Advances in Biological Chemistry
Vol.3 No.3(2013), Article ID:33580,9 pages DOI:10.4236/abc.2013.33039
Mitochondrial DNA sequence analysis in Anabantoidei fish

School of Science and Technology, Tel-Hai Academic College, Upper Galilee, Israel
Email: gad@migal.org.il
Copyright © 2013 G. Degani. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 23 March 2013; revised 4 May 2013; accepted 19 May 2013
Keywords: Anabantoidei; Belontiidae; Betta; Colisa; Cytochrome b; 12S
ABSTRACT
The decline of ornamental fish populations in their native habitats due to over-fishing has brought about the development of ornamental fish aquaculture, to which fish of the Anabantoidei suborder are important contributors. The genetic variations among species of this suborder were examined by mitochondrial gene sequencing analysis using the cytochrome b and 12S genes. According to the cytochrome b gene, the most similar strains were Trichogaster trichopterus (gold) and Trichogaster trichopterus (blue) (100%). Trichogaster leerii was less similar to them (86.0%), and an even lower similarity was found between the species T. trichopterus and Trichogaster labiosus (85.6%). The least similarity was observed between Betta betta and the genera Colisa (50.2%) and Trichogaster (60.1%). The phylogenetic trees of 12S and cytochrome b were very similar. According to 12S, the similarity between Trichogaster species was high (91.4% - 100%), and between species of this genus and Colisa lalia, it was lower (88.4%). In conclusion, sequence analyses, based on cytochrome b and 12S can provide a useful tool for the study of the systematic relationships between species belonging to the Anabantoidei suborder. Moreover, it may support other parameters used in systematics. Thus, these genes may be applied as genetic markers in aquaculture.
1. INTRODUCTION
The decline of ornamental fish populations in their native habitats, mainly in tropical areas (due to overfishing for their sale on the tropical fish market), has led to their introduction into aquaculture and to the development of this relatively new industry. The Anabantoidei suborder is an important fish in the ornamental fish industry [1].
In phylogenetic systematics of fish, geographical, morphological, biochemical and molecular biological methods are used [2-5].
Information on the molecular variation of species belonging to the Anabantoidei suborder from the Perciformes order (perch-like) is very limited. The Anabantoidei suborder is characterized by the presence of a chamber above the gills for the retention of air for breathing [6]. The systematics, distribution and biology of these fish have been reviewed by Forselius [7].
The 16 known genera of the Anabantoidei suborder contain about 80 species, they are distributed throughout most of southern Asia, India and central Africa [7,8]. However, there is a lack of agreement concerning the systematic characteristics of this suborder, and many synonymies have been used. According to Vierke [8], taxonomists classify the labyrinth fishes into four families: Anabantidae (genera: Sandelia, Ctenopoma and Anabans); Belontiidae (genera: Trichopsis, Trichogaster, Sphaerichthys, Pseudosphromenus, Parosphromenus, Malpulutta, Helostoma, Ctenops, Colisa, Betta and Belontia); Osphornemidae (genera: Osphronemus); and Helostomatidae (genus: Helostoma).
Twenty-two isozyme systems found in the muscle and liver of 10 species of the Anabantidae family have been investigated by starch gel electrophoresis [3]. In this family, the two species, Trichogaster trichopterus and Trichogaster leerii are morphologically and geographically distributed. The Colisa lalia species does not exhibit an overlapping geographical distribution, but shows a high similarity to Betta betta, which is distributed in the small area covered by the Trichogaster genus. The gene loci of species belonging to the Colisa genus show a higher degree of similarity to those of the Trichogaster species than to those of the Betta species [3]. Population structures of Trichogaster pectoralis collected from five locations in Thailand (Samutprakan, Pitsanulok, Suphanburi, Ubonratchathani and Pattanee) were studied by isozyme analysis [9].
The structure and evolution of fish have been examined and described in terms of their DNA composition (for review [10,11]), microsatellites [12] and guppy (Poecilia reticulata) variation.
The sequence of the growth hormone (GH) has become the marker of choice for genetic studies in many fish species since it is an important hormone. GH has been studied extensively, and many of the teleost GH cDNA nucleotide sequences are available [13-15]. The GH sequence is only one of the molecular parameters that may aid in phylogenetic analysis within the Anabantoidei suborder. This information supports the other parameters and adds to the knowledge on this suborder [5].
The mitochondrial DNA sequence analysis of cytochrome b gene has been employed to study the polygeography of Sandelia capensis and S. bainsii, two fish belonging to the Anabantidae family, located in South Africa [16]. Cytochrome and 12S—a protein-coding mitochondrial gene—is widely used in vertebrates population and evolutionary genetic studies [17-19]. Restriction fragment length polymorphism (RFLP) analysis of mtDNA has proven to be effective in distinguishing between different species of fishes.
The present study examines the genetic differences between species belonging to the Anabantoidei suborder by means of mitochondrial gene sequencing analysis. The species investigated in the present study are: T. trichopterus (blue), T. trichopterus (gold), T. leerii, Trichopterus microlepis, Trichopterus labiosus, C. lalia (Hamilton), C. lalia (cobalt), B. betta and Macropodus opercularis.
2. MATERIALS AND METHODS
2.1. Fish
Different Belontiidae fish species were imported to Israel from southern Asia and India and maintained in a laboratory of MIGAL in northern Israel [3]. The species belonged to the Trichogasternae sub-family: T. trichopterus (Pallas)—blue gourami and T. leerii (Bleeker)— pearl gourami, native to Sumatra, Thailand, Borneo and Malaysia; T. microlepis—native to still or slowly moving waters in Thailand and central Cambodia (widely transported around the world as part of the aquarium fish industry); T. labiosus (C. labiosa)—native to Asia, South Myanmar (also transported around the world as part of the aquarium fish trade industry); C. lalia (Hamilton & Cobalt)—dwarf gourami native to India; and B. betta (Regan)—fighting fish native to Singapore and Thailand and M. opercularis—distributed in China (from Yangtze basin to the south), on Hainan Island, in Taiwan and North Vietnam.
2.2. DNA Extraction and PCR
Total genomic DNA was extracted from fins of adult fish. Extraction was carried out by the QIAamp DNA Mini Kit (Qiagen, Germany) after proteinase K digestion. The primer combinations, Fish ND6F and Betta cabout R and Fish 12F1 and Fish 12F2 (own sequence), were used to amplify the mitochondrial conserved region cytochrome b [20] and portions of the ribosomal gene, 12S [21], respectively (Table 1).
PCR amplification was performed in a 50 μl solution containing 10 mM Tris-HCl, 50 mM KCl, 2.5 mM MgCl2, 1 mM of each dNTP, 0.5 μM of each primer, 10 ng - 500 ng gemonic DNA and 2.5 units of Taq DNA polymerase (Promega, USA). The PCR parameters included 32 cycles of denaturation for 1 min at 94˚C (initial denaturation for 3 min at 94˚C), primer annealing for 1 min at 54˚C and extension for 1 min at 72˚C. Amplification was completed with a final elongation at 72˚C for 5 min. After amplification, the PCR products were separated by electrophoresis on a 1.3% agarose gel and stained with ethidium bromide. Following the excision of the DNA bands from the gel, the DNA was extracted with the Jet Quick Gel Extracting Spin Kit (Genomed, Germany) and suspended in distilled, deionized water. Purified DNA was sequenced directly with an ABI PRISM 3100 Genetic Analyzer (PE Biosystems, USA). All gene segments were sequenced in both directions.
2.3. DNA Sequence Analysis
Multiple sequences of cytochrome b and 12S were aligned and phylogenetic trees constructed by the

Table 1. Primers used for amplification of the mitochondrial cytochrome b and ribosomal 12S genes in gourami fish.
MegAlign program of the DNASTAR software package, which employs the neighbor-joining algorithm of Saitou and Nei [22].
3. RESULTS
Several sets of degenerate oligonucleotides were used to clone DNA fragments encoding portions of the cytochrome b and 12S genes. The results demonstrated the similarity between the gene sequences of the various Belontiidae species, leading us to hypothesize that these genes could serve as molecular markers for the systematic study of Belontiidae reproduction.
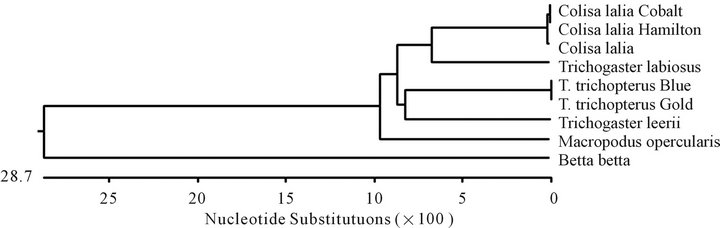
The cytochrome b gene sequences of Anabantoidei fish, which were examined in the present study, are presented in Figure 1, and the phylogenetic tree based on the nucleotide similarity between the sequences constructed with the MegAlign program (DNASTAR) by the Clustral W method is shown in Figure 2. The highest similarity (100%) with respect to the cytochrome b genes was found when the two strains, T. trichopterus (gold)

Figure 1. Comparison of cytochrome b nucleotide sequences in T. trichopterus—blue and gold gourami, T. leerii—pearl gourami, C. lalia (Cobalt and Hamilton)—dwarf gourami, T. labiosus (C. labiosa), and B. betta (Regan)—fighting fish and M. opercularis.
 (a)
(a) (b)
(b)
Figure 2. Composite phylogeny tree (a) constructed from the analysis of cytochrome b sequences of six species: T. trichopterus—blue gourami and gold gouramis, T. leerii—pearl gourami, C. lalia—dwarf gourami, T. labiosus (C. labiosa ), and B. betta (Regan)—fighting fish and M. opercularis. The phylogenetic tree was constructed by the Clustal W method using the MegAlign program (DNASTAR). Branch lengths represent evolutionary distances. Table of identities according to analysis (b).
and T. trichopterus (blue) were compared. Less similarity was detected when the cytochrome b sequence of these two strains was compared with that of T. leerii (86.0% similarity) (Figure 2), and even less similarity was observed between the species T. trichopterus and T. labiosus (85.6%). The cytochrome b sequence of T. labiosus was found to be more identical to sequences of the Colisa genus (88.0%) than to those of the Trichogaster genus. Very little similarity was found between B. betta and the genera Colisa (50.2%) and Trichogaster (60.1%) according to the cytochrome b gene (Figure 2). The similarity value for the cytochrome b gene of M. opercularis was between that of Betta and Colisa.
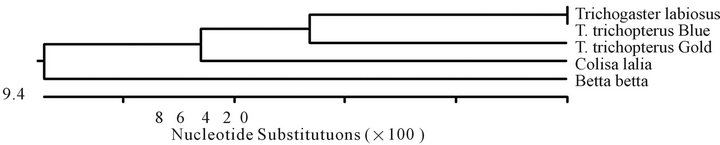
The 12S gene sequences of the species T. trichopterus (gold), T. trichopterus (blue), T. leerii, C. lalia and B. betta are shown in Figure 3, and the nucleotide similarity-phylogenetic tree is presented in Figure 4. The 12S gene phylogenetic tree results were very similar to those of the cytochrome b gene. According to the 12S gene, the sequence similarity between the species belonging to the Trichogaster genus was high (91.4% - 100%), and there was less similarity between this genus and that of C. lalia (88.4%). In addition, less similarity was evident between T. trichopterus and C. lalia than between T. trichopterus and B. betta (82.6% and 84.0%, respectively).
4. DISCUSSION
In this study, mitochondrial gene sequencing analysis of the genes cytochrome b and 12S of the Belontiidae fish family (Anabantoidei sub-order) were examined. These genes could be used as markers to distinguish between the various species in tropical fish aquaculture.
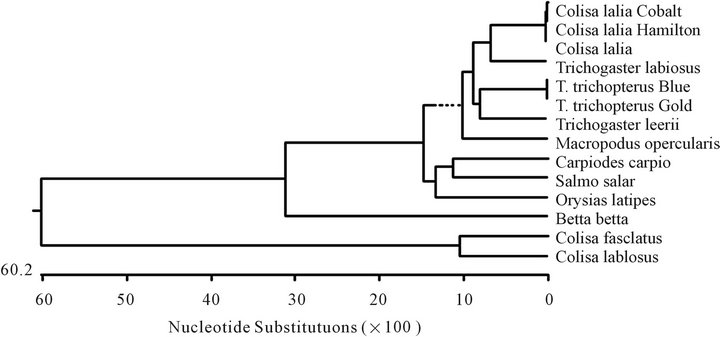
The Anabantoidei fish cytochrome b and 12S gene sequences, that were examined in the present study were compared to those of other fish and presented as a phylogenetic tree of nucleotide similarity constructed with the Missalign program (DNASTAR) by the Clustral W method (Figures 5 and 6). In this comparison, some fish, which do not belong to Anabantoidei, seem to have a high similarity to the Anabantoidei fish.
The above results support those of previous studies, in which 22 isozyme systems in the muscle and liver of 10 Anabantoidae family species were studied using a biochemical indicator of the Belontiidae fish family in

Figure 3. Comparison of the 12S gene nucleotide sequences in Trichogaster trichopterus—blue gourami and gold gourami, T. leerii—pearl gourami, C. lalia—dwarf gourami and B. betta (Regan)—fighting fish.
 (a)
(a)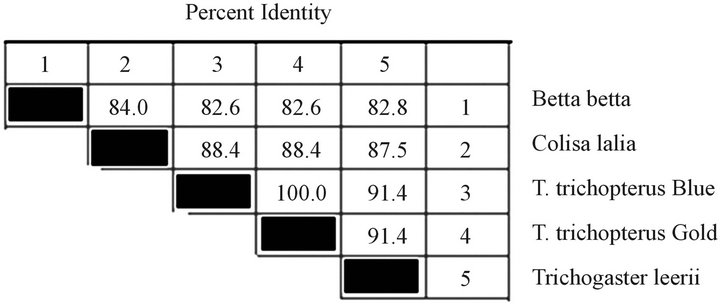 (b)
(b)
Figure 4. Composite phylogeny tree (a) constructed from analysis of 12S sequences of T. trichopterus—blue gourami and gold gouramis, T. leerii—pearl gourami, C. lalia—dwarf gourami and B. betta (Regan)—fighting fish. The phylogenetic tree was constructed by the Clustal W method using the MegAlign program (DNASTAR). Branch lengths represent evolutionary distances. Table of identities according to analysis (b).
 (a)
(a) (b)
(b)
Figure 5. Unrooted phylogenetic tree (a) of the cytochrome b fragment based on nucleotide sequences of various fish species. The length of each pair of branches represents the distance between sequence pairs, while the units at the bottom of the tree indicate the number of substitution events. The phylogenetic tree was constructed by the Clustal W method using the MegAlign program (DNASTAR). Table of identities according to analysis (b).
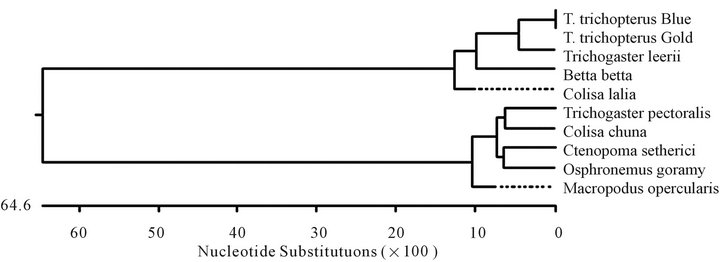 (a)
(a)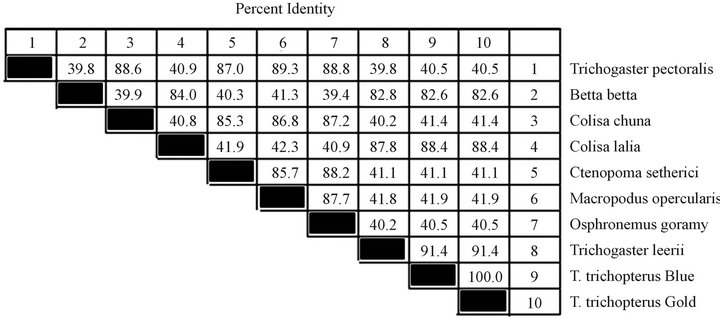 (b)
(b)
Figure 6. Unrooted phylogenetic tree (a) of the 12S fragment based on nucleotide sequences. The length of each pair of branches represents the distance between sequence pairs, while the units at the bottom of the tree indicate the number of substitution events. The phylogenetic tree was constructed by the Clustal W method using the MegAlign program (DNASTAR). Table of identities according to analysis (b).
starch gel electrophoresis [3]. In this family, the two species T. trichopterus and T. leerii were found to be morphologically and geographically distributed. In addition, the C. lalia species did not exhibit an overlapping geographical distribution, but did show a high similarity to B. betta, which is distributed in the small area occupied by the Trichogaster genus. The gene loci of species belonging to the Colisa genus demonstrated a higher degree of similarity to that of the Trichogaster species than to that of the Betta species [3]. In the present study, the results of the mitochondrial gene sequencing analysis of cytochrome b and 12S concur. A high similarity was found between the species belonging to the Trichogaster genus, and less similarity was observed between the species belonging to the Trichogaster genus and the species of the Colisa genus. Here, more species were examined than had been studied by Degani & Veit [3]. The sequence analyses of cytochrome b and 12S seem to be a useful tool for studying the systematic relationships between species belonging to this family and to support the parameters used in systematics.
The GH sequence analysis has been described in a number of fish, but in only a few orders. Knowledge concerning species from the Anabantoidei sub-order is very limited. The cDNA sequence analysis of GH has been demonstrated to be a useful tool for studying the systematic relationship between species belonging to this family, and it seems to support the parameters used in systematics [5]. The results of the present study correspond to results regarding other fish species [11]. The fish species T. leerii and T. trichopterus, which belong to the same genera, demonstrated a very high similarity to each other compared to the species C. lalia and B. betta, which belong to different genera. The gene loci of species belonging to the Colisa genus revealed a high degree of similarity to that of the Trichogaster species. There was less difference between the GH sequence of the Trichopterus and Colisa genera than between their GH sequence and that of the Betta species. From a geographical point of view, Trichopterus and Colisa are sympatric species, while Betta is not [23]. The results of a study by Degani & Veit [3] have led us to the conclusion that high polymorphisms in some enzymes of a number of species can be utilized as a genetic marker for these species and as an aid in aquaculture, when dealing with these systematic groups.
As in most teleost GHs, four cysteine residues were found in cDNA of Trichogaster trichopterus (bgGH). In comparison, five cysteine residues have been described in that of goldfish [24] and in other cyprinids [25]. These cysteine residues are involved in the formation of two disulfide bonds in a pattern analogous to the “big loop”, “little loop” (in human GH) [26], which plays an important role in determining its biological activity [27]. A putative glycosylation site was found in bgGH. The existence of this site is a common feature of many teleost GHs. It has been demonstrated that N-linked glycosylation might serve as a signal for protein transport to the cell surface [28].
In conclusion, this cytochrome b and 12S gene sequence study demonstrates that these genes may serve as a systematics tool marker for differentiating between various species of the Anabantoidei suborder.
REFERENCES
- Degani, G. (2001) Blue gourami (Trichogaster trichopterus) model for labyrinth fish. Laser Pages Publishing, Jerusalem, Israel.
- Liu, J.X., Gao, T.X., Yokogawa, K. and Zhang, Y.P. (2006) Differential population structuring and demographic history of two closely related fish species, Japanese sea bass (Lateolabrax japonicus) and spotted sea bass (Lateolabrax maculatus) in Northwestern Pacific. Molecular Phylogenetics and Evolution, 39, 799-811. doi:10.1016/j.ympev.2006.01.009
- Degani, G. and Veit, M. (1990) Electrophoretic variations of isozyme systems in the muscle and liver of Anabantidae fish. Israeli Journal of Aquaculture, 42, 67-76.
- Feulner, P.G., Kirschbaum, F., Schugardt, C., Ketmaier, V. and Tiedemann, R. (2006) Electrophysiological and molecular genetic evidence for sympatrically occuring cryptic species in African weakly electric fishes (Teleostei: Mormyridae: Campylomormyrus). Molecular Phylogenetics and Evolution, 39, 198-208. doi:10.1016/j.ympev.2005.09.008
- Degani, G., Jackson, K., Yom-Din, S. and Goldberg, D. (2006) cDNA cloning and mRNA expression of growth hormone in Belontiidae (Anabantoidei suborder). Israeli Journal of Aquaculture, 58, 124-136.
- Britz, R. (1994) Ontogenetic feature of Luciocephalus (Perciformes, Anabantoidei) with a revised hypothesis of anabantoid interelationship. Zoological Journal of the Linnean Society, 112, 491-508. doi:10.1111/j.1096-3642.1994.tb00333.x
- Forselius, S. (1957) Studies of anabantid fishes. Parts I, II, III. Zoologiska Bidrag Fran Uppsala, 32, 593-597.
- Vierke, J. (1988) Betta, gouramis and other anabantid labyrinth fishes of the world. TFH Publication, New Jersey.
- Pongthana, N. (2001) Aquaculture genetics research in Thailand. In: Gupta, M.V. and Acosta, B.O. Eds., Fish Genetics Research in Member Countries and Institutions of the International Network on Genetics in Aquaculture, The World Fish Center, Penang, 77-89.
- Degani, G. (2004) Application of RAPD and cytochrome b sequences to the study of genetic variations in F1 and F2 generations of two different strains of guppy (Poecilia reticulata, Peter 1854). Aquaculture Research, 35, 807- 815. doi:10.1111/j.1365-2109.2004.01071.x
- Dores, R.M., Rubin, D.A. and Quinn, T.W. (1996) Is it possible to construct phylogenetic trees using polypeptide hormone sequences? General and Comparative Endocrinology, 103, 1-12. doi:10.1006/gcen.1996.0088
- DeWoody, J.A., Walker, D. and Avise, J.C. (2000) Genetic parentage in large half-sib clutches: Theoretical estimates and empirical appraisals. Genetics, 154, 1907- 1912.
- Anathy, V., Venugopal, T., Koteeswaran, R., Pandian, T. J. and Mathavan, S. (2001) Cloning, sequencing and expression of cDNA encoding growth hormone from Indian catfish (Heteropneustes fossilis). Journal of Bioscience, 26, 315-324. doi:10.1007/BF02703740
- Company, R., Calduch-Giner, J.A., Mingarro, M. and Perez-Sanchez, J. (2000) CDNA cloning and sequence of European sea bass (Dicentrarchus labrax) somatolactin. Comparative Biochemistry and Physiology B Biochemical and Molecular Biology, 127, 183-192.
- Goldberg, D., Jackson, K., Yom-Din, S. and Degani, G. (2004) Growth hormone of Trichogaster trichopterus: cDNA cloning, sequencing and analysis of mRNA expression during oogenesis. Journal of Aquaculture in the Tropics, 19, 215-229.
- Roos, H. (2005) Genetic diversity in anabantids Sandelia capensis and S. vainss: A phylogeogrphic and phlogenetic investigation. Mster of Science University of Pretoria, Pretoria.
- Goldberg, T., Pearlson, O., Nevo, E. and Degani, G. (2007) Mitochondrial DNA analysis of Salamandra infraimmaculata larvae from habitats in northern Israel. Progrese şi Perspective in Medicina Veterinară”—Lucrări Ştiinţific, 50, 23-31.
- Caccone, A., Milinkovitch, M.C., Sbordoni, V. and Powell, J.R. (1997) Mitochondrial DNA rates and biogeography in European newts (genus Euproctus). Systems Biology, 46, 126-144. doi:10.1093/sysbio/46.1.126
- Pearlson, O., Blaustein, L., Goldberg, D. and Degani, D. (2005) Molecular DNA variation and mitochondrial sequence analysis of triturus vittatus vittatus (urodela) from breeding sites at various altitudes in the southern limit of its distribution. FISEB Federation of the Israel Societies for Experimental Biology, 1, 425.
- Riberon, A., Miaud, C., Guyetant, R. and Taberlet, P. (2004) Genetic variation in an endemic salamander, Salamandra atra, using amplified fragment length polymerphism. Molecular Phylogenetics and Evolution, 31, 910- 914. doi:10.1016/j.ympev.2003.10.016
- Ruber, L., Van Tassell, J.L. and Zardoya, R. (2003) Rapid speciation and ecological divergence in the American seven-spined gobies (Gobiidae, Gobiosomatini) inferred from a molecular phylogeny. Evolution, 57, 1584-1598.
- Saitou, N. and Nei, M. (1987) The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4, 406-425.
- Hoedeman, J. (1975) Naturalists’ guide to fresh-water aquarium fish. Sterling Publ. Co., Inc, New York, 971- 991.
- Law, M.S., Cheng, K.W., Fung, T.K., Chan, Y.H., Yu, K.L. and Chan, K.M. (1996) Isolation and characterization of two distinct growth hormone cDNAs from the goldfish, Carassius auratus. Archives of Biochemistry and Biophysics, 330, 19-23. doi:10.1006/abbi.1996.0221
- Chang, Y.S., Liu, C.S., Huang, F.L. and Lo, T.B. (1992) The primary structures of growth hormones of three cyprinid species: Bighead carp, silver carp, and grass carp. General and Comparative Endocrinology, 87, 385-393. doi:10.1016/0016-6480(92)90045-L
- Vestling, M., Murphy, C., Fenselau, C. and Chen, T.T. (1991) Disulfide bonds in native and recombinant fish growth hormones. Molecular Marine Biology and Biotechnology, 1, 73-77
- Lewis, U.J., Singh, R.N., Tutwiler, G.F., Sigel, M.B., VanderLaan, E.F. and VanderLaan, W.P. (1980) Human growth hormone: A complex of proteins. Recent Progress in Hormone Research, 36, 477-508.
- Guan, J.L., Machamer, C.E. and Rose, J.K. (1985) Glycosylation allows cell-surface transport of an anchored secretory protein. Cell, 42, 489-496. doi:10.1016/0092-8674(85)90106-0

