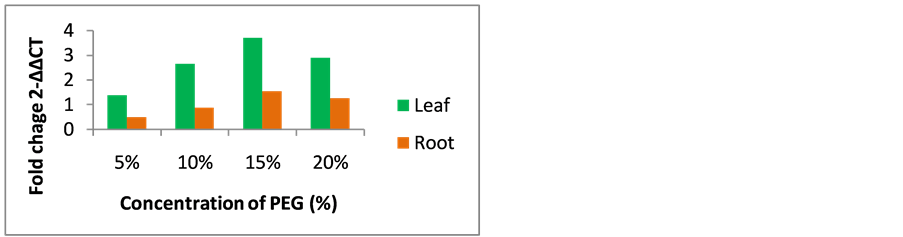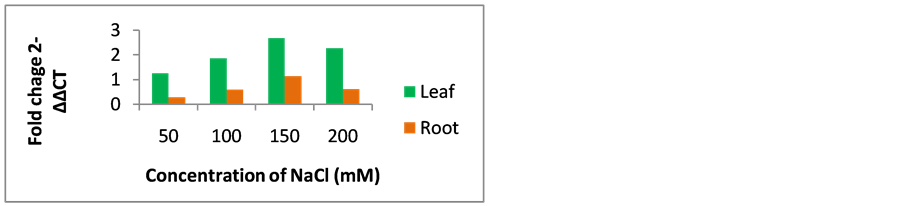American Journal of Plant Sciences
Vol.07 No.03(2016), Article ID:64778,10 pages
10.4236/ajps.2016.73044
Expression Analysis of Aldo-Keto Reductase 1 (AKR1) in Foxtail Millet (Setaria italica L.) Subjected to Abiotic Stresses
Tanguturi Venkata Kirankumar, Kalaiahgari Venkata Madhusudhan, Ambekar Nareshkumar, Kurnool Kiranmai, Uppala Lokesh, Boya Venkatesh, Chinta Sudhakar*
Plant Molecular Biology Laboratory, Department of Botany, Sri Krishnadevaraya University, Anantapuramu, India

Copyright © 2016 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 4 February 2016; accepted 18 March 2016; published 21 March 2016
ABSTRACT
Foxtail millet (Setaria italica L.) is a drought-tolerant millet crop of arid and semi-arid regions. Aldo-keto reductases (AKRs) are significant part of plant defence mechanism, having an ability to confer multiple stress tolerance. In this study, AKR1 gene expression was studied in roots and leaves of foxtail millet subjected to different regimes of PEG- and NaCl-stress for seven days. The quantitative Real-time PCR expression analysis in both root and leaves showed upregulation of AKR1 gene during PEG and salt stress. A close correlation exits between expression of AKR1 gene and the rate of lipid peroxidation along with the retardation of growth. Tissue-specific differences were found in the AKR1 gene expression to the stress intensities studied. The reduction in root and shoot growth under both stress conditions were dependent on stress severity. The level of lipid peroxidation as indicated by MDA formation was significantly increased in roots and leaves along with increased stress levels. Finally, these findings support the early responsive nature of AKR1 gene and seem to be associated at least in part with its ability to contribute in antioxidant defence related pathways which could provide a better protection against oxidative stress under stress conditions.
Keywords:
Aldo-Keto Reductase, Lipid Peroxidation, Reactive Carbonyls, Cellular Homeostasis, Plant Abiotic Stress Response

1. Introduction
Drought and salinity are major constraints on plant productivity, with an evident effect on plant growth. Drought or salt resistance often depends on the ability of the plant to develop adaptive strategies under stress conditions. In order to get resistance plants adapt several antioxidant defense strategies through up-regulation or down- regulation of stress responsive genes [1] . When plants are exposed to high light intensity under different environmental stresses initiates the generation of reactive oxygen species (ROS) in plant cells [2] . Prolonged stress conditions contribute to the disruption of balance between the oxidants and antioxidants, causing oxidative stress. Further, the interaction of ROS with several cellular constituents mainly by damaging nucleic acids, oxidizing proteins and causing lipid peroxidation leads to the production of cellular toxic reactive carbonyls such as Acrolein, 4-hydroxynon-2-enol (HNE) and enhance cellular damage in plants [3] [4] . Scavenging of these carbonyl compounds is essential for maintaining the viability of plants. Several reports have been indicated that aldo-keto reductase genes (AKRs) are stress regulated genes and play a central role in the cellular response to osmotic and oxidative stresses by detoxifying toxic aldehydes generated during stresses in plants and human beings [5] .
AKRs are cytosolic, monomeric, oxidoreductases which catalyse NADPH dependent reduction activity on carbonyl metabolites, belong to the AKR superfamily [6] . AKR family members are characteristic with (α/β)8- barrel motif three dimension structure with a cofactor-binding site and open substrate-binding sites [7] . AKRs are able to metabolize a broad range of substrates such as steroids, sugar, prostaglandins, chalcones, and aliphatic/aromatic aldehyde, as well as some toxins [8] .
The first isolated plant AKR is from barley, showed osmoprotective function by synthesising osmolyte (sorbitol) to acquire desiccation tolerance during embryo development subjected to ABA treatment [9] [10] . Several studies on barley, rice and corn stress inducible AKRs suggested their effective role in detoxification of toxic reactive carbonyls produced during severe oxidative stress. Overproduction of an alfalfa AKR (MsALR) protein in transgenic tobacco plants improves tolerance against variety of oxidative stress induced by methylviologen (MV), heavy metals (CdCl2), osmotic and long periods of drought [11] , cold [12] , UV radiation [13] compared to wild type plants. Heterologous overexpression of OsAKR1 in tobacco plants showed better tolerance against heat [4] . Overexpression of Arabidopsis AKR4C9 in transgenic barley showed enhanced freezing tolerance and higher post-frost regenerative capacity [14] . Peach AKR1 (PpAKR1) overexpression in transgenic Arabidopsis plants indicated its significant role in salt tolerance than wild plants [15] . The direct enzymatic action studies of AKR4C8 and AKR4C9 enzymes from Arabidopsis thaliana supported the AKR active role in detoxification of reactive carbonyls but AKR4C9 enzyme role was specifically observed in MDA and 2-E-Hexenal [16] . All these works suggested the beneficial role of AKR in desiccation, drought, salt, heavy metal, cold stresses by detoxifying cell toxic aldehydes into their respective alcohols (sorbitol). But overproduction of GmAKR1 protein in transgenic soybean hairy roots suggested negative role in inhibiting nodule development [17] .
Malondialdehyde (MDA) is a product of lipid peroxidation and is regarded as a biomarker for evaluation of the damages in plasmalemma and organelle membranes caused by oxidative stress. MDA content in plants increases under environmental stresses. Usually, the better oxidative stress tolerance is accompanied with lower MDA levels. Lipid peroxidation can be estimated as the amount of MDA present in plants as an effect of oxidative damage [18] . Overproduction of OsAKR1 protein in transgenic tobacco plants detected lower levels of MDA and methylglyoxal (MG) in leaf tissues subjected to chemical and heat stress treatments [4] . Based on these observations we can predict the potential role of AKRs in reactive carbonyls detoxification and promoting abiotic stress tolerance as a consequence. However, in plants biological roles of AKR family proteins are still not well defined. Based on these observations, in this study we planned to correlate AKR gene expression with MDA levels in foxtail millet plant tissues.
Foxtail millet is a small diploid (2n = 18), self-pollinating, C4 panicoid short duration drought tolerant crop belongs to the family Poaceae. It is cultivated for food grain, hay and pasture, mainly in arid and semi-arid regions of China, India and Japan are the major foxtail millet growing countries in the world. In India, foxtail millet cultivation is confined to Andhra Pradesh, Karnataka and Tamil Nadu and some parts of Maharashtra [19] . Earlier studies on foxtail millet cultivars indicated that Setaria italica cv. Prasad was a salt tolerant and cv. Lepakshi was a salt susceptible varieties [20] . Moreover, foxtail millet is having a small genome of ~515 Mb and highly conserved genome structure relative to ancestral grass lineages thus it could be considered as an ideal model crop for genetic and molecular studies [21] . Due to the high genetic similarity among cereals, isolated novel stress related genes from foxtail millet will be of great advantage to transfer into other cultivars as well as other crop plants to make them stress tolerant.
In this regard, our present investigation has been made for identifying abiotic stress responsive role of foxtail millet AKR1 gene by qRT-PCR expression in shoot and root tissues under different abiotic stresses. These experimental results, making feasible to use foxtail millet AKR1 gene to develop abiotic stress tolerant crop plants in near future.
2. Materials and Methods
2.1. Plant Materials, Growth Conditions and Stress Treatments
Seeds of foxtail millet (Setaria italica L. cv. Prasad), were procured from Acharya N. G. Ranga regional agricultural research station, Nandyal, Kurnool District, Andhra Pradesh, India. Healthy seeds were surface sterilized with 0.1% mercuric chloride (w/v) solution for 1 minute and thoroughly rinsed thrice with distilled water. Sterilized seeds were spread to germinate on Petri plates lined with filter papers. For stress treatments, seeds were treated with PEG 6000 solutions (5%, 10%, 15% and 20%) for providing osmotic stress, NaCl solutions (50, 100, 150, and 200 mM) for providing salt stress levels. The treatments were characterized as mild, moderate, severe and very stress treatments respectively in PEG and NaCl stress. These seeds containing Petri plates were kept in a growth chamber at mean temperature, 25˚C ± 4.0˚C, relative humidity 60% ± 10.0% for 7 days. The plants were supplemented with water and Hoagland solution on alternate days. Unstressed plants were maintained as control. After stress treatments, root and leaf materials were carefully harvested, immediately flash frozen in liquid nitrogen and stored at −80˚C until RNA isolation. Three independent experiments were conducted for precision and reproducibility, and for each experiment, ~100 mg root and leaf samples were collected by random sampling.
2.2. Growth Measurements
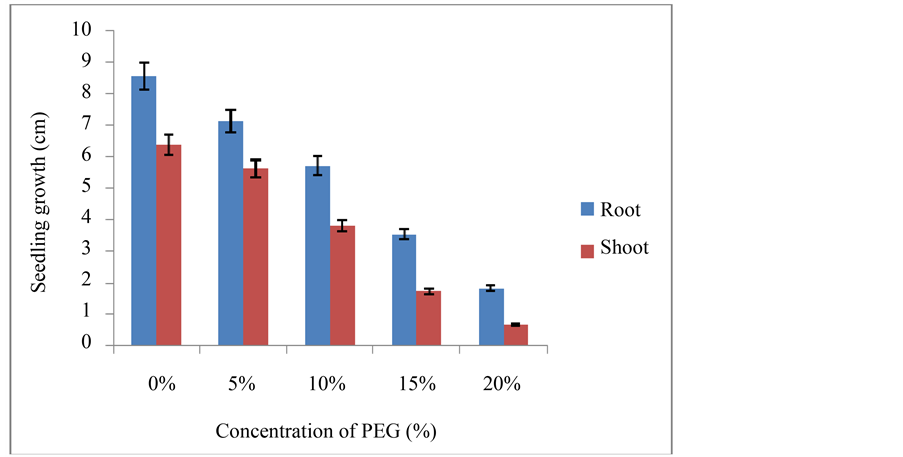
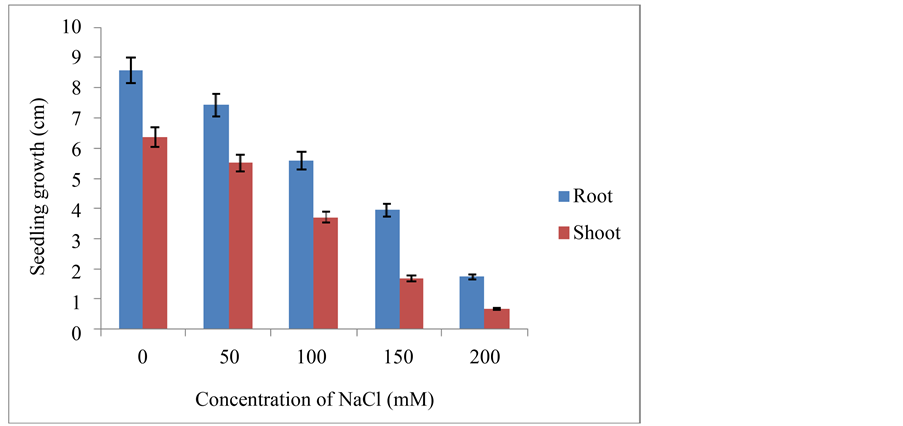
PEG and NaCl stress subjected seven days old seedlings were harvested and their root and shoot growths were measured and recorded. Mean values were depicted in Figure 1 and Figure 2.
2.3. Lipid Peroxidation (MDA) Assay
Lipid peroxidation was determined by measuring the amount of MDA produced by the thiobarbituric acid reaction as described by Heath and Packer [22] . The crude extract was mixed with the same volume of a 0.5% (w/v)
Figure 1. Root and shoot length of foxtail millet seedlings under control and PEG stress (±SD).
Figure 2. Root and shoot length of foxtail millet seedlings under control and NaCl stress (±SD).
thiobarbituric acid solution containing 20% (w/v) trichloroacetic acid. The mixture was heated at 95˚C for 30 min and then quickly cooled in an ice-bath. The mixture was centrifuged at 3000 ×g for 10 min and the absorbance of the supernatant was monitored at 532 nm and 600 nm. After subtracting the non-specific absorbance (600 nm), the MDA concentration was determined by its molar extinction coefficient (155 mM−1∙cm−1).
2.4. RNA Isolation and cDNA Synthesis
Total RNA was isolated by the RNeasy Plant Mini Kit (Qiagen, USA) from frozen leaf and root samples according to manufacturer’s protocol. The concentration of RNA from each samples was determined by UV spectrophotometry at A260, while the quality of total RNAs analyzed by 1% ethidium-bromide agarose-gel electrophoresis. The contaminated genomic DNA was removed by Turbo DNA-free (Ambion, USA) treatment. Then, 5 μg of total RNA was used as a template for cDNA synthesis using M-MLV reverse transcriptase (Fermentas, Germany) with oligo (dT) primer according to the protocol.
2.5. Expression Analysis of Foxtail Millet AKR1 Gene by qRT-PCR
Quantifying gene expression levels is an important research tool to unravel complex regulatory gene networks. Reverse transcription-quantitative real time PCR (qRT-PCR) is a widely used method for gene expression measurements because of its sensitivity, reproducibility and dynamic quantification range [23] . cDNAs were used as templates for qRT-PCR reactions using foxtail millet AKR1 gene specific primers, forward 5’- CTTGCTTGCTGTAGCTCGTC-3’ and reverse 5’-AGGCGTTTTCCCAAGTTTCT-3’. β-actin primers were used as internal controls in this reaction [24] .
The RT-PCR reaction was carried in StepOne RT-PCR machine (Applied Biosystems, USA) conditions followed by 40 cycles of 1min at 95˚C; 1min at 57˚C and 1min at 72˚C. The final extension was carried at 72˚C for 10 min, 1 cycle. After 40 cycles, the specificity of the amplifications was checked by heating from 60˚C to 95˚C with a ramp speed of 1.9˚C∙min−1, resulting in melting curves. Triplicate measurements were carried out to determine the mRNA abundance of each gene in each sample. Data analysis was performed using SDS 2.2.1 software (Applied Biosystems, USA). Amplification curves were analyzed with a normalized reporter (Rn: the ratio of the fluorescence emission intensity of the SYBR Green to the fluorescence signal of the passive reference dye) threshold of 0.2 to obtain the CT values (threshold cycle). Data were normalized to reference gene actin, ΔCT = CT (gene)-CT (Actin). Its expression was measured with three replicates in each PCR run, and the mean CT was used for relative expression analyses. The fold change value was calculated using the expression 2−ΔΔCT, where ΔΔCT represents, ΔCT treatment-ΔCT control. The results obtained were transformed to log2 scale [24] .
2.6. Data Analysis
All data were analyzed using the SPSS (Statistical Package for the Social Sciences) version 16.0. Data presented here are mean values and standard deviation (±SD). One-way ANOVA was carried out using Post hoc multiple comparison from the Duncan’s test at a significance level of p < 0.05.
3. Results
3.1. Effect of PEG and NaCl Stress on Plant Growth
At the whole plant level the effect of stress is usually perceived as a decrease in photosynthesis and growth, and is associated with alteration in carbon and nitrogen metabolism. The total seedlings growth rate was gradually decreased with increasing severity of PEG and NaCl stress concentrations and was dependent on intensity of stress. Significant decrease in the seedling growth was observed at 15% PEG and 150 mM NaCl stresses respectively (Figure 1 and Figure 2). The percent decrease in seedling growth was between 75% to 80% under 20% PEG and 200 mM NaCl stress conditions respectively at the end of experimentation, which were lethal to seedling growth. Likewise, many researchers reported that seedling growth in salt- and stress-containing medium was less than that in the control medium [25] - [27] . This reduced growth under stress has been ascribed either due to osmotic or ionic effects; inhibition of cell division and cell elongation processes associated with the growth of seedling and the decrease in plastic extensibility of the growing cell walls [25] .
3.2. Effect of PEG and NaCl Stresses on MDA Accumulation in Plant Tissues
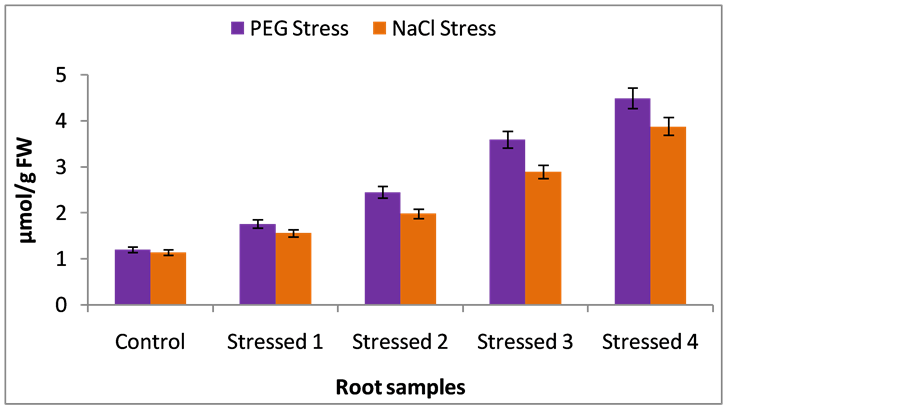
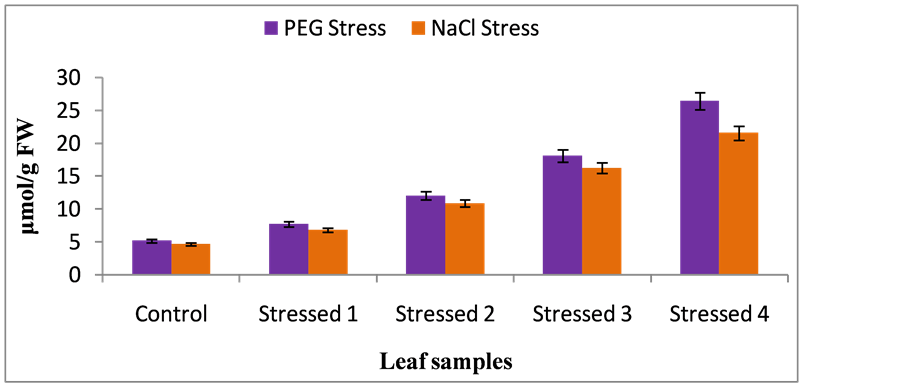
Environmental stresses insists extensive lipid peroxidation, which has often been used as indicator of stress induced oxidative damage in membranes [28] . Total MDA content was estimated in control, PEG-stressed and NaCl-stressed seven days old seedlings root and leaf samples. The total MDA content was significantly increased in root and leaf samples of stressed seedlings over control seedlings under all stress regimes. A linear increase in MDA accumulation was observed with increasing PEG and NaCl stresses. MDA levels were ~8-fold and ~10-fold increase was seen in PEG and NaCl stressed leaf and root samples of foxtail millet seedlings respectively, than control root and leaf samples of foxtail millet seedlings at the end of experimentation (Figure 3 and Figure 4). Similarly, increase in MDA in response to drought and salt stress was also reported by many workers [26] . The increase in MDA levels under stress would result in membrane lipid peroxidation by means of ROS production [29] . The formation of ROS enhances lipid peroxidation at the cellular level and the rate of such enhancement relates to plant species and the severity of stress [30] .
Figure 3. Malondialdehyde (MDA) content in root samples (nmol/g FW) of foxtail millet cultivar under control, PEG-stress and NaCl-stress. The data represent the mean ± SD (n = 3) of three different experiments and the same letters above the bars are not significantly different at p < 0.05 (DMR test).
Figure 4. Malondialdehyde (MDA) content in leaf samples (nmol/g FW) of foxtail millet cultivar under control, PEG-stress and NaCl-stress. The data represent the mean ±SD (n = 3) of three different experiments and the same letters above the bars are not significantly different at p < 0.05 (DMR test).
3.3. Effect of PEG and NaCl Stress on Expression of Foxtail Millet AKR1 Gene in Plant Tissues
The qRT-PCR of foxtail millet AKR1 analysis showed significant expression in leaf and root tissues. In PEG stress conditions, foxtail millet AKR1 transcript expression showed 1.4, 2.6, 3.65 and 2.9-folds expression in 5% PEG, 10% PEG, 15% PEG and 20% PEG stressed leaf samples respectively, whereas 0.5, 0.9, 1.5 and 1.3- folds AKR1 expression was observed in 5% PEG, 10% PEG, 15% PEG and 20% PEG stressed root samples respectively (Figure 5). Foxtail millet AKR1 transcript expression was showed 1.3, 1.9, 2.6 and 2.3-folds in 50 mM NaCl, 100 mM NaCl, 150 mM NaCl and 200 mM NaCl stressed leaf samples respectively, 0.2, 0.55, 1.1 and 0.6-folds expression was observed in 50 mM NaCl, 100 mM NaCl, 150 mM NaCl and 200 mM NaCl stressed root samples respectively (Figure 6).
RT-PCR expression profile of foxtail millet AKR1 mRNA showed that green parts of foxtail millet plants especially leaves have high level of AKR1 mRNA abundance, consistent with the increasing osmotic, dehydration and salt stress treatments. Significant mRNA abundance was seen at 15% PEG, and 150 mM NaCl stress conditions in plant tissues. The stimulation and synthesis of foxtail millet AKR1 gene under abiotic stress conditions that may be an indication of AKR1 gene physiological role in plants defense. The decreased AKR transcript expression supports the lethal stages of plant under severe PEG and NaCl stresses.
4. Discussion
In this present work, at first we observed reduced growth in seven days old foxtail millet seedlings under PEG and salt stress treatments. Growth reduction is proportionate to the increased severity of NaCl and PEG stress conditions in root and shoot lengths in seedlings. It is well documented that abiotic stress cause an important modification in gene expression in plants. Such modification may lead to accumulation or depletion of certain metabolites, and synthesis of new set of proteins that are specific to the particular type of stress which possibly adapt plants to the stressful environment by physiological and biochemical adjustments.
A number of studies demonstrated that ROS scavenging mechanisms have an important role in protecting plants against high light induced cellular damages under different abiotic stresses [31] . Environmental stresses stimulates the ROS production as a consequence imbalance between ROS and scavenging enzymes occurs that promotes extensive lipid peroxidation, which has often been used as indicator of stress induced oxidative damage in membranes [28] . Lipid peroxidation majorly occur in the vicinity of biomembrane having polyunsaturated fatty acids may lead to the formation of lipid hydroperoxides and diffuse throughout chloroplast forms reactive carbonyl products such as MDA, MG, acrolein, hexenal etc. [32] . Parallel to these observations, in the present study we observed a significant increase in MDA content in roots and leaf samples of stressed foxtail
Figure 5. Quantification of foxtail millet AKR1 expression level in leaf and root PEG subjected samples by comparative CT.
Figure 6. Quantification of foxtail millet AKR1 expression level in leaf and root NaCl stressed samples by comparative CT.
millet seedlings. However a significant increase in MDA content was recorded in 20% PEG and 200 mM NaCl stress treatments. The degree of increase was dependent on severity of stress in foxtail millet cultivar. Increase in MDA content in response to drought and salt stress in plant tissues was also reported by many workers [26] . The increase in MDA levels under stress would result in membrane lipid peroxidation by means of ROS production [29] . Lipid peroxidation brings about the loss of cell membrane integrity leading to electrolyte leakage and chlorophyll breakdown by reducing the rate of photosynthesis [33] . Lipid peroxidation can occur in both chloroplasts and mitochondria [34] .
Aldo-keto reductase (AKRs) are stress regulated genes and play a central role in detoxification of toxic aldehydes like MDA, Methylglyoxal, acrolein etc., generated during water stress [5] . AKR protein was detected in vegetative tissues of rice mainly by following dehydration, salt-stress and exogenous ABA treatments [8] [35] . In present study, qRT-PCR expression analysis of foxtail millet AKR1 showed that AKR1 transcript abundance was shown high in green parts of foxtail millet seedlings especially leaves. AKR1 mRNA abundance was consistent with the increasing osmotic (PEG) and salt stress treatments. Because leaves are directly exposed to sunlight under different abiotic stresses thus may cause the generation of ROS leading to higher lipid peroxidation in leaves compared to roots as a consequence of toxic reactive carbonyls accumulation. As a part plant defense foxtail millet AKR1 gene may be up-regulated to detoxify these reactive carbonyls into their corresponding alcohols in the leaves.
The decreased AKR transcript expression supports the drastic increase in MDA levels along with lethal stages of plant under 20% PEG and 200 mM NaCl stresses respectively at the end of experimentation. These results support the argument for a mechanism of water stress tolerance operating at the cellular level and a very strong correlation between AKR1 gene expression and the ability of foxtail millet tissue to grow on media amended with NaCl and PEG. Very severe stress treatments produced a significant increase in leaf MDA. As a part of stress tolerance a substantial increase in mRNA abundance of AKR1 was observed in increasing severity of PEG and NaCl stresses. However our results support the publicly available data [8] [14] [16] . Moreover our results indicated the early responsive and positively up-regulation nature of foxtail millet AKR1 gene in contribution of plant defense under osmotic (PEG) and NaCl stress conditions. Perhaps their products could be involved in various metabolic functions to promote stress tolerance conditions in foxtail millet associated at least in part to enhance of ROS scavenging capacity. This protective effect appears to prevent cellular damage and allow seedlings to retain high levels of metabolic activity and growth under environmental stresses.
5. Conclusion
Based on the above conditions, we can predict that foxtail millet AKR1 gene is a promising stress responsible gene to modulate and enhance the stress tolerance in major crops. In conclusion, we made an initial attempt to identify the abiotic stress responsive foxtail millet AKR1 gene further it would be feasible in developing stress tolerant crops in a part of agronomical importance.
Acknowledgements
The financial support from the UGC, Government of India, New Delhi in the form of research grants to Chinta Sudhakar is gratefully acknowledged.
Cite this paper
Tanguturi Venkata Kirankumar,Kalaiahgari Venkata Madhusudhan,Ambekar Nareshkumar,Kurnool Kiranmai,Uppala Lokesh,Boya Venkatesh,Chinta Sudhakar, (2016) Expression Analysis of Aldo-Keto Reductase 1 (AKR1) in Foxtail Millet (Setaria italica L.) Subjected to Abiotic Stresses. American Journal of Plant Sciences,07,500-509. doi: 10.4236/ajps.2016.73044
References
- 1. Gill, S.S. and Tuteja, N. (2010) Reactive Oxygen Species and Antioxidant Machinery in Abiotic Stress Tolerance in Crop Plants. Plant Physiology and Biochemistry, 48, 909-930.
http://dx.doi.org/10.1016/j.plaphy.2010.08.016 - 2. Yamauchi, Y., Hasegawa, A., Taninaka, A., Mizutani, M. and Sugimoto, Y. (2011) NADPH-Dependent Reductases Involved in the Detoxificaiton of Reactive Carbonyls in Plants. The Journal of Biological Chemistry, 286, 6999-7009.
http://dx.doi.org/10.1074/jbc.M110.202226 - 3. Colrat, S., Latche, A., Guis, M., Pech, J.C., Bouzayen, M., Fallot, J. and Roustan, J.P. (1999) Purification and Characterization of a NADPH-Dependent Aldehyde Reductase from Mung Bean That Detoxifies Eutypine, a Toxin from Eutypa lata. Plant Physiology, 119, 621-626.
http://dx.doi.org/10.1104/pp.119.2.621 - 4. Turoczy, Z., Kis, P., Torok, K., Cserhati, M., Lendvai, A., Dudits, D. and Horvath, G.V. (2011) Overproduction of a Rice Aldo-Keto Reductase Increases Oxidative and Heat Stress Tolerance by Malondialdehyde and Methylglyoxal Detoxification. Plant Molecular Biology, 75, 399-412.
http://dx.doi.org/10.1007/s11103-011-9735-7 - 5. Hyndman, D., Bauman, D.R., Heredoa, V.V. and Penning, T.M. (2003) The Aldo-Keto Reductase Superfamily Homepage. Chemico-Biological Interactions, 143, 621-631.
http://dx.doi.org/10.1016/S0009-2797(02)00193-X - 6. Bohren, K.M., Bullock, B., Wermuth, B. and Gabbay, K.M. (1989) The Aldo-Keto Reductase Superfamily: cDNAs and Deduced Amino Acid Sequences of Human Aldehyde and Aldose Reductase. The Journal of Biological Chemistry, 264, 9547-9551.
- 7. Mindnich, R.D. and Penning, T.M. (2009) Aldo-Keto Reductase (AKR) Superfamily: Genomics and Annotation. Genome Review, 3, 362-370.
- 8. Narawongsanont, R., Kabinpong, S., Auiyawong, B. and Tantitadapitak, C. (2012) Cloning and Characterization of AKR4C14, a Rice Aldo-Keto Reductase, from Thai Jasmine Rice. The Protein Journal, 31, 35-42.
http://dx.doi.org/10.1007/s10930-011-9371-8 - 9. Bartels, D., Engelhardt, K., Roncarati, R., Schneider, K., Rotter, M. and Salamini, F. (1991) An ABA and GA Modulated Gene Expressed in the Barley Embryo Encodes in Aldose Reductase Related Protein. The EMBO Journal, 5, 1037-1043.
- 10. Roncarati, R., Salamini, F. and Bartels, D. (1995) An Aldose Reductase Homologous Gene from Barley: Regulation and Function. The Plant Journal, 7, 809-822.
http://dx.doi.org/10.1046/j.1365-313X.1995.07050809.x - 11. Oberschall, A., Deak, M., Torok, K., Sass, L., Vass, I., Kovacs, I., Feher, A., Dudits, D. and Horvath, G.V. (2000) A Novel Aldose/Aldehyde Reductase Protects Transgenic Plants against Lipid Peroxidation under Chemical and Drought Stress. The Plant Journal, 24, 437-446.
http://dx.doi.org/10.1046/j.1365-313x.2000.00885.x - 12. Hegedus, A., Erdei, S., Janda, T., Toth, E., Horvath, G. and Dudits, D. (2004) Transgenic Tobacco Plants Overproducing Alfalfa Aldose/Aldehyde Reductase Show Higher Tolerance to Low Temperature and Cadmium Stress. Plant Science, 166, 1329-1333.
http://dx.doi.org/10.1016/j.plantsci.2004.01.013 - 13. Hideg, E., Nagy, T., Oberschall, A., Dudits, D. and Vass, I. (2003) Detoxification Function of Aldose/Aldehyde Reductase during Drought and Ultra Violet-B (230-320 nm) Stresses. Plant, Cell and Environment, 26, 513-522.
http://dx.doi.org/10.1046/j.1365-3040.2003.00982.x - 14. Eva, C., Zelenyanszki, H., Farkas, R.T. and Tamas, L. (2014) Transgenic Barley Expressing the Arabidopsis AKR4C9 Aldo-Keto Reductase Enzyme Exhibits Enhanced Freezing Tolerance and Regenerative Capacity. South African Journal of Botany, 93, 179-184.
http://dx.doi.org/10.1016/j.sajb.2014.04.010 - 15. Kanayama, Y., Mizutani, R., Yaguchi, S., Hojo, A., Ikeda, H., Nishiyama, M. and Kanahama, K. (2014) Characterization of an Uncharacterized Aldo-Keto Reductase Gene from Peach and Its Role in Abiotic Stress Tolerance. Phytochemistry, 104, 30-36.
http://dx.doi.org/10.1016/j.phytochem.2014.04.008 - 16. Simpson, P.J., Tantitadapitak, C., Reed, A.M., Mather, O.C., Bunce, C.M., White, S.A. and Ride, J.P. (2009) Characterization of Two Novel Aldo-Keto Reductases from Arabidopsis: Expression Patterns, Broad Substrate Specificity, and an Open Active-Site Structure Suggest a Role in Toxicant Metabolism Following Stress. Journal of Molecular Biology, 392, 465-480.
http://dx.doi.org/10.1016/j.jmb.2009.07.023 - 17. Hur, Y., Shin, K., Kim, S., Nam, K.H., Lee, M., Chun, J. and Cheon, C. (2009) Overexpression of GmAKR1 a Stress-Induced Aldo-Keto Reductase from Soybean, Retards Nodule Development. Molecules and Cells, 27, 217-223.
http://dx.doi.org/10.1007/s10059-009-0027-x - 18. Bailly, C., Benamar, A. and Corbineau, Y. (1996) Changes in Malondialdehyde Content and in Superoxide Dismutase, Catalase and Glutathione Reductase Activities in Sunflower Seeds as Related to Deterioration during Accelerated Aging. Physiolagia Plantarum, 97, 104-110.
http://dx.doi.org/10.1111/j.1399-3054.1996.tb00485.x - 19. Yi, F., Xie, S., Liu, Y., Qi, X. and Yu, J. (2013) Genome-Wide Characterization of microRNA in Foxtail Millet (Setaria italica). BMC Plant Biology, 13, 212-226.
- 20. Veeranagamalllaiah, G., Ranganayakulu, G.S., Thippeswamy, M., Sivakumar, M., Eswaranarayana Reddy, K., Pandurangaiah, M., Sridevi, V. and Sudhakar, C. (2009) Aldose Reductase Expression Contributes in Sorbital Accumulation and 4-Hydroxynon-2-enal Detoxification in Two Foxtail Millet (Setaria italica L.) Cultivars with Different Salt Stress Tol-erance. Plant Growth Regulation, 59, 137-143.
http://dx.doi.org/10.1007/s10725-009-9396-6 - 21. Chen, Z., Chen, M., Xu, Z.S., Li, L.C., Chen, X.P. and Ma, Y.Z. (2014) Characteristics and Expression Patterns of the Aldehyde Dehydrogenase (ALDH) Gene Superfamily of Foxtail Millet (Setaria italica L.). PLOS ONE, 9, 7.
http://dx.doi.org/10.1371/journal.pone.0101136 - 22. Heath, R.L. and Packer, L. (1968) Photoperoxidation in Isolated Chloroplasts: I. Kinetics and Stoichiometry of Fatty acid Peroxidation. Archives in Biochemistry and Biophysics, 125, 189-198.
http://dx.doi.org/10.1016/0003-9861(68)90654-1 - 23. Dekkers, B.J.W., Willems, L., Bassel, G.W., Marieke, R.P., Veldkamp, V.B., Ligterink, W., Hilhorst, H.W.M. and Bentsink, L. (2012) Identification of Reference Genes for RT-qPCR Expression Analysis in Arabidopsis and Tomato Seeds. Plant Cell Physiology, 53, 28-37.
http://dx.doi.org/10.1093/pcp/pcr113 - 24. Caldana, C., Scheible, W.R., Roeber, B.M. and Ruzicic, S. (2007) A Quantitative RT-PCR Platform for High-Through-put Expression Profiling of 2500 Rice Transcription Factors. Plant Methods, 3, 1.
http://dx.doi.org/10.1186/1746-4811-3-7 - 25. Veeranagamallaiah, G., Chandraobulreddy, P., Jyothsnakumari, G. and Sudhakar, C. (2007) Glutamine Synthetase Expression and Pyrroline-5-Carboxylate Reductase Activity Influence Proline Accumulation in Two Cultivars of Foxtail Millet (Setaria italica L.) with Differential Salt Sensitivity. Environmental and Experimental Botany, 60, 239-244.
http://dx.doi.org/10.1016/j.envexpbot.2006.10.012 - 26. Kusvuran, S., Ellioltioglu, S. and Polat, Z. (2013) Antioxidative Enzyme Activity, Lipid Peroxidation, and Proline Accumulation in the Callus Tissues of Salt and Drought Tolerant and Sensitive Pumpkin Genotypes under Chilling Stress. Horticulture, Environment and Biotechnology, 54, 319-325.
http://dx.doi.org/10.1007/s13580-013-1042-6 - 27. Lata, C. (2015) Advances in Omics for Enhancing Abiotic Stress Tolerance in Millets. Proceedings of Indian National Science Academy, 81, 397-415.
- 28. Hernandez, J.A. and Almansa, M.S. (2002) Short-Term Effects of Salt Stress on Antioxidant Systems and Leaf Water Relations of Pea Leaves. Physiologia Plantarum, 115, 251-257.
http://dx.doi.org/10.1034/j.1399-3054.2002.1150211.x - 29. Sairam, R.K. and Tyagi, A. (2004) Physiology and Molecular Biology of Salinity Stress Tolerance in Plants. Current Science, 86, 407-421.
- 30. Wang, X. and Hun, J. (2009) Changes of Proline Content Antioxidation Isoforms in Two Alfalfa Cultivars under Salt Stress. Agricultural Sciences in China, 8, 431-440.
http://dx.doi.org/10.1016/S1671-2927(08)60229-1 - 31. Suzuki, N. and Mittler, R. (2006) Reactive Oxygen Species and Temperature Stresses: A Delicate Balance between Signaling and Destruction. Physiologia Plantarum, 126, 46-51.
http://dx.doi.org/10.1111/j.0031-9317.2005.00582.x - 32. Yamunchi, Y., Hasegawa, A., Mizutani, M. and Sugimoto, Y. (2012) Chloroplastic NADPH-Dependent Alkenal/One Oxidoreductase Contributes to the Detoxification of Reactive Carbonyls Produced under Oxidative Stress. Federation of European Biochemical Societies Letters, 586, 1208-1213.
http://dx.doi.org/10.1016/j.febslet.2012.03.013 - 33. Reddy, D.Y., Reddy V.R. and Anbumozhi, V. (2003) Physiological Responses of Groundnut (Arachis hypogea L.) to Drought Stress and Its Amelioration: A Critical Review. Plant Growth Regulation, 41, 75-88.
http://dx.doi.org/10.1023/A:1027353430164 - 34. Bowler, C., Montagu, M.V. and Inze, D. (1992) Superoxide Dismutase and Stress Tolerance. Annual Review of Plant Physiology and Molecular Biology, 43, 83-116.
http://dx.doi.org/10.1146/annurev.pp.43.060192.000503 - 35. Sree, B.K., Rajendrakumar, S.V. and Reddy, A.R. (2000) Aldose Reductase in Rice (Oryza sativa L.): Stress Response and Developmental Specificity. Plant Science, 160, 149-157.
http://dx.doi.org/10.1016/S0168-9452(00)00376-9
Abbreviations
AKR (Aldo-Keto Reductase),
ROS (Reactive Oxygen Species),
qRT-PCR (Quantitative Real-Time PCR),
PEG (Poly Ethylene Glycol),
MDA (Malondialdehyde),
NADPH (Nicotinamide Adenine Dinucleotide Phosphate).
NOTES
*Corresponding author.