure. Nevertheless, the very different profiles of the curves in Figure 4 (RMSD per residue) suggest there are no conformational similarities between them. The results of the thanatin/thany2 pair also suggest that some differences in the mutant/wild peptide structures have occurred. Moreover, some instability in the thany2 structure are easily detected in the RMSDs (Figure 5) in the 20 - 35
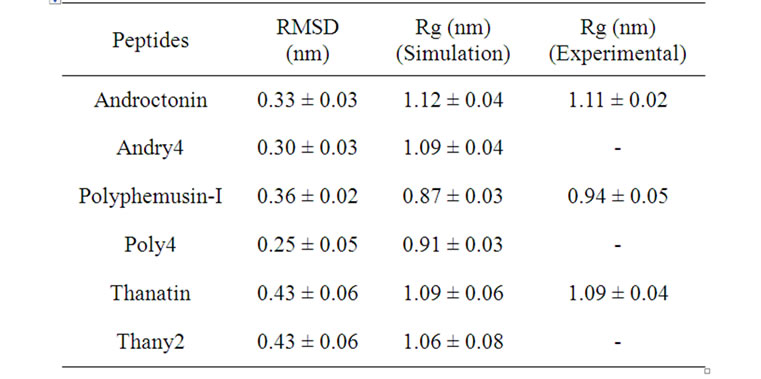
Table 2. Average RMSD and Rg for the wild peptides and their mutants.
ns region. The Rg values (Table 2) are very close to the experimental Rg value calculated from the structures contained in the PDB file. The RMSD and the Rg data suggest a structural similarity between thanatin and thany2, as well as the maintenance of the β-hairpin structure by the mutant. The RMSD per residue in Figure 6 shows that the profiles for thanatin and thany2 are slightly different, suggesting that they have different conformations.
3.2. The Hydrogen Bonds
The simulation results concerning the three peptidemutant pairs confirm that a peculiar pattern of crossstrand HBs contributes with their respective mutants. The HB between the residues Val3 and Arg23 that is present in androctonin, is absent in andry4 (Table 3). The lack of this HB allows increased motion of the Val3 residue in the mutant, as can be seen from the RMSD per residue (Figure 2). The same flexibility does not happen with the Arg23 significantly to the stabilization of the β-hairpin-like structure adopted by androctonin, polyphemusin-I, and thanatin in aqueous solution. This same peculiarity was observed in our previous work [25] with the simulations of gomesin and protegrin-1 in aqueous media. Tables 3, 4, and 5 summarize the results for the cross-strand HBs, together residue because, in andry4, this residue forms an HB with the Ser2 residue, as shown in Table 3. In general, HB occurrence is larger in andry4 due to the absence of disulfide bridges. These results draw attention to the significant role of the cross-strand HBs in the maintenance of the structural stability of the mutants despite the absence of the disulfide bridges. Indeed, it has been experimentally observed that the location of the disulfide bridges within the β-hairpin structure determines the β-hairpin stability only when the disulfide bonds link originally non-hydrogen-bonded residues facing each other [26]. An exception is the HB between the Arg5 and Thr21 residues, since its occurrence decreases as a consequence of the
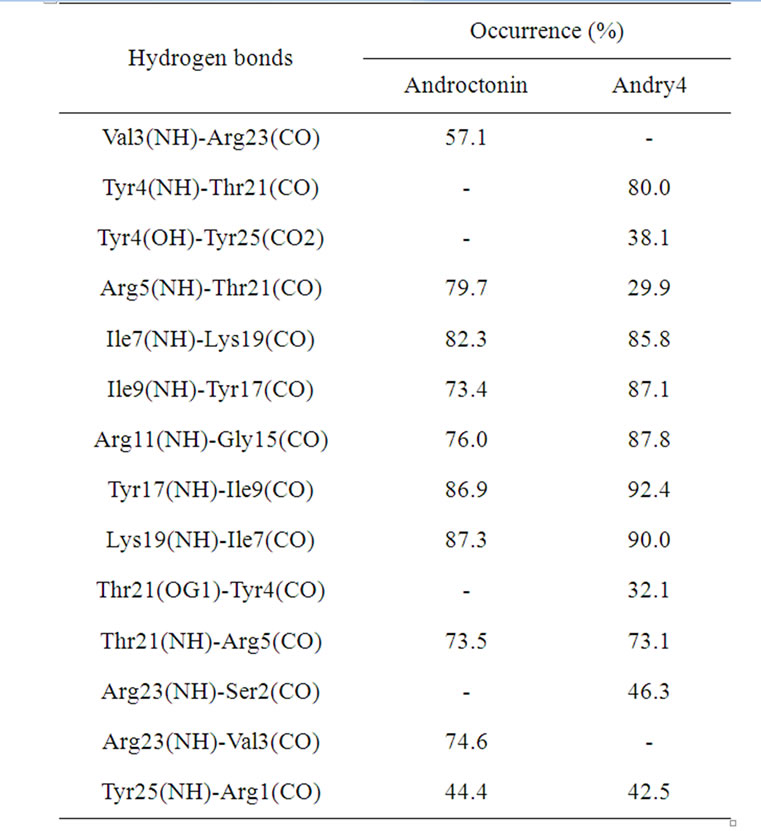
Table 3. Androctonin and andry4 hydrogen bonds determined by molecular dynamics simulations.
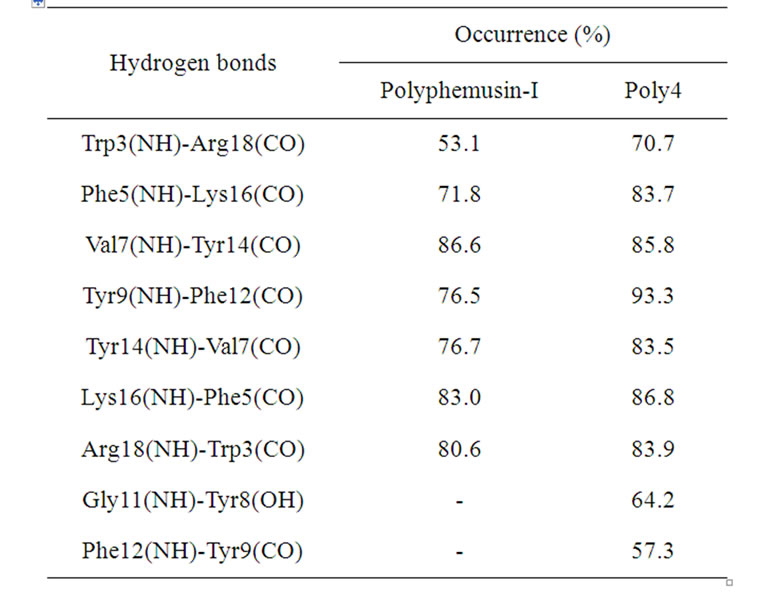
Table 4. Hydrogen bonds found in the polyphemusin-I and poly4 simulations.
HB formed between the Tyr4 and Thr21 residues in andry4. Three extra HBs between the Tyr4/Thr21, Tyr4/ Tyr25, and Arg23/Ser2 residues, which are not present in androctonin, also contribute to the maintenance of the β-hairpin structure in andry4. The Tyr4 residue uses the NH and the OH (from the aromatic ring) to form HB simultaneously with Thr21 and Tyr25 residues. All the HBs found in polyphemusin-I are also present in poly4
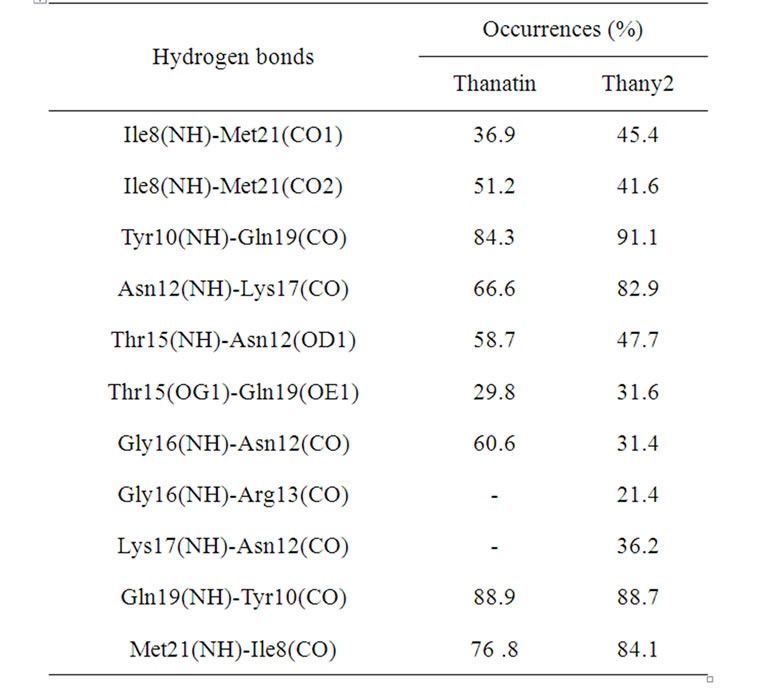
Table 5. Hydrogen bonds found in the thanatin and thany2 simulations.
(Table 4), albeit with increased occurrences. In addition, two extra HBs are also found in poly4, between the Gly11/Tyr8 and Phe12/Tyr9 residues, promoting a relevant decrease in motion in the region of the turn (Tyr8, Tyr9, Arg10, Gly11, and Phe12), as corroborated by the RMSD per residue in Figure 4. In the absence of the disulfide bridges, the occurrence of the HB between Tyr9(NH)-Phe12(CO) increases from 76.5% in polyphemusin-I to 93.3% in poly4 (Table 4). In addition, formation of another HB between Phe12(NH)-Tyr9(CO), which is not present in polyphemusin-I, is observed, and it contributes even more to the reduction in motion, as detected by the RMSD per residue. These two new HBs play an important role in the stabilization of the folding region and are probably responsible for the differences in the observed RMSD behaviors.
Finally, in the pair thanatin/thany2, the same thanatin HBs were found in the thany2 peptide, Table 5. However, the occurrence of some HBs decreases, as in the case of the HB between the Ile8/Met21 residues, which is formed sometimes between the Ile8 NH group or alternatively with one of the CO groups of Met21. These interactions are not simultaneous, so the sums of their occurrences for thanatin (36.9 + 51.2, Table 5) and for thany2 (45.4 + 41.6, Table 5) result in a total of 88.1% and 87.0%, respectively. These values are very close. The case of the HB between Thr15 and Asn12 is different since the reduction of its occurrence occurs because the Asn12 residue forms an HB with the Lys17 residue in thany2, which does not take place in the thanatin peptide. The turn connecting the two strands of the β-sheet is composed by six residues (Asn12, Arg13, Arg14, Thr15, Gly16, and Lys17). Mandard et al. [13] reported that this six-membered hairpin conformation is stabilized by an HB between the residues Asn12-Lys17 and by two distorted C = O(i) – NH(i + 3) HBs between the Asn12- Thr15 and the Arg13-Gly16 residues. In the present MD simulations, the same HBs were found, except the one between the residues Arg13-Gly16. However, this HB was found in the simulation of thany2. Therefore, the lack of the disulfide bridge in thany2 was compensated by the increase in the occurrence of the pre-existing HBs and by the new HBs such as the ones between the residues Gly16-Asn13 and Lys17-Asn12. Accordingly, in the present case, the removal of the disulfide bridges and the introduction of tyrosine residues not only essentially preserve almost all the original HBs, but also promote a generalized increase in the HB occurrences, thereby contributing to the stabilization of the β-hairpin structure as a compensation for the lack of the disulfide bridges.
3.3. The Ramachandran Angles
The Ramachandran angles for the three peptides and their mutants were also calculated. Tables 6, 7, and 8 show the maxima in the Ramachandran angle histograms for the pair androctonin/andry4, polyphemusin-I/poly4, and thanatin/thany2, respectively. Experimental data for gomesin [27] assert that both dihedral angles Φ and Ψ for Lys8 are positive, while both Φ and Ψ of the neighbor Gln9 are negative. The same motif was encountered in our previous simulations of gomesin [25], (Φ, Ψ) Lys8 = (+50˚, +75˚), and protegrin-1, (Φ, Ψ) Arg10 = (+55˚, +75˚) peptides. These values correspond to the maxima in the histograms of the Ramachandran angles and not to their averages. Interestingly, both residues occupy the position where the chain starts to fold to form the typical hairpin β-strand. Moreover, in these peptides, some Φ and Ψ angles present two preferential values [25]. These remarks support the premise that such peptides exhibit more than one meta-stable conformation in aqueous solution, which may be correlated with the broad-spectrum of their antibiotic activeties. Actually, the alternative conformations must reflect some flexibility of the structure, which may be responseble for increasing the interaction efficiency between the peptides and different kinds of cellular membrane [25]. The same motif was also observed in the simulations of androctonin, polyphemusin-I, and thanatin. However, the Φ and Ψ angles of the androctonin and thanatin peptides are both negative, while in polyphemusin-I they are both positive. Some residues present a multimodal distribution of the Ramachandran angles, displaying two or more peaks (see Figure 7 for instance), suggesting that, despite the presence of the disulfide bridges, these pep
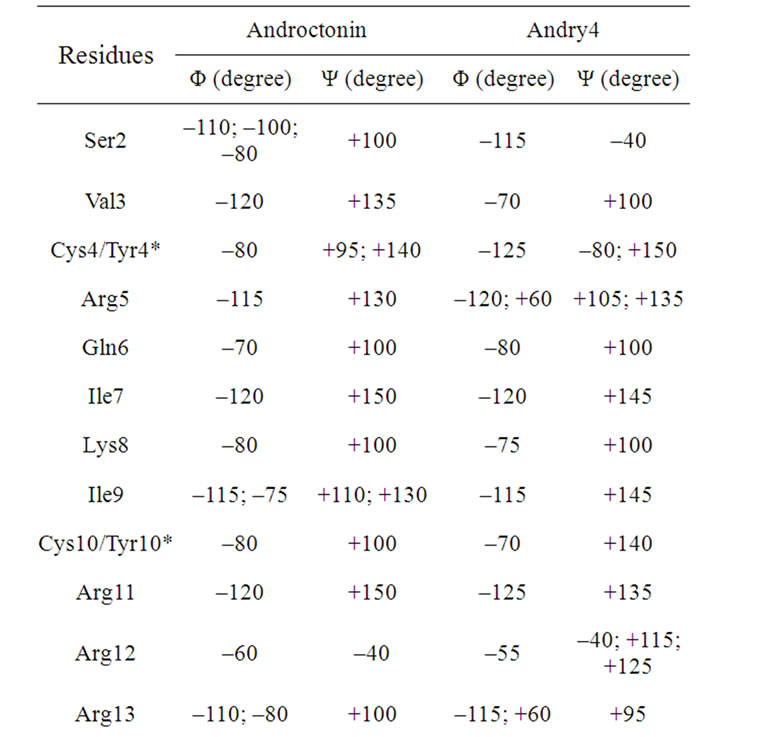
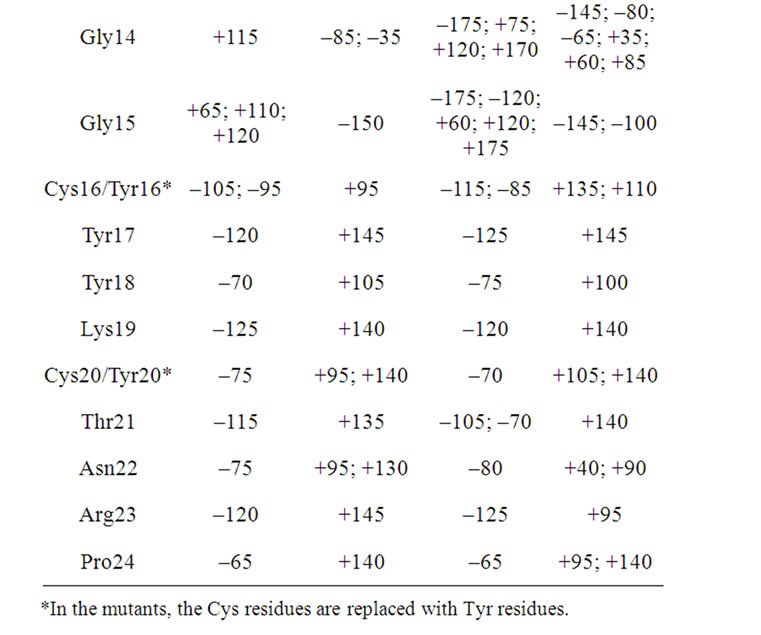
Table 6. The maxima of the Ramachandran angles distributions for the androctonin and andry4 peptides.
tides possess some flexibility, which may be correlated with their broad spectra of action. The maxima of the histograms of the Ramachandran angles for the androctonin Arg12 residue are (Φ, Ψ) Arg12 = (–60˚, –40˚), while in andry4 this motif is observed in the Ser2 residue at (Φ, Ψ) Ser2 = (–115˚, –40˚), Table 6. The polyphemusin-I Arg10 residue presents (Φ, Ψ) Arg10 = (+55˚, +80˚), Table 7. The thanatin Arg13, Arg14, and Thr 15 residues present (Φ, Ψ) Arg13 = (–60˚, –35˚), (Φ, Ψ) Arg14 = (–60˚, –40˚), and (Φ,Ψ) Thr15 = (–95˚, –35˚),
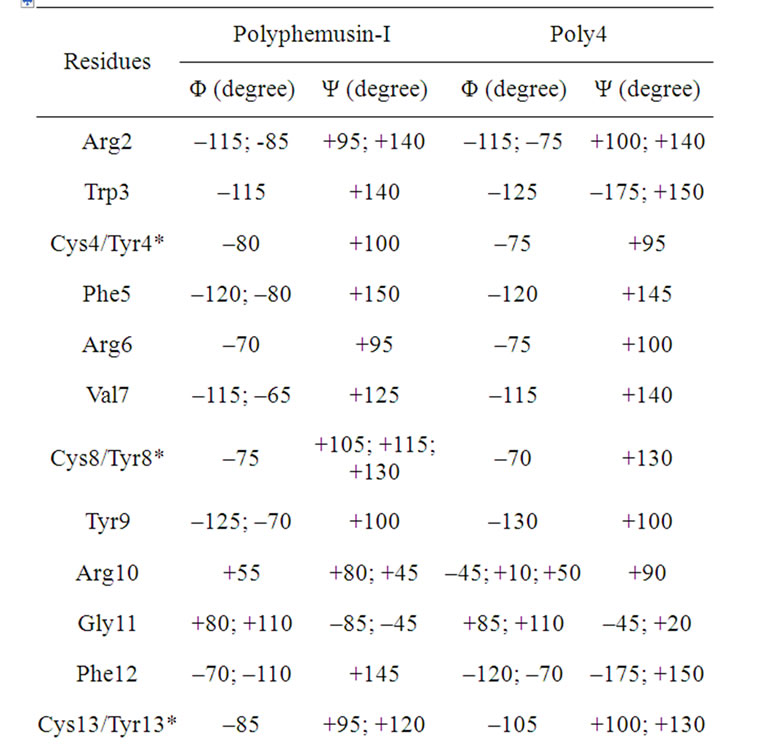

Table 7. The maxima of the Ramachandran angles distributions for the polyphemusin-I and poly4 peptides.
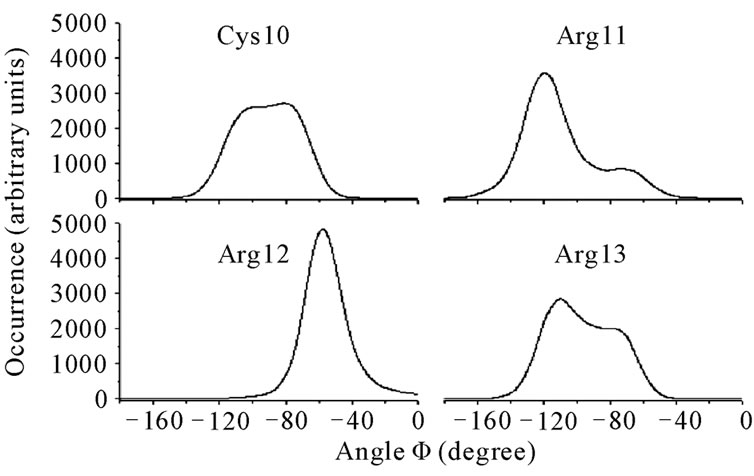
Figure 7. Histograms of the Ramachandran Φ angles for the residues Cys10, Arg11, Arg12, and Arg13 in androctonin.
respectively, while thany2 Ser2 residue has (Φ, Ψ) Ser2 of (–65˚/–120˚, –40˚), Table 8. As reported for the simulations of the gomesin and protegrin-1 peptides [25], the residues number 12 occupy the position where the chain starts to fold. In the case of thanatin, besides the Arg13 residue, where the chain starts to fold, the Φ and Ψ angles of the residues Arg14 and Thr15 are also negative.
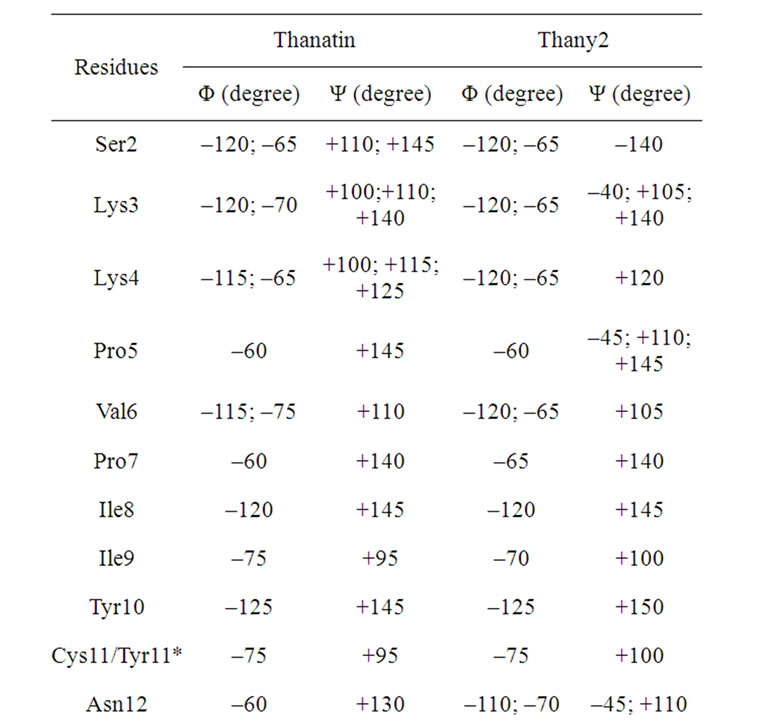
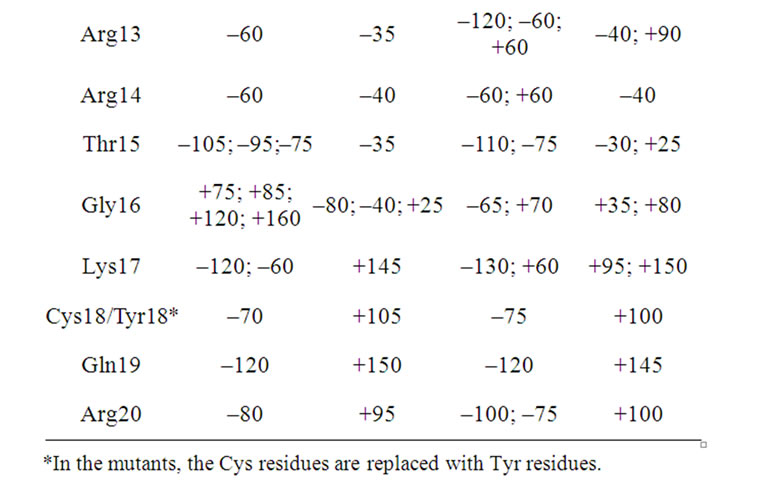
Table 8. The maxima of the Ramachandran angles distributions for the thanatin and thany2 peptides.
*In the mutants, the Cys residues are replaced with Tyr residues.
3.4. Interatomic Distances
The distances between the Cα atoms of the cysteine or tyrosine residues were measured for the three peptides and their respective mutants. The average distances between the Cα atoms of the cysteine residues are due to the covalent disulfide bridges linking antiparallel β-strands [28]. The mean distances between the Cα atoms of the cysteine residues in the simulations of the wild peptides are in very good agreement with the experimental ones (mean value in the PDB files), as seen in Table 9. The relatively larger value of the Cα(Cys4)- (Cys20) mean distance in the androctonin peptide, as reported by Mandard et al. [11], allows the formation of
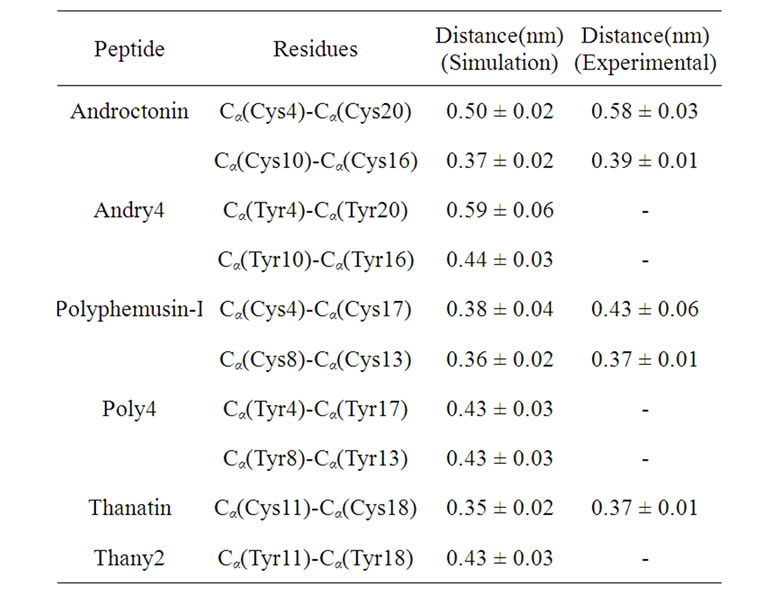
Table 9. Mean distance between the Cα of the Cys residues in the wild peptides and between the Cα of the Tyr residues in the mutants.
the Arg5(CO)-Thr21(NH) HB across the S-S bridge. The Cα(Tyr4)-Cα(Tyr20) average distance (0.59 nm) in andry4 is very similar to the experimental data (0.58 nm) for androctonin. However, the magnitude of the standard deviation (0.06 nm) indicates a considerable fluctuation around the mean value as a result of some flexibility of this region. The Cα(Tyr10)-Cα(Tyr16) average distance (0.44 nm) in andry4 is larger than the corresponding Cα(Cys10)-Cα(Cys16) average distance in androctonin (0.39 nm in experimental data and 0.37 nm in simulation data), showing that this part of the molecule is more separated in andry4 than in androctonin. The distances between Cα found in the simulation of polyphemusin-I are also in good agreement with the experimental ones, considering the magnitude of the standard deviations. The Cα(Tyr4)-Cα(Tyr17) mean distance in poly4 is exactly the same as the experimental value for the Cα(Cys4)-Cα(Cys17) mean distance in the polyphemusin-I peptide, but the standard deviation (0.03 nm) is smaller than the experimental one (0.06 nm), indicating that polyphemusin-I has more flexibility in this region compared with poly4. The Cα(Tyr8)-Cα(Tyr13) mean distance is slightly larger than the corresponding Cα(Cys8)-Cα(Cys13) mean distance in polyphemusin-I, showing that the two sheets in poly4 are more separated than those in the same region in polyphemusin-I. The thanatin Cα(Cys11)-Cα(Cys18) mean distance (simulation data) is very close to the experimental data. The withdrawal of the disulfide bridge in the thany2 peptide causes a small increase in this distance (0.43 nm). All these results suggest that the mutants present good structural similarities with the respective wild peptides. However, the differences in the Cα distances suggest the wild peptides and the mutants do not have exactly the same conformations.
In order to obtain more information about the differences in the secondary structures of the mutants and of the respective wild peptide, some Cα distances between neighbor residues were measured. Large values could mean a loss of the original β-hairpin structure. Figure 8 shows the distributions of the Cα distances for neighboring residues in the androctonin and andry4 peptides (plot A and plot B, respectively). Two kinds of profiles can be observed: one kind with a set of curves between 0.30 and 0.40 nm and a second group with a set of curves appearing between 0.50 and 0.55 nm (Figure 8(a)). This pattern of two kinds of distributions results from the androctonin β-hairpin structure, which is anti-parallel. The same pattern can be observed for andry4 in Figure 8(b), indicating that the mutant does not lose the anti-parallel structure. However, it can be seen that the extremities of the molecule are more separated in andry4 compared with androctonin, as can be seen in Figure 8(b) by the shift to larger values of the distribution curves of the Cα(Ty4)-Cα(Asn22) and Cα(Arg5)-Cα(Thr21) distances (black and red lines, respectively). The distance between the Cα(Tyr10)-Cα(Tyr16) residues in andry4 is larger than that between the corresponding Cα(Cys10)- Cα(Cys16) residues in androctonin (violet line in Figure 8), due to the replacement of the Cys residues with Tyr residues. Once these residues in andry4 do not form an HB, the removal of the disulfide bridge allowed for a slightly increase in the Cα distance. The Cα(Ile7)- Cα(Lys19) distance in andry4 (Figure 8(b) yellow line) is slightly smaller than in androctonin because of the deletion of the disulfide bridges in andry4, which promotes an increase in the HB occurrences shown in Table 3 (82.3% and 85.8% for androctonin, and 87.3% and 90.0% for andry4), thereby resulting in the approximation of these residues. Figure 9 presents the distributions of the Cα distances of neighbor residues in the polyphemusin-I and poly4 peptides (plots A and B, respecttively). Here we also observe two kinds of profiles where one set of curves is found between 0.30 and 0.40 nm and a second set of curves appears between 0.50 and 0.60 nm (Figure 9(a)). The distances between Cα(Tyr4)- Cα(Tyr17) and Cα(Tyr8)-Cα(Tyr13) in poly4 (Figure 9(b), black line, and cyan line respectively) are a little bit larger compared with the corresponding distances Cα(Cys4)-Cα(Cys17) and Cα(Cys8)-(Cys13) in the polyphemusin-I peptide (Figure 8(a), black line and cyan line respectively). The removal of the disulfide bridges and the absence of HB between these residues in poly4 allow these residues to be a little more distant from each other, resulting in small conformational changes. The distance between Cα(Tyr9)-Cα(Phe12) decreases. In this
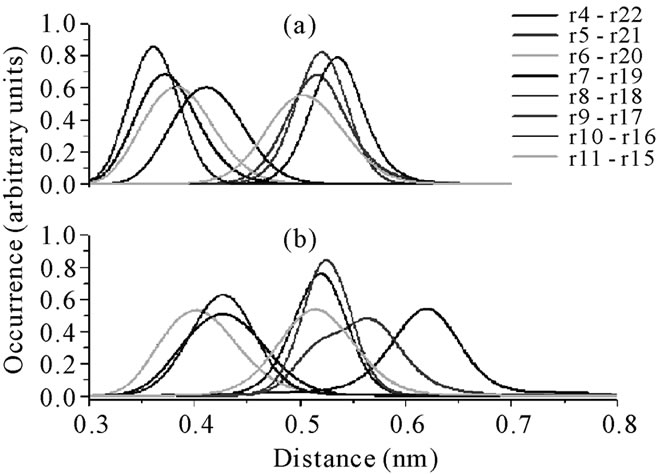
Figure 8. The distributions of the Cα distances of neighbor residues: (a) androctonin. r = residue; r4 = Cys4; r5 = Arg5; r6 = Gln6; r7 = Ile7; r8 = Lys8; r9 = I le9; r10 = Cys10; r11 = Arg11; r15 = Gly15; r16 = Cys16; r17 = Tyr17; r18 = Tyr18; r19 = Lys19; r20 = Cys20; r21 = Thr21; r22 = Asn22; (b) andry4. r = residue; r4 = Tyr4; r5 = Arg5; r6 = Gln6; r7 = Ile7; r8 = Lys8; r9 = Ile9; r10 = Tyr10; r11 = Arg11; r15 = Gly15; r16 = Tyr16; r17 = Tyr17; r18 = Tyr18; r19 = Lys19; r20 = Tyr20; r21 = Thr21; r22 = Asn22.
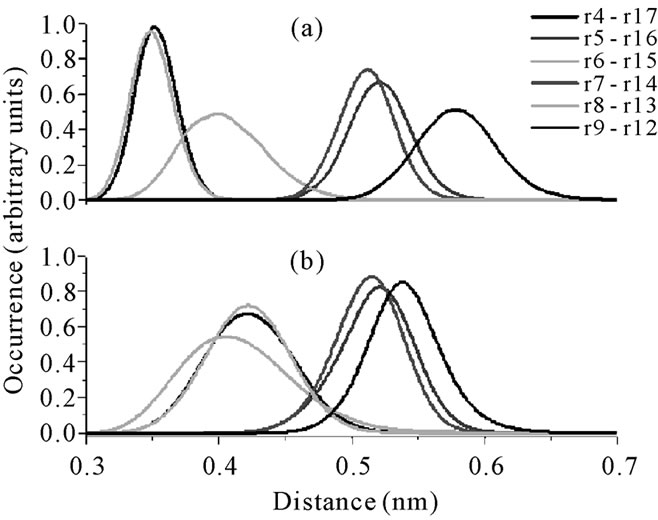
Figure 9. The distributions of the Cα distances of neighbor residues in: (a) polyphemusin-I. r = residue; r4 = Cys4; r5 = Phe5; r6 = Arg6; r7 = Val7; r8 = Cys8; r9 = Tyr9; r12 = Phe12; r13 = Cys13; r14 = Tyr14; r15 = Arg15; r16 = Lys16; r17 = Cys17; (b) poly4. r = residue; r4 = Tyr4; r5 = Phe5; r6 = Arg6; r7 = Val7; r8 = Tyr8; r9 = Tyr9; r12 = Phe12; r13 = Tyr13; r14 = Tyr14; r15 = Arg15; r16 = Lys16; r17 = Tyr17.
case, the absence of the disulfide bridges promoted an increase in the HB occurrence (see Table 4), from 76.5% in polyphemusin-I to 93.3 in andry4, allowing them to get closer. Besides, residues Tyr9 and Phe12 in poly4 form two hydrogen bonds, between Tyr9(NH)- Phe12(CO) and Phe12(NH)-Tyr9(CO). Nevertheless, poly4 continues exhibiting the two sets of curves, characterizing that the mutant does not lose its anti-parallel structure.
Figure 10 shows the distributions of some Cα distances

Figure 10. The distributions of the Cα distances of neighbor residues: (a) thanatin. r = residue; r8 = Ile8; r9 = Ile9; r10 = Tyr10; r11 = Cys11, r12 = Asn12; r13 = Arg13; r16 = Gly16; r17 = Lys17; r18 = Cys18; r19 = Gln19; r20 = Arg20; r21 = Met; (b) thany2. r = residue; r8 = Ile8; r9 = Ile9; r10 = Tyr10; r11 = Cys11, r12 = Asn12; r13 = Arg13; r16 = Gly16; r17 = Lys17; r18 = Cys18; r19 = Gln19; r20 = Arg20; r21 = Met.
of neighbor residues in the thanatin and thany2 peptides (plots A and B, respectively). Two kinds of profiles can be observed: one with a set of curves between 0.30 and 0.40 nm and a second with a set of curves between 0.50 and 0.55 nm (Figure 10(a)). The widening of the Cα(Arg13)-Cα(Gly16) distance distribution curve in thanatin (orange line in Figure 10(a)) is a consequence of the flexibility of the region. The Cα(Tyr11)-Cα(Tyr18) distance distribution in thany2 (cyan line in Figure 10(b)) shifts to larger values compared with the corresponding Cα(Cys11)-Cα(Cys18) distance distribution in thanatin (cyan line in Figure 10(a)), because of the absence of the disulfide bridge in the mutant. In addition, a displacement of the distance distribution curves of the Cα(Asn12)-Cα(Lys17) and Cα(Arg13)-Cα(Gly16) residues (violet and orange lines, respectively, in Figure 10(b)) to smaller values is observed, compared with the same residues of thanatin (Figure 10(a)). This is related to the increase in HB occurrence (from 66.6% in thanatin to 82.9% in thany2, Table 5) and by the new HB between Lys17(NH)-Asn12(CO), which was not present in thanatin. The approximation of the Cα(Arg13)- Cα(Gly16) residues is explained by the formation of the Gly6(NH)-Arg13(CO) HB, which is not present in thanatin (Table 5).
These interatomic distance data have shown that the deletion of the disulfide bridges promotes small conformational changes in the mutant molecules without loss of the initial β-hairpin structure.
3.5. Side Chain Interactions
In order to better understand the contribution of the side chain to the maintenance of the β-hairpin structure of the mutants, the interaction energies between the side chains of the residues were investigated by calculating the interaction energy between the side chain of one residue with the side chain of the others. Only the attractive energies greater than 10 kJ·mol–1 were considered. Attractive interactions between the side chains of the residues Gln6-Tyr18 and Lys8-Tyr18 were found in androctonin apart from hydrogen bonds and the disulfide bridges. Once attractive energies between these residues were detected, the next step was to calculate the average distance between their side chain. The distance between the side chain of the residues Gln6-Tyr18 was calculated considering the center of mass of the group O=C-NH2 (OE1 CD NE2 HE21 HE22 in GROMOS96 notation[18]) in the Gln6 residue and the center of mass of the aromatic ring in the Tyr18 residue. In the case of the Lys8 and Tyr18 residues, the center of mass of the group -NH2 (HZ2 NZ HZ1 in GROMOS96 notation) of the Lys8 residue and the center of mass of the aromatic ring in the Tyr18 residue were considered. The average distances and energies are listed in Table 10. In the mutant andry4, the side chain of the residues Gln6-Tyr18 and Lys8-Tyr18 interact attractively with almost the same average energy as in androctonin (Table 10). This is in good agreement with the interatomic distance data, which reveal no relevant change in the average distance between the residues Lys8-Tyr18 in androctonin and andry4 (olive line in Figures 8(a) and (b)). In addition, another attractive interaction, which did not exist in androctonin, arises between the side chains of the residues Tyr10-Tyr16 exactly where the replacement of Cys10 and Cys16 with Tyr10 and Tyr16 respectively occurred. This is also in good agreement with the interatomic distance data, once the side chain of the Tyr residues are longer than the side chain of the Cys residues, promoting an increase in the average Cα distance between these residues. Here the results suggest a π-π-like interaction. These side chain interactions acting together to the hydrogen bonds are contributing to the maintenance of the andry4 β-hairpin structure.
For polyphemusin-I, only the side chain of the residues Tyr9-Tyr14 interact attractively in a π-π-like interaction. The average distances and energies are listed in Table 11. Therefore, we have found that the attractive interaction between these residues, together with the disulfide bridges and the hydrogen bonds, are responseble for the stabilization of the polyphesumin-I in aqueous media. In poly4, the side chain of the residues Tyr9-Tyr14 do not interact, but attractive interactions arise between the side chains of the residues Tyr4-Arg15, Tyr4-Tyr17, Arg6-Tyr13, and Tyr8-Tyr13. Table 11 shows the average distances and energies for the side
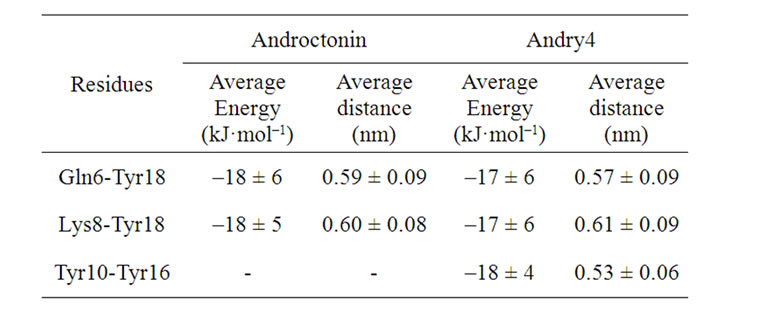
Table 10. Average energy and distance between the interacting side groups in androctonin and andry4.
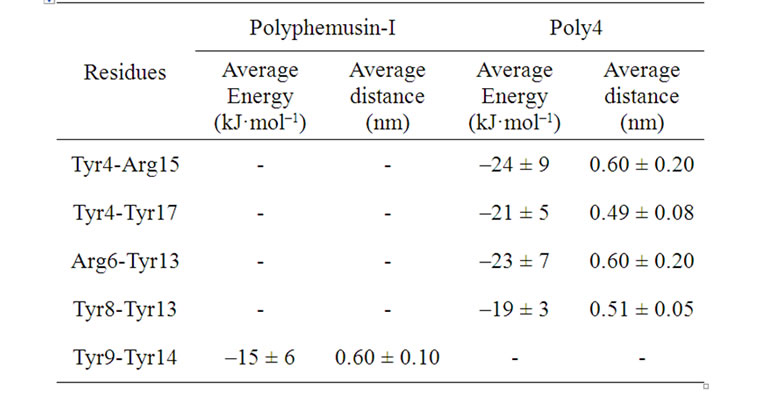
Table 11. Average energy and distance between the interacting side groups in polyphemusin-I and poly4.
chain of these residues. The center of mass of the group –C(NH2)2 (HH11 HH12 NH1 CZ NH2 HH21 HH22 in GROMOS96 notation) in the side chain of the Arg15 and Arg6 residues and the center of mass of the aromatic ring in the Tyr4, Tyr8, Tyr13, and Tyr17 residues were considered in the calculation of the average distances between them. Residues Tyr4-Arg15 and Arg6-Tyr13 interact through a cation-π-like interaction, while residues Tyr4-Tyr17 and Tyr8-Tyr13 interact by a π-π-like interaction. These new interactions, together with the hydrogen bonds, contribute to the maintenance of the β-hairpin structure in poly4, also collaborating with the decreased motion of these residues, as observed in the RMSD per residue in Figure 4. However, while the β-hairpin structure is kept, it can be seen that conformational changes take place (Figure 9). No relevant attractive interactions between the side chains were found in thanatin and thany2.
3.6. The Average Structures
The experimental structures found in the PDB files for androctonin, polyphemusin-I, and thanatin can be used to determine a kind of average structure for each corresponding peptide. Figures 11, 12, and 13 depict the superposition of the average structures of the wild peptides and their respective mutants. Figure 11 presents the superposition of the main chain of androctonin (green) and

Figure 11. Androctonin (green) and andry4 (blue), fit of main chain.

Figure 12. Polyphemusin-I (green) and poly4 (blue). Fit of the main chain.
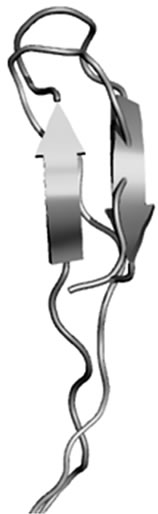
Figure 13. Thanatin (green) and thany2 (blue). Fit of the main chain.
andry4 (blue), revealing their striking structural similarity, which is confirmed by the low RMSD values listed in Table 12. The “tail” of the molecules consisting of the Nand C-terminal extensions presents high RMSD values, confirming that these regions are disordered in aqueous media, as reported by Mardard et al. [11]. The strong structural similarities between androctonin and andry4 are due to a set of facts acting simultaneously in the molecules such as HB formation and side chain interactions, thereby resulting in similar structures.
Figure 12 shows the superposition of the main chain for polyphemusin-I (green) and poly4 (blue), where the β-hairpin structure of polyphemusin-I has been kept in poly4. Nevertheless, the superposition of the main chains of polyphemusin-I and poly4 are not as good as in the case of androctonin/andry4, since slightly different conformations are observed, as confirmed by the RMSD values listed in Table 13. The molecules have conformational similarities only in the region between residues Cys4 (Tyr4 in poly4) and Cys8 (Tyr8 in poly4), where the RMSD is low (0.159 nm), Table 13. These differences may be due to the HB and to the side chain interactions that exist in poly4 and are absent in polyphemusin-I. Figure 13 depicts the superposition of the main chain of thanatin (green) and thany2 (blue). This pair of peptides also have great structural similarities, once the β-hairpin secondary structure is kept in thany2. However, they present low conformational similarities, as seen in Figure 13 and confirmed by the RMSD values of the three regions of the molecules, listed in Table 14. The HBs are not able to maintain the conformational similarities without the cooperation of the side chain attracttive interactions, which were not found in the present case.

Table 12. RMSD of the fit of the main chain of androctonin and andry4.
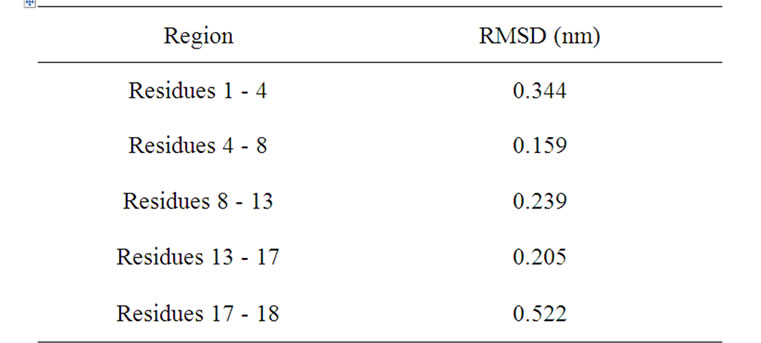
Table 13. RMSD of the fit of the main chain of polyphemusin-I and poly4.

Table 14. RMSD of the fit of the main chain of thanatin and thany2.
4. DISCUSSION
The simulations of the mutants andry4, poly4, and thany2 demonstrate the importance of the cross-strand hydrogen bonds and the side chain interactions (cation-π like and π-π like) for the maintenance of the structural stability in aqueous solution, since most of the HBs were preserved and strengthened (higher occurrence). In addition to the hydrophobic effect, these attractive interacttions are responsible for the structural stability of the mutants. Moreover, the present simulation studies show that the β-hairpin structure in androctonin and polyphemusin-I are stabilized not only by the disulfide bridges and the hydrogen bonds, but also by attractive interacttions between the side chains (cation-π-like and π-π-like). The residues Cys and Tyr are sterically very distinct, but both are hydrophobic, exhibiting amphipathy values of 3.8 and 21.3 respectively, as reported by Mazzé et al. [29]. Besides the hydrophobic effect, both residues interact attractively with the solvent through short range interactions, but they are stronger for Tyr than for Cys because in the former case such interactions are roughly proportional to the number of atoms in the residue [29], and the hydroxyl polar group in Tyr increases such attractive interactions. In addition, the aromatic rings of the Tyr residues interact attractively not only with other Tyr residues, but also with the side chain of the arginine, lysine, and glutamine residues. The net result is an effecttive stability of the structure around the Tyr residues. All these results explain the resulting structural stability of the mutants and, because of the extra hydrophobic packing factor introduced by Tyr residues, they also suggest that the mutants andry4, poly4, and thany2 may accomplish a completely successful folding process, since the covalent disulfide bonds are not necessarily required for correct folding in many cases [30]. The three peptides androctonin, polyphemusin-I, and thanatin present some residues with Ramachandran (Φ,Ψ) angles with more than one preferential angle pair (multimodal distribution), endowing them with structural flexibility despite the disulfide bridges and cross-strand HBs and allowing for systematic and specific conformational exchanges, which may help explain their broad-spectrum of action. They still present the unusual combination of positive or negative Φ and Ψ angles which are located in the turn region (the head) of their respective β-hairpin motifs. Therefore, the Arg12 residue of the androctonin peptide presents both Φ and Ψ angles with negative values. The Φ and Ψ angles of the polyphemusin-I Arg10 residue are both positive, while the Φ and Ψ angles of the three residues Arg13, Arg14, and Thr15 in thanatin are negative: these residues are located in the turn region. The Φ and Ψ of the same residues in the respective mutants undergo a qualitative change, assuming a combination of positive and negative values, respectively. This unusual motif is present in all the three peptides studied in this work and also in the peptides gomesin and protegrin-1 [25]. The interpretation of the consequences of such subtle behavior is that it may promote an increase, or decrease, in the peptide activity, thereby affecting their antimicrobial properties and/or their hemolytic effect. Such an observation deserves a specific experimental investigation.
5. CONCLUSIONS
The influence of disulfide bridges on the structures of the androctonin, polyphemusin-I, and thanatin antimicrobial peptides was investigated by substituting all the cysteine with tyrosine residues. Our results show that no major changes in the β-hairpin structures of the mutants are observed, although conformational changes have been noted. About the conformation, there are signifycant differences between the conformation of the mutants (mainly poly4 and thany2) and the wild peptides because of several factors that act simultaneously, such as hydrogen bonds, side chain interactions (cation-π-like and π-π-like), and hydrophobic effect, which all play important roles and must be considered. Our MD simulations show that, in these cases, the disulfide bridges are not crucial to the maintenance of the β-hairpin structure of the mutants. However, the consequences of these mutations on the antimicrobial activity and on the hemolytic effect of these peptides must be experimentally investigated.
6. ACKNOWLEDGEMENTS
The authors are thankful to Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) for financial support.
REFERENCES
- Hawkey, P.M. (2008) The growing burden of antimicrobial resistance. Journal of Antimicrobial Chemotherapy, 62, i1-i9. doi:10.1093/jac/dkn241
- Mulvey, M.R. and Simor, A.E. (2009) Antimicrobial resistance in hospitals: How concerned should we be? Canadian Medical Association Journal, 180, 408-415. doi:10.1503/cmaj.080239
- Prates, M.V. and Bloch Júnior, C. (2000) Peptídeos antimicrobianos: Uma alternativa no combate a microorganismos resistentes. Biotecnologia, Ciência & Desenvolvimento, 17, 30-36.
- Fazio, M.A., Oliveira, V.X., Bulet, P., Miranda, M.T., Daffre, S. and Miranda, A. (2006) Structure-activity relationship studies of gomesin: Importance of the disulfide bridges for conformation, bioactivities and serum stability. Biopolymers, 84, 205-218. doi:10.1002/bip.20396
- Ramamoorthy, A., Thennarasu, S., Tan, A., Gottipati, K., Sreekumar, S., Heyl, D.L., An, F.Y. and Shelburne, C.E. (2006) Deletion of all cysteines in tachyplesin I abolishes hemolytic activity and retains antimicrobial activity and lipopolysaccharide selective binding. Biochemistry, 45, 6529-6540. doi:10.1021/bi052629q
- Che, Q., Zhou, Y., Yang, H., Li, J., Xu, X. and Lai, R. (2008) A novel antimicrobial peptide from amphibian skin secretions of Odorrana grahami. Peptides, 29, 529-535. doi:10.1016/j.peptides.2008.01.004
- Daffre, S., Miranda, A., Miranda, M.T.M., Bulet, P., Silva, P.I. Jr., Machado, A., Fogaça, A.C., Lorenzini, D.M., Periera, L.S., Fázio, M.A., Esteves, E. and Burgierman, M.R. (2001) Peptídeos antibióticos produzidos por aracní- deos. Biotecnologia Ciência & Desenvolvimento, 23, 48-55.
- Chan, D.I., Prenner, E.J. and Vogel, H.J. (2006) Tryptophan and arginine-rich antimicrobial peptides: Structures and mechanisms of action. Biochimica et Biophysica Acta, 1758, 1184-1202. doi:10.1016/j.bbamem.2006.04.006
- Zasloff, M. (2002) Antimicrobial peptides of multicellular organisms. Nature, 415, 389-395. doi:10.1038/415389a
- Ehret-Sabatier, L., Loew, D., Goyffon, M., Fehlbaum, P., Hoffmann, J.A., Van Dorsselaer, A. and Bulet, P. (1996) Characterization of novel cysteine-rich antimicrobial peptides from scorpion blood. Journal of Biological Chemistry, 271, 29537-29544. doi:10.1074/jbc.271.47.29537
- Mandard, N., Sy, D., Maufrais, C., Bonmatin, J.M., Bulet, P., Hetru, C. and Vovelle, F. (1999) Androctonin, a novel antimicrobial peptide from scorpion Androctonus australis: Solution structure and molecular dynamics simulations in the presence of a lipid monolayer. Journal of Biomolecular Structure and Dynamics, 17, 367-380.
- Powers, J.P.S., Rozek, A. and Hancock, R.E.W. (2004) Structure-activity relationships for the b-hairpin cationic antimicrobial peptide polyphemusin I. Biochimica et Biophysica Acta, 1698, 239-250. doi:10.1016/j.bbapap.2003.12.009
- Mandard, N., Sodano, P., Labbe, H., Bonmatin, J.M., Bulet, P., Hetru, C., Ptak, M. and Vovelle, F. (1998) Solution structure of thanatin, a potent bactericidal and fungicidal insect peptide, determined from proton two-dimensional nuclear magnetic resonance data. European Journal of Biochemistry, 256, 404-410. doi:10.1046/j.1432-1327.1998.2560404.x
- Fehlbaum, P., Bulet, P., Chernysh, S., Briand, J.P., Roussel, J.P., Letellier, L., Hetru, C. and Hoffmann, J.A. (1996) Structure-activity analysis of thanatin, a 21-residue enducible insect defense peptide with sequence homology to frog skin antimicrobial peptides. Proceedings of the National Academy of Sciences, 93, 1221-1225.
- Rao, A.G. (1999) Conformation and antimicrobial activeity of linear derivatives of tachyplesin lacking disulfide bonds. Archives of Biochemistry and Biophysics, 361, 127-134. doi:10.1006/abbi.1998.0962
- Guex, N. and Peitsch, M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis, 18, 2714-2723. doi:10.1002/elps.1150181505
- Van der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A.E. and Berendsen, H.J.C. (2005) GROMACS: Fast, flexible and free. Journal of Computational Chemistry, 26, 1701-1718. doi:10.1002/jcc.20291
- Van Gunsteren, W.F. (1996) Biomolecular simulation: The GROMOS96 manual and user guide. Verlag der Fachvereine Hochschulverlag AG an der ETH Zurich, Zürich.
- Berendsen, H.J.C., Grigera, J.R. and Straatsma, T.P. (1987) The missing term in effective pair potentials. Journal of Physical Chemistry, 91, 6269-6271. doi:10.1021/j100308a038
- Hess, B., Bekker, H., Berendsen, H.J.C. and Fraaije, J.G.E.M. (1997) LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry, 18, 1463-1472. doi:10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.3.CO;2-L
- Miyamoto, S. and Kollman, P.A. (1992) Settle—An analytical version of the Shake and Rattle algorithm for rigid water models. Journal of Computational Chemistry, 13, 952-962. doi:10.1002/jcc.540130805
- Berendsen, H.J.C., Postma, J.P.M., Van Gunsteren, W.F., Dinola, A. and Haak, J.R. (1984) Molecular-dynamics with coupling to an external bath. Journal of Chemical Physics, 81, 3684-3690. doi:10.1063/1.448118
- Darden, T., York, D. and Pedersen, L. (1993) Particle Mesh Ewald—An N-Log(N) method for Ewal sums in large systems. Journal of Chemical Physics, 98, 10089- 10092. doi:10.1063/1.464397
- Van der Spoel, D., Lindahl, E., Hess, B., Van Buuren, A.R., Apol, E., Meulenhoff, P.J., Tieleman, D.P., Sijbers, A.L.T.M., Feenstra, K.A., Van Drunen, R. and Berendsen, H.J.C. (2005) Gromacs User manual version 3.3. http://www.gromacs.org/
- Castro, J.R.M., Fuzo, C.A. and Degrève, L. (2008) The role of the disulfide bridges in the 3D structures of the antimicrobial peptides gomesin and protegrin-1: A molecular dynamics study. Genetics and Molecular Research, 7, 1070-1088. doi:10.4238/vol7-4gmr507
- Santiveri, C.M., Leon, E., Rico, M. and Jimenez, M.A. (2008) Context-dependence of the contribution of disulfide bonds to beta-hairpin stability. Chemistry—A European Journal, 14, 488-499. doi:10.1002/chem.200700845
- Mandard, N., Bulet, P., Caille, A., Daffre, S. and Vovelle, F. (2002) The solution structure of gomesin, an antimicrobial cysteine-rich peptide from the spider. European Journal of Biochemistry, 269, 1190-1198. doi:10.1046/j.0014-2956.2002.02760.x
- Srinivasan, N., Sowdhamini, R., Ramakrishnan, C. and Balaram, P. (1990) Conformations of disulfide bridges in proteins. International Journal of Peptide and Protein Research, 36, 147-155. doi:10.1111/j.1399-3011.1990.tb00958.x
- Mazzé, F.M., Fuzo, C.A. and Degrève, L. (2005) A new amphipaty scale. Determination of the scale from molecular dynamics data. Biochimica et Biophysica Acta, 1747, 35-46. doi:10.1016/j.bbapap.2004.09.019
- Eisenberg, D. and Maclachlan, A.D. (1986) Solvation energy in protein folding and binding. Nature, 319, 199- 203. doi:10.1038/319199a0

