American Journal of Plant Sciences
Vol.05 No.20(2014), Article ID:50091,10 pages
10.4236/ajps.2014.520314
Combined Selection in Backcross Population of Papaya (Carica papaya L.) by the Mixed Model Methodology
Helaine Christine Cancela Ramos1*, Messias Gonzaga Pereira1, Alexandre Pio Viana1, Lucas Nunes da Luz1, Deisy Lúcia Cardoso1, Geraldo Antônio Ferreguetti2
1Laboratory of Plant Breeding, Centre for Agricultural Sciences and Technology, North Fluminense State University (UENF), Campos dos Goytacazes, Brazil
2Caliman Agrícola S.A., Linhares, Brazil
Email: *helainecr@uenf.br
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 15 July 2014; revised 17 August 2014; accepted 4 September 2014
ABSTRACT
The selection of superior genotypes based on the simultaneous response to several characteristics of agronomic importance is a key strategy to overcome the scarcity of available varieties of papaya. This study aimed to apply the combined selection by using distinct selection indexes based on both the genetic values obtained by the REML/BLUP methodology and the real measured values to select agronomically superior genotypes of papaya within backcross progenies. The combined selection was carried out based on genetic and phenotypic values, original and standardized, multiplied by the agronomic weights. The results of the analysis of genetic parameters indicate that the evaluated progenies have expressive genetic variability for the considered traits, and that there are real possibilities of genetic progress with the selection. Among the analyzed indexes, the one based on standardized genetic value presented greater consistency in the ranking of genetic material, demonstrating the advantage of data standardization. Five progenies belonging to the BC1 generation, and five to the BC3 generation were selected using this index. A total of 27 plants agronomically superior were selected within the top five progenies and recommended for generation advance, 23 being selected by combined selection and 4 using the direct selection for the four mainly characters in papaya breeding program: production, pulp and fruit firmness and soluble solids. Beyond the selection of superior genotypes for the development of future inbred lines, this study also allowed defining the best strategy to apply the combined selection in papaya using predicted breeding values obtained by BLUP. This strategy may allow higher accuracy in the selection process, thus increasing the chances of success of the breeding programs.
Keywords:
Papaya Tree, Backcross, Selection Index, Mixed Model, REML/BLUP

1. Introduction
Despite its major socioeconomic significance and the fact that it is one of the main fruits that are sources of income to Brazil, narrow genetic variability has been observed in commercial plantations of papaya in almost all producing regions, revealing the reduced number of choices of commercial varieties [1] -[4] . Part of this problem would be solved if more studies were carried out to identify and select agronomically superior genotypes from segregating populations. Genetic breeding programs for this crop have been conducted by the North Fluminense State University (UENF) in an attempt to change this situation [5] - [9] . The main objective is to develop agronomically superior genetic materials that may provide good returns for producers and meet the demand of both domestic and foreign markets.
The development of superior genetic materials in any breeding program necessarily depends on a simultaneous favorable response for several traits of agronomic importance. The selection index methodology is the most appropriate procedure for the selection of genotypes based on multiple traits. This procedure was originally proposed by Smith [10] and Hazel [11] , aiming to select multiple characters simultaneously. A great number of selection indexes to be used in breeding programs are available in the literature. In general, these indexes are built from estimates of genetic parameters and phenotypic averages obtained by the analysis of variance [12] . Since one of the primary objectives of breeding programs is the prediction of the genetic value of the evaluated individuals [13] [14] , the methodologies for mixed models emerge as an important selection procedure, involving the estimation of the variance components by the restricted maximum likelihood method (REML), while the prediction of the genotypic values was estimated by the best linear unbiased prediction method (BLUP), which provided a more accurate selection process [13] .
The first applications of mixed models were carried out more than fifty years ago and were presented by Henderson [15] . This methodology allows an accurate prediction of genotypic performance using covariance structures that consider genetic correlations and information from relatives through the coefficient of kinship, which allowed the potential use of this procedure to select superior genotypes [16] . This methodology has become popular in recent years due to the incorporation of efficient algorithms in statistical programs widely available [17] .
Although the REML/BLUP methodology is not extensively used in plant breeding as it is in animals, its application has become increasingly common in agricultural science [17] . The analytical procedures of mixed models have been widely applied in breeding of forest species and perennial crops [13] [18] , because it is an estimation procedure, mainly for unbalanced data, which allows the analysis of characteristics of low heritability [19] . However, this application is not so common in annual crops [14] [20] .
Therefore, this study aimed to estimate genetic parameters and achieve the genetic values for the evaluated traits using the mixed model methodology and to apply the combined selection methodology and the direct selection based on both standardized and non-standard phenotypic and genetic values. In addition, it aimed to compare selection efficiency using these indexes and to select the best progenies, and within them, the best genotypes for the generation advance in the papaya breeding program of UENF.
2. Material and Methods
2.1. Genetic Material and Experiment Design
Hermaphrodite plants were evaluated in 32 progenies. Of these, 26 are progenies derived from backcross (BC), being sixteen derived from the first generation (BC1), one from the second (BC2) and nine from the third generation (BC3) of recurrent crossing using the Cariflora as the recurrent parent. The other treatments consisted of controls Golden variety, “SS783” (donor parent of hermaphroditism), SS72/12 variety, UC 01 hybrid, BC3(2)XSS72/12 and BC3(3)XSS72/12, included in the study for comparison with segregating generations.
The progenies evaluated in this study are the result of selection for morphoagronomic attributes over the generation of backcross program of UENF followed by several generations of self-fertilization in order to develop superior inbred lines with different genomic proportion of recurrent parent. The main purpose of this backcross program was to convert the dioecious sex of parent Cariflora to gynoecious-andromonoecious with the transfer of the chromosomal region that determines hermaphroditism donated by elite genotype “SS783”.
The recurrent genitor (Cariflora) is a dioecious selection and thus has a segregating behavior. It has yellow flesh, moderate fruit firmness and good flavor. The fruits weigh around 1.67 kg with a rounded shape and large internal cavity. On the other hand, the donor genitor (SS783) is an elite homozygous cultivar, belonging to the “Solo” group, showing fruits with a pear shape, red flesh, good quality, and average weight of 0.52 kg. The cross between these genotypes results in vigorous and productive hybrid [5] .
The experiment was conducted in February 2008 in the commercial field of the Caliman Agrícola S/A company, Romana Farm, located in Linhares (19˚23'28"S and 40˚04'20"W, 33 meters asl), in the Espírito Santo State. It was used a randomized complete block design with two replications, consisting of 32 treatments and plots of 15 plants, spaced 3.60 m between rows and 1.80 m between plants in the row.
Due to the segregation ratio between female and hermaphrodite plants of 2:1 [21] , generated by self-fertiliza- tion, the number of evaluated genotypes was less than the total of plants in the experiment (960), given that only hermaphrodite plants were evaluated for presenting fruit with commercially accepted pattern. Thus, the assess- ments began with the measurement of morphoagronomic traits in 580 hermaphrodite plants. However, due to the loss of plants caused by disease incidence, the evaluations were completed in 360 hermaphrodite plants, which are used in the analysis performed in this study.
The fertilization, management, control of pests and diseases and cultural practices were the same used in commercial plantations of the company.
2.2. Traits Evaluated and Selection Index
Sixteen morphoagronomic traits were evaluated 150, 240 and 270 days after transplanting for the prediction of genetic values, as demonstrated by Silva and collaborators [22] . The combined selection of the sixteen characteristics was performed by establishing weights associated with agronomic values, as described below: plant height—PH (1), height of insertion of the first fruit—HIFF (−10), stem diameter—SD (5), number of total fruits —NTFr (50), number of carpelloid fruits—NCFr (−20), number of pentandric fruits—NPFr (−20), fruitless leaf axils—FLLA (−20), number of commercial fruits—NcoFr (100), average fruit weight—FrW (1), plant production—PROD (100), soluble solids content—SS (100), fruit firmness—FF (100), pulp firmness—PF (100), diameter of the fruit—DIAM (1), the fruit length—LENG (1) and average thickness of the pulp—TP (70). These weight ratios were established experimentally, based on the importance of the traits evaluated at agronomic level [22] . This index was used for selection of papaya progenies with success [9] .
The combined selection was carried out based on both the individual plants and the progeny means, to identify the best genotypes. To this end, the (original and standardized) genetic and phenotypic values were multiplied by the agronomic weights described above, and the results were compared to assess the degree of coincidence between these methodologies and the effectiveness of these indexes to indicate superior genetic materials. The indexes used were constructed as follows:
Equation (1). Genotypic and phenotypic index based on mixed model.
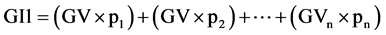
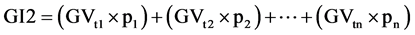
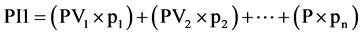
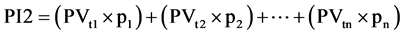
where GV is the predicted genotypic value, PV is the phenotypic value or measured value, p is the agronomic weight established for each variable and GVt and PVt are the genotypic and phenotypic values standardized by the following equation: , where Xg is the value measured in the individual for the variable X,
, where Xg is the value measured in the individual for the variable X, 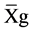 is the general average of the variable, and
is the general average of the variable, and 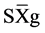 is the standard deviation.
is the standard deviation.
Two additional indexes were used to identify the best progenies, namely, the multiplicative index (MI) [23] [24] , and the rank sum (RSI) [25] , as presented below:
Equation (2). Genotypic selection by MI and RSI indexes.
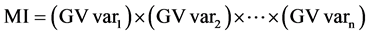
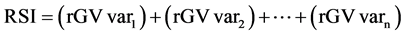
where GV is the predicted genotypic value; r is the position in which the genotype is presented in the ranking for each evaluated traits (var).
2.3. Statistical Analysis
The data were analyzed by the mixed model methodology via the REML/BLUP procedure, using randomized complete block design. Thus, the estimation of genetic parameters was performed via REML (restricted maximum likelihood), and the genotypic values, or genotypic averages, were estimated by the BLUP (best linear unbiased predictor) using the Selegen-REML/BLUP software system [26] . The following statistical model was used for the genetic evaluation of the data:
Equation (3). Mixed model equation.
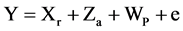
where Y is the data vector, r is the vector of the replication effects (regarded as fixed) plus the overall average, a is the vector of the individual additive genetic effects (regarded as random), p is the vector of plot effects, e is the vector of errors or residues (random). The capital letters (X, Z and W) represent the incidence matrices for the referred effects.
Since the genetic material used in this study derives primarily from self-fertilization, the analyses were performed using the 59 statistical model of the Selegen software system, which is the one closest to the actual genetic structure of the analyzed progenies. This model is designed to evaluate individuals in F3 progenies of autogamous plants (or S1 of allogamous plants), considering multiple observations per plot.
The procedures required for the construction of selection indexes were performed by the Selegen REML/ BLUP [27] and Microsoft Office Excel 2007 software systems. A selection intensity of 30% was carried out to indicate the best progenies, and 25% for the selection of superior genotypes within the selected progenies. In papaya crop are considered superior those genotypes with high productivity, low incidence of deformed fruit, high fruit and pulp firmness, and high soluble solids content, as well as other attributes.
We estimate genetic gain using the following estimator:
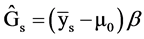 ,
,
where 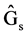 = Genetic gain;
= Genetic gain;  = Selection differential;
= Selection differential; 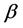 = Regression coefficient of additive genetic values on phenotypic values.
= Regression coefficient of additive genetic values on phenotypic values.
3. Results and Discussion
The Table 1 shows the genetic parameters estimated for the evaluated traits in this study and the genotype averages. Based on the estimates of individual genotypic coefficient of variation (CVgi%), which present the percentage of the overall average for the quantity of the existing genetic variation, we observed values ranging from low to high (4.96 to 90.83). Except for the variables PH, SD, SS, FF, DIAM and TP, which presented low values for CVgi, the other evaluated traits presented values ranging from moderate to high (10.29 to 90.83). It demonstrates that, although the population has already gone through several cycles of selection, it is still suitable for breeding. In other words, genetic progress can be achieved with the adoption of appropriate selection procedures. The greatest genetic variation was observed for pentandric fruits (90.83), which indicates the possibility of selecting genotypes with lower expression of this type of fruit abnormality. On the other hand, a slight genetic progress is expected for fruit firmness, due to the low variation observed among the surveyed progenies (4.96).
Estimates of the coefficient of residual variation (CVe%) ranged from 6.48 to 109.82. In general, we can infer that the values of CVe remained at acceptable levels for the field trial, except for carpelloid (46.72), pentandric (109.82) fruits and pulp firmness (54.37). The high values presented indicate low accuracy and lower experimental precision. High values of CVe have also been associated with the large size of the experiments, different
Table 1. Genetic parameters estimated for the 16 traits assessed by the REML/BLUP procedure, using individual data.
Va: genetic variance among progenies, which is equal to the additive genetic variance plus (1/4) of the dominance genetic variance; Vf: individual phenotypic variance; h2b = h2: individual heritability in the broad sense among progenies; h2aj: individual heritability in the broad sense among progenies, adjusted for the plot effects; c2prc = c2: coefficient of determination of the plot effects; h2mp: heritability of the average of the progenies; Acprog: accuracy of the selection of progenies; CVgi%: individual additive genetic coefficient variation coefficient; CVe%: residual variation coefficient; CVr: relative variation coefficient (CVgi/CVe). PH: plant height; HIFF: height of insertion of the first fruit; SD: stem diameter; NTFr: number of total fruits; NCFr: number of carpelloid fruits; NPFr: number of pentandric fruits; FLLA: fruitless leaf axilst; NCoFr: number of commercial fruits; FrW: average fruit weight; PROD: plant production; SS: soluble solids content; FF: fruit firmness; PF: pulp firmness; DIAM: diameter of the fruit; LENG: the fruit length; and TP: average thickness of the pulp.
responses of genotypes to stress from high temperatures and drought, incidence of pests and diseases, and stress caused by wind and pruning [28] . High estimates of CVe for NCFr and NPFr have been observed in studies with papaya [29] , being indicated the sum of the values of two variables to form a single trait (deformed fruits) [30] , thus reducing the estimates of CVe to acceptable values.
The magnitude of the relationship between CVg and CVe allows inferences about accuracy and genotypic evaluation. Thus, the coefficient of relative variation (CVr = CVg/CVe) presented values ranging from 0.55 (DIAM) to 0.94 (TP). According to Venkovsky [31] , CVr values close to the unit are suitable for experiments in corn, which is corroborated by Resende and Duarte [32] , who report that CVr values equal to 1.0 are suitable because they provide inferences with high and very high accuracy and precision. In the present study, CVr values equal to one unity were not observed, but, for the variables PH, HIFF, NTFr, NCoFr, FrW, SS and TP, the values are very close to the unity, which can be considered a favorable condition for the success of the selection. According to Resende and Duarte [32] , a conclusion about whether CVr values are appropriate or not should be constructed together with the number of replications, since it is directly related to experimental accuracy.
The quality of genotypic assessment should be preferably inferred based on accuracy [33] . This parameter refers to the correlation between the actual genotypic value of the genetic material and that estimated or predicted from the information provided by field experiments and it increases as the absolute deviations among these values decrease. In our study, accuracy values ranged from 0.62 to 0.80, which are considered from moderate to high [32] . In studies aiming to associate alternative values of the variation coefficient to accuracies was found that data obtained from experiments with two to four replications will not probably reach the desired accuracy values, namely, above 90% [32] . According to the authors, under this situation (low number of replications), accuracy above 90% is only possible for characters with high heritability, which is unlikely to be found, given the quantitative nature and low heritability of the major traits of interest in breeding.
The estimates of individual heritability in the broad sense (h2b) were considered low for all evaluated variables. The highest values for h2b were observed for plant height, height of first fruit, commercial fruit, soluble solids and pulp thickness, while the lowest values were observed for production, fruit and pulp firmness, and fruit diameter. This result shows that much of the variation observed for these characteristics are due to environmental factors, indicating that more stringent selection methods need to be applied to obtain satisfactory genetic gains. The standard deviations for each estimate of h2b ranged from low to moderate. According to Pedrozo et al. [12] , this standard deviation value reveals that heritability estimates are statistically different from zero.
One possible cause for the low estimates of heritability is the narrow genetic base of the population, since the progenies come from an initial crossing between two parents, also being present in the first, second and third generation of self-fertilization. It is also important to consider that selection was also carried out in previous generations, which favors the reduction of the genetic variability available in the present generation. In other words, in each cycle of generation advance, only superior genotypes will remain with the practice of selection. Even so, real possibilities of genetic progress are still observed.
When considering the average level of heritability in families (h2mp), magnitudes significantly higher are observed, as well as values on average go up to four times higher than individual heritability. In a study aiming to obtain predicted values in rubber, the authors pointed out that, under these conditions of heritability magnitudes, selection can be more effective using information from families [18] . The highest estimates for h2mp were observed for plant height, height of first fruit, commercial fruits, average weight, soluble solids and pulp thickness, while the lowest values were observed for pulp firmness and fruit diameter. According to Falconer [34] , heritability is a property not only of a character, but also of the population and environmental circumstances to which individuals are subject, and its value may be affected by changes in any of the components of genetic and phenotypic variances.
The coefficients of coincidence in the ranking of progenies by multiplicative index (MI), index of rank sums (RSI), genotypic index (GI1), standardized genotypic index (GI2), phenotypic index (PI1) and standardized phenotypic index (PI2) and direct selection for yield per plant, applying a selection pressure of 30%, were compared and their efficiency was analyzed (Table 2). When only the coincidence among the indexes is considered, it seems that these coefficients ranged from low to high. Since the coefficient of coincidence reveals the agreement in the selection result among indexes, it is observed that the lowest agreement was found between MI and RSI (0.2), while the largest one was observed for GI1 and PI1 (1.0), which demonstrates that the last two indexes indicated the same progenies as superior.
In general, the multiplicative index (MI) was not consistent when ranking families based on all the evaluated traits, which makes it inadequate for selecting superior genetic materials. RSI was more consistent for use in selection than MI, but it failed to select some potential progenies. GI1 and PI1 presented moderate consistency since the ordering of the best progenies was mainly based on the number of commercial fruits and production. Therefore, some selected progenies did not present favorable values for the other evaluated traits. This condition is not desirable in papaya breeding, since production is not the only trait of interest. In other words, genetic material is considered superior when good performance is coupled with good fruit quality, and other relevant traits. In this sense, the GI2 and PI2 indexes were the most consistent by allowing the indication of productive progenies that also presented satisfactory values for the content of soluble solids, firmness and deformation of fruit, avoiding, for example, the effect of scale. These indexes showed high correlation in selection (0.9), which also
Table 2. Coefficient of coincidence between five selection indexes, besides direct selection for PROD, used to identify the best progenies, considering a selection pressure of 30%.
RSI: rank sum index; MI: multiplicative index; GI1: genotypic index; GI2: genotypic index with standardized values; FI1: phenotypic index; FI2: phenotypic index with standardized values; PROD: plant production.
occurred between RSI and them, but in lower magnitude (0.8). The high agreement found between the GI1 and PI1 indexes and between GI2 and PI2 corroborates the accuracy values found in this study.
The analysis of the efficiency of MI, RSI and classical (CI) indexes in the selection of superior genotypes of sugar cane using the REML/BLUP methodology demonstrated that MI was the most efficient for selection due to its higher indirect gain for BRIX production per hectare (TBH), a key trait in the breeding of this crop [12] . Considering only the MI and RSI indexes, in the present study, RSI presented higher efficiency, which is corroborated by the higher agreement in selection between the GI2 and PI2 indexes. Perhaps the disagreement between the two works is found in the desired results. In other words, this study tried to select genotypes with satisfactory superiority for various characters, and not just for a main trait. This demonstrates that besides the genetic structure of the materials analyzed, the efficiency of the selection methods depends on the purpose of the breeding program.
Besides the selection agreement among the indexes, the agreement between these indexes and direct selection for production was also analyzed. The highest coefficient of coincidence was presented by the GI1 and PI1 indexes (1.0), while among the other indexes, this coefficient was only average (0.5 to 0.6). This result is in accordance with the expectations, since the selection with the GI1 and PI1 indexes had production as a trait of higher weight in ranking of the genotypes, while the other indexes provide better weight distribution among the other attributes of interest. In other words, this result demonstrates that by using the original data to form an index, the trait with the highest phenotypic value has greater weight in the selection. In the present study, production is the trait of higher measured value.
Although high correlation was observed between the GI2 and PI2 indexes, only the information from the GI2 was used to perform the selection, since it provides estimates free from environmental effects and predicts the gains and the new averages of the selected material. Thus, in the next three tables we highlight the genetic gains and new averages predicted for the four key characteristics for the papaya breeding programs based on GI2.
Table 3 show the genetic gains and new averages predicted in the selection of superior progenies. It was observed that the ten superior progenies have presented gains ranging from 5.26% to 16.86% for production, from 0.53% to 9.24% for soluble solids, 1.46% to 4.65% for fruit firmness, and 1.50% to 8.12% for pulp firmness. These values are considered satisfactory in view of the average gains observed for each trait. The largest average gains were observed for production, and increases in the averages of 11.03%, 3.90%, 2.64% and 5.13% are expected for the variables PROD, SS, FF and PF, respectively.
The indexes were also used in this study for the selection of superior genotypes within the best progenies. However, at this stage, only the GI1, GI2, PI1 and PI2 indexes were used, since the MI and RSI indexes (obtained by the REML/BLUP methodology) only allow progeny selection. Considering a selection pressure of
Table 3. Genetic gains and new averages predicted in four traits of great significance in papaya breeding for the progenies selected by the GI2 index.
Gs (%): gain in percentage;
25%, in general, the results related to the agreement between the selection strategies were similar to those obtained when considering the selection of the best progenies. There is variation in the level of agreement between the different selection indexes in the evaluated progenies, demonstrating that coincidence in the selection depends significantly on the genetic constitution of the evaluated individuals, as described by Pedrozo et al. [12] .
When considering the selection of superior genotypes within the best progenies (Table 4), there is a slight increase in the gains predicted in the selection of genotypes in relation to the analysis based on the average of the progenies, considering the four characters of greater interest for papaya breeding (PROD, SS, FF and PF). It reveals the presence of significant variability within the progenies, which favors the achievement of genetic progress by selection. On average, the largest predicted gain was observed for production (14.2%), followed by pulp firmness (5.3%), soluble solids (5.3%) and fruit firmness (3.1%). These values were two times higher than the average of the controls for production, pulp and fruit firmness, being only slightly higher for soluble solids. It is also observed that, out of the selected genotypes, 43.5%, 30.4%, 52.2% and 39.1% presented higher predicted gains than the best control for PROD, SS, FF and PF, respectively (data not showed).
This methodology provides the ranking of potential genotypes for selection, exploring all the genotypic variation between and within progenies [33] , considering, however, each analyzed variable separately. This procedure has been effective in studies on beans [20] , rubber [18] , sugarcane [35] [36] , soybean [37] and acerola fruit [38] , but it is not appropriate to select promising genotypes for several attributes simultaneously. Therefore, the
Table 4. Genetic gains and new averages predicted for four new traits of great significance for papaya breeding, considering the genotypes selected within the evaluated progenies.
Gs (%): gain in percentage;
Table 5. Direct selection for four traits of agronomic importance for papaya breeding.
a: additive value; VG: genetic value; Gs: gain with selection; Gs (%): percentage gain;
present research aimed to use genotypic values for the construction of indexes, thus allowing the use of new strategies for the selection of superior genotypes.
In addition to the genotypes selected by combined selection, the direct selection of PROD, SS, FF and PF was also performed to maintain genotypes sources of production, total soluble solids and internal and external firmness in the breeding population. Thus, Table 5 presents the gains and the new averages predicted for the selection of superior genotypes for each trait mentioned above. It is observed that the predicted gains were significantly higher than those considering combined selection, which was already expected. The combined selection index was applied in papaya considering six morphoagronomic traits and direct selection for production/plant, in the selection of superior genotypes in segregating population [22] . The comparison between the two used selection strategies indicated higher gains for direct selection. However, the authors point out that despite the lower gain, combined selection generates higher expectation of success in future generations because it considers agronomic variables both favorable and unfavorable to the cultivation of papaya.
Based on the standardized genetic index, 27 plants were selected for generation advance. Twenty-three progenies were selected within the superior progenies by combined selection, and four, by direct selection for PROD, SS, FF and PF. However, it is important to highlight that the estimated genetic parameters and the efficiency of the indexes used in the selection are inherent to the population involved and to the experimental conditions established in this study.
4. Conclusion
Considering the purpose of simultaneous selection, we found that the selection index GI2 (using standardized genetic values) was more consistent both in the indication of the best progenies and in the identification of potential genotypes to form the next breeding generation. This shows the advantage of standardizing genetic data and values in the evaluation and selection of superior genotypes. Thus, it is evident that this strategy may allow higher accuracy in the selection process, thus increasing the chances of success of the breeding programs.
Acknowledgements
We thank the Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) for providing the Master’s scholarship, and the Empresa Caliman Agrícola S/A (Caliman) for the financial and logistical support.
References
- Sharon, D., Hillel, J., Vainstein, A. and Lavi, U. (1992) Application of DNA Fingerprints for Identification and Genetic Analysis of Carica papaya and Other Carica Species. Euphytica, 62, 119-126. http://dx.doi.org/10.1007/BF00037937
- Stiles, J.I., Lemme, C., Sondur, S., Morshidi, M.B. and Manshardt, R. (1993) Using Randomly Amplified Polymorphic DNA for Evaluating Genetic Relationships among Papaya Cultivars. Theoretical Applied Genetics, 85, 697-701.
- Kim, M.S., Moore, P.H., Zee, F., Fitch, M.M.M., Steiger, D.L., Paull, R.E., Drew, R.A., Sekioka, T. and Ming, R. (2002) Genetic Diversity of Carica papaya as Revealed by AFLP Markers. Genome, 45, 503-512. http://dx.doi.org/10.1139/g02-012
- Ruggiero, C., Marin, S.L.D. and Durigan, J.F. (2011) Mamão, Uma História de Sucesso. Revista Brasileira de Fruticultura, S1,76-82. http://dx.doi.org/10.1590/S0100-29452011000500011
- Marin, S.L.M., Pereira, M.G., AmaralJúnior, A.T., Martelleto, L.A.P. and Ide, C.D. (2006) Heterosis in Papaya Hybrids from Partial Diallel of “Solo” and “Formosa” Parents. Crop Breeding and Applied Biotechnology, 6, 24-29. http://dx.doi.org/10.12702/1984-7033.v06n01a04
- Silva, F.F., Pereira, M.G., Campos, W.F., Damasceno Júnior, P.C., Pereira, T.N.S., Souza Filho, G.A., Ramos, H.C.C., Viana, A.P. and Ferreguetti, G.A. (2007) DNA Marker-Assisted Sex Conversion in Elite Papaya Genotype (Carica papaya L.). Crop Breeding Applied Biotechnology, 7, 52-58. http://dx.doi.org/10.12702/1984-7033.v07n01a08
- Damasceno Junior, P.C., Costa, F.R., Pereira, T.N.S., Freitas Neto, M. and Pereira, M.G. (2009) Karyotype Determination in Three Caricaceae Species Emphasizing the Cultivated Form (C. papaya L.). Caryologia, 62, 10-15. http://dx.doi.org/10.1080/00087114.2004.10589660
- Ramos, H.C.C., Pereira, M.G., Silva, F.F. and Viana, A.P. (2011) Seasonal and Genetic Influences on Sexual Expression in Segregating Papaya Population Derived From Backcross. Crop Breeding Applied Biotechnology, 11, 97-105. http://dx.doi.org/10.1590/S1984-70332011000200001
- Pinto, F.O., Luz, L.N., Pereira, M.G., Cardoso, D.L. and Ramos, H.C.C. (2013) Metodologia dos Modelos Mistos para Seleção Combinada em Progênies Segregantes de Mamoeiro. Revista Brasileira de Ciências Agrárias, 8, 211-217. http://dx.doi.org/10.5039/agraria.v8i2a2409
- Smith, H.F. (1936) A Discriminant Function for Plant Selection. Annals of Eugenics, 7, 240-250. http://dx.doi.org/10.1111/j.1469-1809.1936.tb02143.x
- Hazel, L.N. (1943) The Genetic Basis for Constructing Selection Indexes. Genetics, 28, 476-490.
- Pedrozo, C.A., Benites, F.R.G., Barbosa, M.H.P., Resende, M.D.V. and Silva, F.L. (2009) Efficiency of Selection Indexes Using the REML/BLUP Procedure in Sugarcane Breeding. Scientia Agraria, 10, 31-36.
- Resende, M.D.V. (2002) Genética Biométrica e Estatística no Melhoramento de Plantas Perenes. Embrapa Informação Tecnológica, Brasília.
- Piepho, H.P., Mohring, J., Melchinger, A.E. and Buchse, A. (2008) BLUP for Phenotypic Selection in Plant Breeding and Variety Testing. Euphytica, 161, 209-228. http://dx.doi.org/10.1007/s10681-007-9449-8
- Henderson, C.R. (1975) Best Linear Unbiased Estimation and Prediction under a Selection Model. Biometrics, 31, 423-447. http://dx.doi.org/10.2307/2529430
- Crossa, J., Burgueño, J., Comelius, P.L., McLaren, G., Trethowan, R. and Krishnamachari, A. (2006) Modeling Genotype × Environment Interaction Using Additive Genetic Covariances of Relatives for Predicting Breeding Values of Wheat Genotypes. Crop Science, 46, 1722-1733. http://dx.doi.org/10.2135/cropsci2005.11-0427
- Wolf, J., Duchateau, L. and Schrevens, E. (2008) Dealing with Source of Variability in the Data-Analysis of Phenotyping Experiments with Transgenic Rice. Euphytica, 160, 325-337. http://dx.doi.org/10.1007/s10681-007-9526-z
- Costa, R.B., Resende, M.D.V., Gonçalves, P.S., Arruda, E.J., Oliveira, L.C.S. and Bortoletto, N. (2002) Prediction of Genotypic Values for Yield in Rubber Tree-Clone Test Trials Using REML/BLUP Procedure. Crop Breeding and Applied Biotechnology, 2, 579-586.
- Nunes, J.A.R., Ramalho, M.A.P. and Ferreira, D.F. (2008) Inclusion of Genetic Relationship Information in the Pedigree Selection Method Using Mixed Models. Genetics and Molecular Biology, 31, 73-78. http://dx.doi.org/10.1590/S1415-47572008000100015
- Chiorato, A.F., Carbonell, S.A.M., Dias, L.A.S. and Resende, M.D.V. (2008) Prediction of Genotypic Values and Estimation of Genetic Parameters in Common Bean. Brazilian Archives of Biology and Technology, 51, 465-472. http://dx.doi.org/10.1590/S1516-89132008000300005
- Storey, W.B. (1958) Modifications of Sex Expression in Papaya. Horticultural Advance, 2, 49-60.
- Silva, F.F., Pereira, M.G., Ramos, H.C.C., Damasceno Júnior, P.C., Pereira, T.N.S., Gabriel, A.P.C., Viana, A.P. and Ferreguetti, G.A. (2008) Selection and Estimation of the Genetic Gain in Segregating Generations of Papaya (Carica papaya L.). Crop Breeding Applied Biotechnology, 8, 1-8. http://dx.doi.org/10.12702/1984-7033.v08n01a01
- Elston, R.C. (1963) A Weight-Free Index for the Purpose of Ranking of Selection with Respect Several Traits at a Time. Biometrics, 19, 85-97. http://dx.doi.org/10.2307/2527573
- Subandi, W., Compton, A. and Empig, L.T. (1973) Comparison of the Efficiencies of Selection Indices for Three Trails in Two Variety Crosses of Corn. Crop Science, 13, 184-186.
- Mulamba, N.N. and Mock, J.J. (1978) Improvement of Yield Potential of the Eto Blanco Maize (Zea mays L.) Population by Breeding for Plant Traits. Egyptian Journal of Genetics and Cytology, 1, 40-51.
- Resende, M.D.V. (2007) Matemática e Estatística na Análise de Experimentos e no Melhoramento Genético. Embrapa Floresta, Colombo, Brasil.
- Resende, M.D.V. (2002) Software SELEGEN—REML/BLUP. Embrapa Florestas, Colombo, Brasil.
- Ferrão, R.G., Cruz, C.D., Ferreira, A., Cecon, P.R., Ferrão, M.A.G., Fonseca, A.F.A., Carneiro, P.C.S. and Silva, M.F. (2008) Parâmetros Genéticos em Café Conilon. Pesquisa Agropecuária Brasileira, 43, 61-69. http://dx.doi.org/10.1590/S0100-204X2008000100009
- Silva, F.F., Pereira, M.G., Ramos, H.C.C., Damasceno Júnior, P.C., Pereira, T.N.S., Viana, A.P., Daher, R.F. and Ferreguetti, G.A. (2008) Estimation of Genetic Parameters Related to Morphoagronomic and Fruit Quality Traits of Papaya. Crop Breeding and Applied Biotechnology, 8, 65-73. http://dx.doi.org/10.12702/1984-7033.v08n01a09
- Ramos, H.C.C., Pereira, M.G., Gonçalves, L.S.A., Berilli, A.P.C.G., Pinto, F.O. and Ribeiro, E.H. (2012) Multivariate Analysis to Determine the Genetic Distance among Backcross Papaya (Carica papaya) Progenies. Genetics and Molecular Research, 11, 1280-1295. http://dx.doi.org/10.4238/2012.May.14.2
- Venkovsky, R. (1987) Herança Quantitativa. In: Paterniani, E. and Viegas, G.P., Eds., Melhoramento e Produção do Milho, Fundação Cargill, Campinas, 420 p.
- Resende, M.D.V. and Duarte, J.B. (2007) Precisão e Controle de Qualidade em Experimentos de Avaliação de Culti- vares. Pesquisa Agropecuária Tropical, 37, 182-194.
- Resende, M.D.V. (2007) SELEGEN-REML/BLUP: Sistema Estatístico e Seleção Genética Computadorizada via Modelos Lineares Mistos. Embrapa Florestas, Colombo, Brasil.
- Falconer, D.S. (1987) Introdução à Genética Quantitativa. Universidade Federal de Viçosa, Viçosa, 279 p.
- Barbosa, M.H.P., Resende, M.D.V., Bressiani, J.A., Silveira, L.C.I. and Peternelli, L.A. (2005) Selection of Sugarcane Families and Parents by Reml/Blup. Crop Breeding and Applied Biotechnology, 5, 443-450. http://dx.doi.org/10.12702/1984-7033.v05n04a10
- Resende, M.D.V. and Barbosa, M.H.P. (2006) Selection via Simulated BLUP Based on Family Genotypic Effects in Sugarcane. Pesquisa Agropecuária Brasileira, 41, 421-429. http://dx.doi.org/10.1590/S0100-204X2006000300008
- Carvalho, A.D.F., Neto, R.F. and Geraldi, I.O. (2008) Estimation and Prediction of Parameters and Breeding Values in Soybean Using REML/BLUP and Least Squares. Crop Breeding and Applied Biotechnology, 8, 219-224. http://dx.doi.org/10.12702/1984-7033.v08n03a06
- Paiva, J.R., Cordeiro, E.R., Corrêa, M.C.M. and Resende, M.C.V. (2007) Acerola Plant Selection and Breeding Value Prediction in Second Selection Cycle Progenies. Crop Breeding and Applied Biotechnology, 7, 125-132. http://dx.doi.org/10.12702/1984-7033.v07n02a03
NOTES
*Corresponding author.



