American Journal of Plant Sciences
Vol.06 No.13(2015), Article ID:59203,10 pages
10.4236/ajps.2015.613211
Glyphosate Resistance in Giant Ragweed (Ambrosia trifida L.) from Mississippi Is Partly Due to Reduced Translocation
Vijay K. Nandula1*, Alice A. Wright1, Christopher R. Van Horn2, William T. Molin1, Phil Westra2, Krishna N. Reddy1
1Crop Production Systems Research Unit, U.S. Department of Agriculture-Agricultural Research Service, Stoneville, MS, USA
2Department of Bioagricultural Sciences and Pest Management, Colorado State University, Fort Collins, CO, USA
Email: *vijay.nandula@ars.usda.gov
Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 23 July 2015; accepted 22 August 2015; published 27 August 2015
ABSTRACT
A giant ragweed population from a glyphosate-resistant (GR) soybean field in Mississippi, USA was suspected to be resistant to glyphosate. Greenhouse and laboratory studies were conducted to confirm and quantify the magnitude of glyphosate resistance in a resistant biotype selected from this population and to elucidate possible physiological and molecular mechanisms of glyphosate resistance. Glyphosate dose response studies indicated that ED50 (effective dose required to reduce plant growth by 50%) values for glyphosate-resistant (GR-MS) and glyphosate-susceptible (GS-MS) biotypes, based on percent injury, were 0.52 and 0.34 kg ae/ha glyphosate, respectively, indicating a 1.5-fold level of resistance in GR-MS. The absorption pattern of 14C-glyphosate in the two giant ragweed biotypes was similar throughout the measured time course of 168 h after treatment (HAT). The amount of 14C-glyphosate that translocated out of treated leaves of the GR- MS and GS-MS plants was similar up to 24 HAT. However, the GS-MS biotype translocated more (71% and 76% of absorbed at 48 and 96 HAT, respectively) 14C-glyphosate than the GR-MS biotype (44% and 66% of absorbed at 48 and 96 HAT, respectively) out of the treated leaf. No target site mutation was identified at the Pro106 location of the EPSPS gene of the GR-MS biotype. The mechanism of resistance to glyphosate in giant ragweed from Mississippi, at least, is due to reduced glyphosate translocation.
Keywords:
Absorption, EPSPS, Giant Ragweed, Glyphosate, Herbicide Resistance, Translocation

1. Introduction
Glyphosate, a nonselective, broad-spectrum, systemic, postemergence herbicide, has been used extensively throughout the world in both crop and noncrop lands since its commercialization in 1974. With the introduction of glyphosate-resistant (GR) crops in the mid-1990s, glyphosate has been used selectively and predominantly for weed control in GR crops. The widespread adoption of GR crops around the world with the associated use of repeat glyphosate applications has resulted in the evolution of several GR weed biotypes. As of July 2015, resistance to glyphosate was confirmed in 32 weed species, including giant ragweed (Ambrosia trifida L.) [1] . Giant ragweed, resistant to glyphosate, has been reported in Arkansas, Indiana, Iowa, Kansas, Kentucky, Minnesota, Mississippi, Missouri, Nebraska, Ohio, Tennessee, and Wisconsin in the US and in Ontario, Canada [1] - [7] .
Giant ragweed is an erect summer annual weed that is commonly found in ditches, stream and creek banks, and roadsides. It has gained importance as an agronomic weed due to its competitive ability in row crops. For instance, interference from giant ragweed at densities of 1 to 13.8 plants/m2 reduced corn (Zea mays L.) yield by 14% to 90% [8] . Several traits of sweet corn ears were affected by competition from giant ragweed [9] . Cotton (Gossypium hirsutum L.) lint yield was reduced by 50% when giant ragweed was present at a density of 0.26 plants/m crop row [10] .
A state-wide survey was conducted in the summer of 2009 in Mississippi to document existing GR weed populations in agronomic crops. As a part of this survey, a grower from Tunica County in northwestern Mississippi reported lack of control of a giant ragweed population from glyphosate applications in GR soybean (Glycine max (L.) Merr.). The objectives of this research were to confirm and quantify the magnitude of glyphosate resistance in giant ragweed from northwestern Mississippi and to elucidate the physiological and molecular mechanisms of glyphosate resistance in this giant ragweed.
2. Materials and Methods
2.1. Seed Collection, Storage, Germination, Planting, Growth, and Herbicide Treatment Conditions
In the summer of 2009 mature giant ragweed plants suspected (apical and/or one or more lateral/terminal meristems uninjured and actively growing) to be resistant to glyphosate were identified in a field in Tunica County, Mississippi that had been continuously planted to GR soybean in 2009 and the preceding six years. Six giant ragweed plants were collected along with roots and transplanted to 10 L pots containing field soil and allowed to grow outdoors until they set seed. Giant ragweed inflorescences containing seeds were collected, kept separate for each individual plant (not bulked), and air-dried in a greenhouse (25˚C/20˚C day/night, 12-h photoperiod under natural sunlight conditions) for 7 d, cleaned, and stored at 0˚C until further use. Seeds from the above six plants were planted at 1.25-cm depth in 50-cm by 20-cm by 6-cm plastic trays with holes containing a commercial potting mix (Metro-Mix 360, Sun Gro Horticulture, Bellevue, WA, USA), watered, and incubated at 2˚C to 8˚C for 30 d. Thereafter seed trays were returned to above mentioned greenhouse conditions supplemented with high pressure sodium lights providing 400 mmol/m2/s. Two weeks after emergence, giant ragweed seedlings were transplanted into 6-cm by 6-cm by 6-cm pots containing the potting mix. Plants were fertilized once with a nutrient solution (Miracle-Gro, The Scotts Company LLC, Marysville, OH, USA) containing 200 mg/L each of N, P2O5, and K2O one week after transplanting and sub-irrigated as needed, thereafter. All herbicide treatments were applied with a moving nozzle sprayer equipped with 8002E nozzles (Spraying Systems Co., Wheaton, IL, USA) delivering 140 L/ha at 280 kPa to giant ragweed plants that were 15-cm tall and at the three-leaf (node) stage. Percent control [visible estimate of injury on a scale of 0 (no injury) to 100 (plant death)] was recorded 4 weeks after treatment (WAT). Seed from a designated glyphosate-susceptible (GS-MS) giant ragweed biotype was collected from a fence row along a non-agricultural wooded area in southern Bolivar County, Mississippi, processed, and screened as described for the suspected GR plants. The natural conditions at sites from where the parent GR and GS giant ragweed seed was collected were similar (data not shown) and were not expected to have an impact on the experimental studies. All studies were conducted from 2011 to 2013 at the Jamie Whitten Delta States Research Center of USDA-ARS in Stoneville, Mississippi.
2.2. Screening with a Discriminating Glyphosate Dose
In preliminary resistance screening studies, 25 plants each from seed of six original field-collected plants, suspected to be GR, and GS-MS, were treated with a 0.84 kg ae/ha rate of glyphosate (potassium salt, Roundup WeatherMAX®, Monsanto Company, St. Louis, MO, USA) (data not shown). A single plant survived 4 WAT, matured, and produced the second generation seed. Additional screening experiments indicated that all the second generation plants survived a glyphosate treatment of 0.84 kg/ha (data not shown). This second generation GR seed, henceforth referred as GR-MS biotype, was used in all subsequent studies.
2.3. Glyphosate Dose Response
GR-MS and GS-MS giant ragweed plants were treated with glyphosate at 0, 0.21, 0.42, 0.84, 1.68, and 3.36 kg/ha, representing rates overlapping the 1X field application rate of 0.84 kg∙ha−1. The above range of rates was determined based on the preliminary screening results. Percent visual injury ratings (0 = no effect on growth, 100 = complete kill) were recorded 4 WAT. There were four replications per treatment and the experiment was conducted two times.
2.4. 14C-Glyphosate Absorption, Translocation, and Phosphor Imaging
GR-MS and GS-MS giant ragweed plants were treated with glyphosate as described before, except that one of the fully expanded leaves at the second node was covered with a water-resistant paper sleeve. This sleeve was removed immediately after herbicide treatment for subsequent (within 30 min of overspray) application of solutions containing 14C-glyphosate (14C-methyl labeled with 2.0 GBq/mmol specific activity, 99.5% radiochemical purity in an aqueous stock solution of 7.4 MBq/mL, American Radiolabeled Chemicals, Inc., St. Louis, MO, USA). A solution containing glyphosate at a final concentration equivalent to 0.84 kg/ha in 140 L was prepared using 14C-glyphosate, a commercial formulation of glyphosate, and distilled water. A 10-µL volume of the treatment solution, containing 5 kBq of 14C-glyphosate, was applied to the adaxial surface of the non sprayed third true leaf of 15-cm tall plants in the form of 25 droplets with a micro applicator.
Plants were harvested at 1, 4, 8, 24, 48, 96, and 168 h after 14C-glyphosate treatment (HAT) for absorption and translocation measurements. The treated leaf was removed and immersed in 10 mL 10% methanol in a glass vial and gently shaken for 20 s to remove nonabsorbed 14C-glyphosate remaining on the leaf surface. The washed leaf was rewashed with an additional 10 mL of 10% methanol. Two 1-ml aliquots of each leaf wash were mixed with 10 mL scintillation cocktail (Ecolume, ICN, Costa Mesa, CA, USA) to measure nonansorbed 14C-glyphosate.
Phosphorimaged (see below) plants were carefully removed from the mounting paper and divided into treated leaf, shoot above treated leaf (SATL), shoot below treated leaf (SBTL), and roots for measuring translocation. The plant parts were wrapped in a single layer of tissue paper (Kimwipes, Kimberly-Clark Corporation, Roswell, GA), placed in a glass vial, and oven dried at 60˚C for 24 h. Oven-dried plant samples were combusted in a biological oxidizer (Packard Instruments Company, Downers Grove, IL) and the evolved 14CO2 was trapped in 10 mL Carbosorb E (Packard BioScience Company, Meridian, CT, USA) and 10 mL Permafluor E+ (Packard BioScience). Radioactivity from leaf washes and oxidations was quantified using liquid scintillation spectrometry. The average recovery of applied 14C-glyphosate was 93%, based on the sum of the radioactivity measured in all plant parts (absorption, expressed as percent of applied 14C) and leaf washes. Total radioactivity recovered in all plant parts except the treated leaf was designated as translocated 14C and expressed as percent of absorbed. There were four replications per treatment (harvest) time per plant type (GR-MS/GS-MS) and the experiment was conducted once.
GR-MS and GS-MS plants were processed for phosphorimaging prior to translocation analysis. The treated leaves from the giant ragweed plants were removed at respective harvest times to wash off unabsorbed radioactivity and set aside. The remaining above ground part of the plant was excised from roots and mounted on a 27 by 21.25 cm piece of plain white paper. Shoot parts were evenly spread and kept in place with thin strips of clear office tape. Care was taken to avoid contact of the washed treated leaf with other parts of the plant. Roots were gently rinsed with water to remove soil, blotted dry with paper towels, and mounted on a separate sheet as with the shoot. The mounted plant parts were pressed between one or more layers of newspaper, and bound with two hard cardboard sections. The assembled plant press was held together with large binder clips and stored at −20˚C for later drying. The plant samples were dried in a gravity convection oven at 60˚C for 24 h. Phosphorimaging was used to develop an image of the plant samples. After cooling the dried sample to room temperature, the plant was placed in a 20 by 40 cm exposure cassette (GE Healthcare Bio-Sciences Corp., Piscataway, NJ, USA) and brought into contact with a storage phosphor screen (BAS IPSR 2025 E, GE Healthcare Bio-Sciences Corp.) under diffused lighting. The apparatus was placed in a dark cabinet for 24 h. A phosphorimager (Typhoon FLA 7000, GE Healthcare) was used to detect distribution of 14C-glyphosate and develop an image.
2.5. EPSPS Sequence Analysis
Leaf tissue was removed from three each of GR-MS and GS-MS plants (15-cm tall and at the three-leaf (node) stage, nontreated with glyphosate) for RNA extraction. The tissue was incubated in RNAlater® (Life Technologies, Grand Island, NY, USA) for five hours at 4˚C. The tissue was transferred to a 2 mL microcentrifuge tube containing 450 µL RLT buffer (Qiagen, Valencia, CA, USA) and a 6.35 mm ball bearing (Boca Bearing, Boynton Beach, FL, USA). The tissue was homogenized by placing the tubes in a 5 G high speed paint shaker (Fluid Management, Wheeling, IL, USA) for 6 min. The lysate was transferred to a fresh tube and the RNA extracted using an RNeasy plant mini-kit (Qiagen, Valencia, CA, USA). The quality and quantity of the RNA was determined by gel electrophoresis, A260, and A260/280 ratios. A 197 bp fragment of the EPSPS containing the Pro 106 codon was amplified using 1-step RT-PCR master mix (Fisher, Pittsburgh, PA, USA). The Pro 106 locus, standardized to Arabidopsis thaliana EPSPS sequence, represents the amino acid proline at location 106 in wild type/sensitive/susceptible biotypes of selected weeds. Amino acid substitutions (http://weedscience.org/mutations/mutationdisplayall.aspx?MOAID=12), due to mutations, in place of proline (and in addition, threonine at position 102 in goosegrass (Eleucine indica L.)] result in biotypes resistant to glyphosate. Primer sequences used were AtF1- 5’ ACATGCTTGGGGCTCTAAGAA 3’ and AtR1-5’ TTGAATTACCACCAGCAGCGGT 3’. The reaction was prepared as follows: 25 - 50 ng RNA, 5 µmol forward and reverse primers, 1× Ready mix, 0.5 µL RT enhancer, 0.4 µL enzyme mix, and water to 10 µL. Cycle conditions were as follows: 50˚C for 15 min, 95˚C for 2 min, 30 cycles of 95˚C for 20 s, 56˚C for 30 s, and 72˚C for 1 min, and 72˚C for 5 min. For TOPO TA cloning, a second round of PCR was performed using the TAKARA LA PCR kit ver 2.1 (Fisher, Pittsburgh, PA, USA). Reactions were prepared as follows: 0.5 µL of the initial PCR reaction, 4 µmol forward and reverse primers, 2.5 mM Mg2+, 1x buffer, 400 µM dNTPs, 2.5 U of polymerase, and water to 25 µL. Cycle conditions were as follows: 94˚C for 1 min, 30 cycles of 94˚C for 30 s, 56˚C for 30 s, and 72˚C for 1 min, and 72˚C for 5 min. PCR products were gel extracted using a GenElute Gel Extraction kit (Sigma Aldrich, St Louis, MO, USA). The gel extracted products were examined by gel electrophoresis to assess quality. PCR products were ligated to cloning vector using a TA Cloning® kit with pCRTM2.1 vector (Life Technologies, Grand Island, NY, USA). Reactions were prepared as follows: 3 to 1 ratio of insert to vector (25 ng), 1× buffer, 5 units of ligase, and water to 10 µL. Reactions were incubated at room temperature for 1 hr. Chemically competent TOP10 cells were prepared and transformed according to previously described protocols [11] . The transformants were screened by PCR amplification of the insert using the primers and cycle conditions described above. Glycerol stocks of positive transformants were prepared by adding 800 µL of an overnight culture to 200 µL of 80% glycerol and were stored at −80˚C. Three positive clones per ragweed biotype (GR-MS and GS-MS) were selected for sequencing. Cultures were prepared by inoculating 500 µL LB media (1% w/v tryptone, 0.5% w/v yeast extract, and 0.5% w/v NaCl) with 50 µg/mL amp with 10 µL of an overnight culture. The cultures were submitted to the Genomics and Bioinformatics Research Unit, USDA-ARS, in Stoneville, MS, USA for plasmid isolation and sequencing. Both DNA strands were covered in sequencing. The sequence data were analyzed and aligned using Geneious software [12] (Biomatters Ltd., Auckland, New Zealand).The purpose of the EPSPS sequence analysis is to acquire knowledge on the presence or absence of mutation-based target-site resistance in the GR-MS biotype of giant ragweed.
2.6. Statistical Analysis
All experiments were conducted using a completely randomized design. Data from all experiments, with the exception of EPSPS sequence analysis, were analyzed by ANOVA via the PROC GLM statement using SAS software (version 9.2, SAS Institute, Inc., Cary, NC, USA). No significant experiment effect was observed in repeated experiments; therefore, data from experiments were pooled. Nonlinear regression analysis was applied to fit a sigmoidal log-logistic curve of the form:
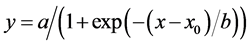 (1)
(1)
where a is an asymptote, x and x0 are the upper and lower response limits with the latter approaching 0, and b is the slope of the curve around x0, to relate the effect of glyphosate dose on giant ragweed control and HAT on 14C-glyphosate absorption and translocation. Equation parameters were computed using SigmaPlot (version 11.0, Systat Software, Inc., San Jose, CA, USA). Treatment means in selected experiments were separated using Fisher’s Protected LSD test at P = 0.05.
3. Glyphosate Dose Response
Response of giant ragweed biotypes to glyphosate dose is presented in Figure 1. ED50 (dose required to reduce plant growth by 50%) values for the GR-MS and GS-MS biotypes, based on percent control, were 0.52 and 0.34 kg ae/ha glyphosate, respectively, indicating a 1.5-fold level of resistance. This resistance level is lower than previously reported cases of GR giant ragweed, 2.1- to 6.2-fold from Ohio [2] , 5.3-fold from Tennessee [3] , and 2.3- to 7.2-fold from Arkansas [6] . GR giant ragweed from Ontario, Canada required 1.66 kg/ha of glyphosate to cause a 50% reduction in growth [7] , which is more than three times that required by the GR-MS biotype from Mississippi. The GR-MS and GS-MS plants survived glyphosate up to 0.84 and 0.42 kg/ha, respectively.
Two types of phenotypic responses are commonly associated with GR giant ragweed accessions treated with glyphosate. First, a rapid necrotic response results in death of mature leaves following a contact herbicide-like injury, but growing points remain uninjured [13] [14] . Second, involves symptoms similar to those displayed by species susceptible to glyphosate, whereby treated plants cease growth for two weeks, turn mildly chlorotic, and resume growth [13] [14] . The symptoms produced by the GR-MS biotype were similar to the slow response phenotype. The rapid necrotic response has been partly attributed to hydrogen peroxide (Tardif, personal communication) and salicyclic acid (Van Horn and Westra, personal communication) formation. Transcriptome analysis of GR and GS giant ragweed biotypes from the Midwest US indicated a rapid response similar to pathogen induced reaction in the former and a slower stress response in the latter [15] .
3.1. 14C-Glyphosate Absorption, Translocation, and Phosphor Imaging
The absorption pattern of 14C-glyphosate in the two giant ragweed biotypes was similar throughout the time course of 168 HAT (Figure 2). Glyphosate absorption measured at 24 and 72 HAT was similar between GR and GS giant ragweed biotypes from Arkansas [6] , analogous to the findings reported here.
Figure 1. Glyphosate dose response on control (0 = no effect on growth, 100 = complete kill) of glyphosate-resistant (GR-MS, closed circles) and -susceptible (GS-MS, open circles) giant ragweed biotypes, 4 weeks after treatment. Vertical bars represent standard error of mean (n = 8).
Figure 2. Absorption of 14C-glyphosate in glyphosate-resistant (closed circles) and -susceptible (open circles) giant ragweed biotypes. Vertical bars represent standard error of mean (n = 4).
The amount of 14C-glyphosate that translocated out of the treated leaves of the GR-MS and GS-MS plants was similar up to 24 HAT (Figure 3). However, the GS-MS biotype translocated more (71% and 76% of absorbed at 48 and 96 HAT, respectively) 14C-glyphosate than the GR-MS biotype (44% and 66% of absorbed at 48 and 96 HAT, respectively) out of the treated leaf, with translocated amounts leveling off to similar levels in both biotypes by 168 HAT (69% and 68% of absorbed in GR-MS and GS-MS, respectively). The translocation levels out of the treated leaf by 168 HAT observed here (68 to 69% of absorbed across the biotypes) are similar to the levels for giant ragweed (71% of absorbed at 168 HAT) previously reported [16] .
Distribution of absorbed 14C-glyphosate in the GR-MS and GS-MS biotypes is summarized in Table 1. The quantity of 14C-glyphosate that remained in the treated leaf was higher in the GR-MS biotype (55% of absorbed) compared to the GS-MS biotype (30% of absorbed) at 48 HAT. The amounts of 14C-glyphosate that accumulated in the shoot above treated leaf (SATL) were lower in the GR-MS biotype (14% and 17% of absorbed at 48 and 96 HAT, respectively) compared to the GS-MS biotype (35% and 37% of absorbed at 48 and 96 HAT, respectively). Similarly, the GR-MS biotype retained lower levels of 14C-glyphosate (8% of absorbed) than the susceptible GS-MS biotype (16% of absorbed) at 48 HAT in SBTL. No difference in the accumulation of translocated 14C-glyphosate in roots of the two biotypes was found, but the resistant GR-MS had higher levels (4% of absorbed) compared to 1% of absorbed in the GS-MS at 1 HAT. This difference in the roots is unlikely to have any effect on the overall physiology of the plants.
Phosphorimaging was used to generate autoradiographs of GR-MS and GS-MS plants treated with 14C-gly- phosate, shown in Figure 4, to visualize 14C-glyphosate translocation patterns. 14C-glyphosate seemingly moved out of the treated leaf faster in susceptible plants than the resistant plants, with discernible accumulation of 14C-glyphosate in the GS-MS biotype but not the GR-MS biotype at 4 HAT. Thereafter, 14C-glyphosate was detectable in all tissues other than the treated leaf at all harvest times in both biotypes. The intensity of the autoradiograph of the shoot was perceptibly deeper for the GS-MS biotype compared to the GR-MS biotype 48 HAT. This observation is also reflected in the translocation and distribution data, where the GS-MS accumulated more 14C-glyphosate than the GR-MS at 48 HAT (Figure 4).
Lower levels of glyphosate accumulation in the GR-MS biotype compared to the GS-MS biotype most likely comprise the resistance mechanism to glyphosate in giant ragweed from Mississippi. Similar findings were reported for glyphosate-resistant rigid ryegrass (Lolium rigidum Gaud.) [17] [18] , Italian ryegrass [19] [20] , horseweed (Conyza canadensis (L.) Cronq.) [21] - [23] , hairy fleabane (Conyza bonariensis (L.) Cronq.) [24] , and tall waterhemp (Amaranthus tuberculatus (Moq.) Sauer) [25] where the resistant accessions accumulated less glyphosate compared to their respective susceptible equivalents. Conversely, there was no differential translocation of glyphosate between GR and GS accessions of common ragweed [26] and giant ragweed [6] , both from Arkansas.
3.2. EPSPS Sequencing
The conserved region of EPSPS containing the Pro106 location was sequenced from three plants each of GR-MS and GS-MS. Alignment of the sequences from the six plants showed that the target site mutation responsible for glyphosate resistance was absent in the resistant GR-MS (Figure 5). This indicates that a mechanism other than a point mutation in the conserved region of EPSPS is responsible for glyphosate resistance in GR-MS.
4. Conclusion
In summary, low level glyphosate resistance has been documented in a giant ragweed accession from Mississippi.
Figure 3. Translocation of 14C-glyphosate in glyphosate-resistant (closed circles) and susceptible (open circles) giant ragweed biotypes. Vertical bars represent standard error of mean (n = 4).
Table 1. Distribution of 14C-glyphosate in glyphosate-resistant and -susceptible giant ragweed biotypes. abc
aAbbreviations: GR-MS: glyphosate-resistant; GS-MS: glyphosate susceptible; ns: not significant; SATL: shoot above treated leaf; SBTL: shoot below treated leaf; TL: treated leaf; bDistribution represents partitioning of absorbed 14C-glyphosate between the treated leaf, shoot above treated leaf, shoot below treated leaf, and root); cLSD (0.05): A number indicates significance at the 5% level of probability and ‘ns’ indicates no significant difference between means within the same column.




Figure 4. Plants ((a), (c), (e), (g)) and corresponding autoradiographs ((b), (d), (f), (h)), respectively, of glyphosate-resistant ((a), (b), (c), (d)) and −susceptible ((e), (f), (g), (h)) giant ragweed plants. Short arrow indicates treated leaf (TL) and dotted arrow shows point of excision of TL. Numbers indicate h after treatment (HAT) with 14C-glyphosate.
Figure 5. Alignment of conserved region of EPSPS from glyphosate resistant (GR-MS1, GR-MS2, GR-MS3) and suscepti- ble (GS-MS1, GS-MS2, GS-MS3) giant ragweed plants. The boxed codons show no mutation at the EPSPS Pro106 position.
The mechanism of resistance, at least in part, is due to reduced translocation of glyphosate and not because of a target site mutation of EPSPS. A very recent review on state of the knowledge on glyphosate resistance mechanisms in plants observes that GR giant ragweed mechanisms are not clearly understood [27] . The research reported here will help clarify glyphosate resistance mechanism in some biotypes of this species.
Cite this paper
Vijay K. Nandula,Alice A. Wright,Christopher R. Van Horn,William T. Molin,Phil Westra,Krishna N. Reddy, (2015) Glyphosate Resistance in Giant Ragweed (Ambrosia trifida L.) from Mississippi Is Partly Due to Reduced Translocation. American Journal of Plant Sciences,06,2104-2113. doi: 10.4236/ajps.2015.613211
References
- 1. Heap, I. (2014) International Survey of Herbicide Resistant Weeds.
http://www.weedscience.org/Summary/home.aspx - 2. Stachler, J.M. (2008) Characterization and Management of Glyphosate-Resistant Giant Ragweed (Ambrosia trifida (L.) and Horseweed [Conyza canadensis (L.) Cronq.]. Ohio State University, Columbus, 124.
- 3. Norsworthy, J.K., Jha, P., Steckel, L.E. and Scott, R.C. (2010) Confirmation and Control of Glyphosate-Resistant Giant Ragweed (Ambrosia trifida) in Tennessee. Weed Technology, 24, 64-70.
http://dx.doi.org/10.1614/WT-D-09-00019.1 - 4. Robertson, R. (2010) Physiological and Biochemical Characterization of Glyphosate Resistant Ambrosia trifida L. Purdue University, West Lafayette, 80.
- 5. Brabham, C.B., Gerber, C.K. and Johnson, W.G. (2011) Fate of Glyphosate-Resistant Giant Ragweed (Ambrosia trifida) in the Presence and Absence of Glyphosate. Weed Science, 59, 506-511.
http://dx.doi.org/10.1614/WS-D-11-00050.1 - 6. Norsworthy, J.K., Riar, D., Jha, P. and Scott, R.C. (2011) Confirmation, Control, and Physiology of Glyphosate-Resistant Giant Ragweed (Ambrosia trifida) in Arkansas. Weed Technology, 25, 430-435.
http://dx.doi.org/10.1614/WT-D-10-00155.1 - 7. Vink, J.P., Soltani, N., Robinson, D.E., Tardif, F.J., Lawton, M.B. and Sikkema, P.H. (2012) Glyphosate-Resistant Giant Ragweed (Ambrosia trifida L.) in Ontario: Dose Response and Control with Postemergence Herbicides. American Journal of Plant Sciences, 3, 608-617.
http://dx.doi.org/10.4236/ajps.2012.35074 - 8. Harrison, S.K., Regnier, E.E., Schmoll, J.T. and Webb, J.E. (2001) Competition and Fecundity of Giant Ragweed in Corn. Weed Science, 49, 224-229.
http://dx.doi.org/10.1614/0043-1745(2001)049[0224:CAFOGR]2.0.CO;2 - 9. Williams II, M.M. and Masiunas, J.B. (2006) Functional Relationships between Giant Ragweed (Ambrosia trifida) Interference and Sweet Corn Yield and Ear Traits. Weed Science, 54, 948-953.
http://dx.doi.org/10.1614/WS-05-187R.1 - 10. Barnett, K.A. and Steckel, L.E. (2013) Giant Ragweed (Ambrosia trifida) Competition in Cotton. Weed Science, 61, 543-548.
http://dx.doi.org/10.1614/WS-D-12-00169.1 - 11. Sambrook, J., Fritsch, E. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual. 2nd edition, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 1.82-1.84.
- 12. Drummond, A., Ashton, B., Buxton, S., Cheung, M., Cooper, A. and Duran, C. et al. (2011) Geneious. v5.4.
http://www.geneious.com - 13. Green, A.C., Tardif, F.J. and Sikkema, P.H. (2012) Determining the Mechanism of Glyphosate Resistance in Giant Ragweed (Ambrosia trifida L.) in Ontario. Proceedings of the Weed Science Society of America, 52, 437.
- 14. Jeffery, T.M., Hall, C., Lawton, M., Sikkema, P. and Tardif F.J. (2013) Investigation into the Molecular and Biochemical Mechanisms of Glyphosate Resistance in Two Populations of Giant Ragweed (Ambrosia trifida). Proceedings of Weed Science Society of America, 53, 315.
- 15. Padmanabhan, K., Best, N.B., Weller, S.C. and Schulz, B. (2014) Transcriptome Analysis of Glyphosate Resistance in Giant Ragweed (Ambrosia trifida). Proceedings of Weed Science Society of America, 52, 158.
- 16. Hoss, N.E., Al-Khatib, K., Peterson, D.E. and Loughin T.M. (2003) Efficacy of Glyphosate, Glufosinate, and Imazethapyr on Selected Weed Species. Weed Science, 51, 110-117.
http://dx.doi.org/10.1614/0043-1745(2003)051[0110:EOGGAI]2.0.CO;2 - 17. Lorraine-Colwill, D.F., Powles, S.B., Hawkes, T.R., Hollinshead, P.H., Warner, S.A.J. and Preston, C. (2003) Investigations into the Mechanism of Glyphosate Resistance in Lolium rigidum. Pesticide Biochemistry and Physiology, 74, 62-72.
http://dx.doi.org/10.1016/S0048-3575(03)00007-5 - 18. Wakelin, A.M., Lorraine-Colwill, D.F. and Preston, C. (2004) Glyphosate Resistance in Four Different Populations of Lolium rigidum Is Associated with Reduced Translocation of Glyphosate to Meristematic Zones. Weed Research, 44, 453-459.
http://dx.doi.org/10.1111/j.1365-3180.2004.00421.x - 19. Perez-Jones, A., Park, K.W., Polge, N., Colquhoun, J. and Mallory-Smith C. (2007) Investigating the Mechanisms of Gyphosate Resistance in Lolium multiflorum. Planta, 226, 395-404.
http://dx.doi.org/10.1007/s00425-007-0490-6 - 20. Nandula, V.K., Reddy, K.N., Poston, D.H., Rimando, A.M. and Duke, S.O. (2008) Glyphosate Tolerance Mechanism in Italian Ryegrass (Lolium multiflorum) from Mississippi. Weed Science, 56, 344-349.
http://dx.doi.org/10.1614/WS-07-115.1 - 21. Feng, P.C.C., Tran, M., Chiu T., Sammons, R.D., Heck, G.R. and CaJacob, C.A. (2004) Investigations into Glyphosate-Resistant Horseweed (Conyza canadensis): Retention, Uptake, Translocation, and Metabolism. Weed Science, 52, 498-505.
http://dx.doi.org/10.1614/WS-03-137R - 22. Koger, C.H. and Reddy, K.N. (2005) Role of Absorption and Translocation in the Mechanism of Glyphosate Resistance in Horseweed (Conyza canadensis). Weed Science, 53, 84-89.
http://dx.doi.org/10.1614/WS-04-102R - 23. Dinelli, G., Marotti, I., Bonetti, A., Minelli, M., Catizone, P. and Barnes J. (2006) Physiological and Molecular Insight on the Mechanisms of Resistance to Glyphosate in Conyza canadensis (L.) Cronq. Biotypes. Pesticide Biochemistry and Physiology, 86, 30-41.
http://dx.doi.org/10.1016/j.pestbp.2006.01.004 - 24. Dinelli, G., Marotti, I., Bonetti, A., Catizone, P., Urbano, J.M. and Barnes, J. (2008) Physiological and Molecular Bases of Glyphosate Resistance in Conyza bonariensis Biotypes from Spain. Weed Research, 8, 257-265.
http://dx.doi.org/10.1111/j.1365-3180.2008.00623.x - 25. Nandula, V.K., Ray, J.D., Ribeiro, D.N., Pan, Z. and Reddy, K.N. (2013) Glyphosate Resistance in Tall Waterhemp (Amaranthus tuberculatus) from Mississippi Is Due to Both Altered Target-Site and Nontarget-Site Mechanisms. Weed Science, 61, 374-383.
http://dx.doi.org/10.1614/WS-D-12-00155.1 - 26. Brewer, C.E. and Oliver, L.R. (2009) Confirmation and Resistance Mechanisms in Glyphosate-Resistant Common Ragweed (Ambrosia artemisiifolia) in Arkansas. Weed Science, 57, 567-573.
http://dx.doi.org/10.1614/WS-08-160.1 - 27. Sammons, R.D. and Gaines, T.A. (2014) Glyphosate Resistance: State of Knowledge. Pest Management Science, 70, 1367-1377.
http://dx.doi.org/10.1002/ps.3743
NOTES

*Corresponding author.






