American Journal of Molecular Biology
Vol.2 No.1(2012), Article ID:16553,7 pages DOI:10.4236/ajmb.2012.21009
Somaclonal variations of soybeans (Glycine Max. L. Merr) stimulated by drought stress based on random amplified polymorphic DNAs (RAPDs)
![]()
1Lab of Molecular Biology, Department of Biology, University of Brawijaya, Malang, Indonesia
2Lab of Plant Physiology, Department of Biology, University of Brawijaya, Malang, Indonesia
3Lab of Plant Taxonomy, Department of Biology, University of Brawijaya, Malang, Indonesia
Email: larasbio@gmail.com, wahyu_widoretno@yahoo.com, indriyani.serafinah04@gmail.com
Received 14 November 2011; revised 8 December 2011; accepted 16 December 2011
Keywords: RAPD; Drought Tolerance; Soybeans; Somaclones
ABSTRACT
In soybeans, drought stress causes 50% yield losses. Breeding for drought tolerance in soybeans has been widely developed using various methods, among which is polyethylene glycol (PEG-6000) induction to simulate drought in vitro. In a previous experiment, three somaclones with different levels of tolerance were generated. The objectives of this research were to determine the RAPD patterns of those somaclones and to investigate the correlation of the RAPD patterns to the drought tolerance characteristics. The results showed eleven RAPD primers capable of amplifying the DNA genome of soybeans, among which four primers were monomorphic and seven were polymorphic. Two of the polymorphic primers, OPK7 and OPK12, are capable of differentiating medium tolerance traits from other traits. Bands that are specific for medium tolerance against drought were 450 bp and 650 bp in size, generated by the OPK7 primer, and the band of 2000 bp, generated by the OPK12 primer. However, there was no band capable of differentiating between sensitive and tolerance varieties/ lines, although some changing of the DNA sequence was detected in this research. This indicates that there are other factors responsible for the expression of drought tolerance.
1. INTRODUCTION
Drought has been reported to cause some problems in plants. In soybeans, drought stress at the generative phase has been reported to decrease seed production up to 5%. Some mechanisms developed by plants to overcome water deficit problems have been reported. Morphological adaptations such as developing of profusely root hairs, rolling of leaves and lengthening of roots, to physiological alterations, such as modifications in carbon partitioning and isotope discrimination, osmotic adjustment, and alterations in rate and efficiency of photosynthesis has been addressed as plants responses toward drought stress [2]. These modifications alter cell structure, activating enzyme complexes that trigger molecular events in cascade, leading to expression of many categories of genes involved in the activation of defense responses [3].
Genes involved in drought tolerance [4-8], can be classified into two groups. The first group consists of genes that code proteins such as chaperones, late embryogenesis abundant (LEA) proteins such as LEA14 [9], osmotin, antifreeze proteins, mRNA-binding proteins such as glycine-rich protein [10], key enzymes for osmolyte biosynthesis, such as galactinol synthase and delta- 1-pyrroline-5-carboxylate synthetase, involved in synthesis of raffinose-family oligosaccharides [11] water channel proteins, such as PIP1b [12], and proline transporters [13,14], detoxification enzymes, and various proteases that are suspected to function in abiotic stress tolerance. The second group is comprised of regulatory proteins, i.e. protein factors involved in further regulation of signal transduction and stress-responsive gene expression. These include various transcription factors, protein kinases, protein phosphatases, enzymes involved in phospholipid metabolism, and other signalling molecules such as calmodulin-binding protein [5,8].
The expression mechanism of some genes was reported to be dependent from Abscisic acid (ABA) which plays a key role in the molecular signal that is triggered by the onset of drought. This phytohormone apparently works as a second messenger after stress perception, inducing stomatal closure and activating several stressrelated genes [15]. On the other hand, some research has shown that there are regulatory systems that are ABAindependent [16]. It was reported that. rd29a in Arabidopsis thaliana, are induced through an ABA-independent pathway. The protein DREB1 (dehydration-responsive element-binding) is a transcription factor that binds to the promoter of genes such as rd29a, thereby inducing expression in response to drought, salinity, or low temperatures.
Research on drought stress mostly concentrated on gene expression, on the other hand study on the effect of drought to gene mutation has not reported. Mutation usually was detected by breeders by observing and selecting several generations for the trait of interest. Development of molecular biology methods gives more straightforward detection methods to the locus responsible for certain traits. Currently, the molecular approaches such as Random Amplified Polymorphic DNA (RAPD), Restriction Fragment Length Polymorphism (RFLP), Simple Sequence Repeat (SSR), Amplified Fragment Length Polymorphism (AFLP), and Single Nucleotide Polymorphism (SNP) [17] are being widely used to detect DNA polymorphism. Among them, RAPD has been proven very useful in the construction of genetic maps of several species and in the construction of genetic markers related to a certain phenotypic trait, especially on using bulk segregant pools [18,19]. RAPD has been reported capable of detecting the relative distance among the member of genus Hibiscus section Furcaria [20]. The genetic diversity of kenaf based on agronomic and RAPD data has been detected [21]. Characterization of flax collection by means of RAPD technique also has been done [19].
Breeding soybeans for drought tolerance is one solution offered by some researchers to counter the problem. Many methods have been developed. In the previous experiment an in-vitro mutation induction using a drought simulation agent, polyetilene glicol (PEG-6000) has been done. Selection of the mutants (somaclones) based on physiological traits in the laboratory level showed that tolerance toward drought stress is related to the increase of proline content. Four somaclones with different levels of tolerance were developed [22-24]. Drought simulation using PEG-6000 to the four somaclones in the greenhouse showed medium tolerance to drought. The aim of the present work was to detect mutations on the soybean somaclones and to detect the relative distance between the somaclones with the initial lines and standard lines using RAPD analysis.
2. EXPERIMENT
2.1. Plant Material and DNA Isolation
Plant materials used were drought-susceptible soybean lines, MSC8606, medium tolerance lines, MLG 2999, and B3731, selected by Research Institute for Legumes and Tuber Crops, Indonesia and the somaclones arising from them, a drought-susceptible somaclone 8B6, and two medium tolerance somaclones, 2S3 and 3SE4, respectively. Those lines and somaclones were compared to the standard variety for drought tolerance, Dieng, and drought susceptible variety, Burangrang. The DNA genome was isolated manually from 100 mg young soybean leaves, followed the CTAB method [25].
2.2. RAPD Analysis and Dendogram Construction
An RAPD-PCR reaction was performed using a reaction mixture consisting of (25 ml ) 10x buffer Ex Taq Polymerase, 2 mM MgCl2, 200 uM dNTP, 10 mM Tris-HCl (pH 8.0) 25 pmol primer, 25 pmol Ex Taq Polymerase, and aquadest. The program was set at 95˚C, 3 minutes preheating, continued with 45 cycles consisting of 94˚C, 1 minute denaturation, 36˚C of 1 minute annealing, and 72˚C for 2 minutes extension, and finally 72˚C for 10 minutes for the last extension and cooling at 4˚C for 5 minutes. One set of RAPD primers, OPK, from Operon Technologies, USA that consist of 20 oligonucleotides was used in this experiment. All the RAPD fragments resulted from polymorphic primers were used for analysis of relative distance between lines using the Clad97 program.
3. RESULT
3.1. RAPD Analysis
An RAPD analysis of the somaclones, their initial lines and the standard varieties using the Operon 10-mer Kit K resulted in 11 primers, OPK-02, OPK-04, OPK-07, OPK- 08, OPK-10, OPK-12, OPK-14, OPK-16, OPK-17, OPK- 19, and OPK-20, capable of amplifying the DNA genome and producing RAPD bands. Seven of them generated polymorphic bands (Figure 1, Table 3).
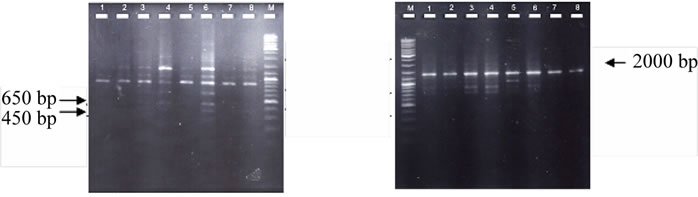
Comparing the individual DNA fragments in those lines shows that not all the primers used are capable of differentiating between different levels of tolerance against drought stress. Only two primers, OPK-07 and OPK-12, produced specific bands for a medium drought tolerant line and somaclones B3731, 3SE4 and 2S3, but did not produce such bands in susceptible lines (Figure 1). Band 650 bp and 450 bp, which were produced by OPK7, and band 2000 bp, produced by OPK12, are capable of distinguishing tolerance to sensitive lines and somaclones.
The percentages of polymorphic fragments versus total fragments varied from 29% (OPK-04) to 100% (OPK-02 and OPK-12) (Table 3). The somaclone variants can be clearly distinguished based on their RAPD banding patterns, ranging from two (OPK-19) to six (OPK-02) (Figure 1 and Table 3). Most somaclone
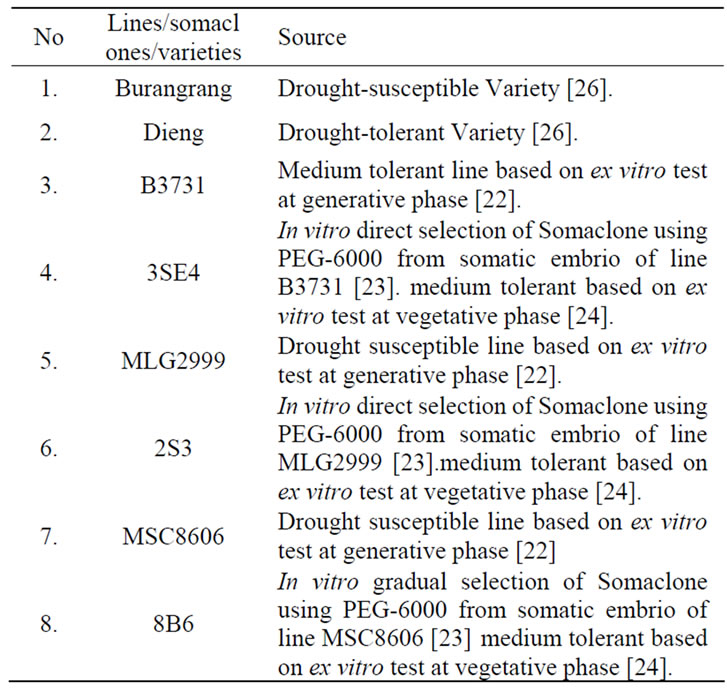
Table 1. Soybean lines, somaclones and varieties used in this experiment.
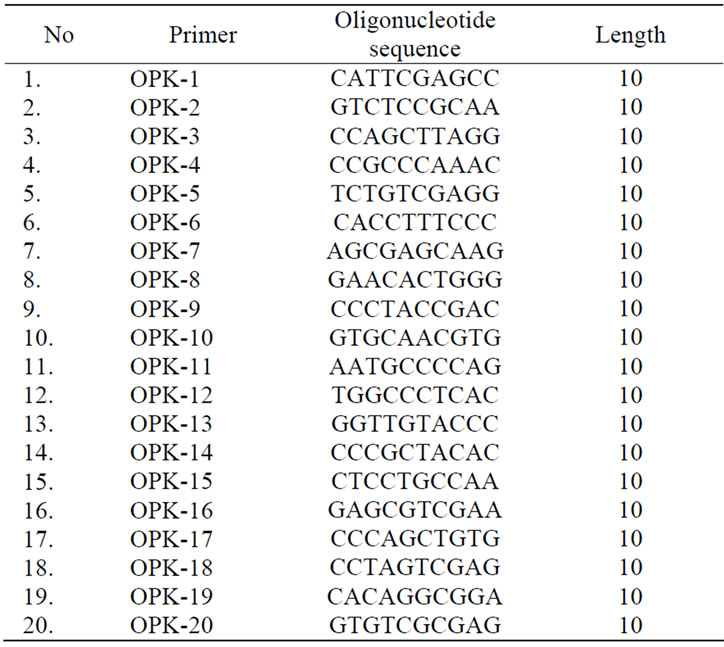
Table 2. Sequence of RAPD primer OPK used in this experiment.
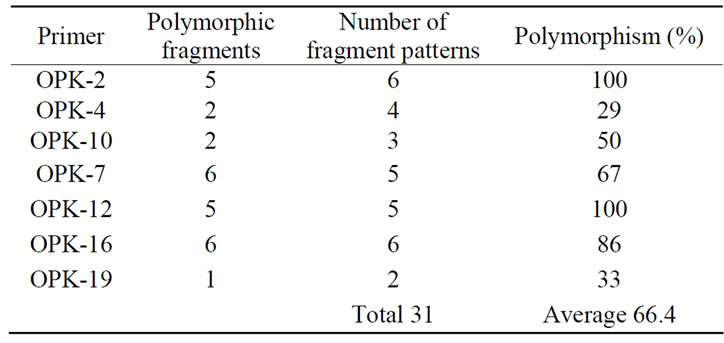
Table 3. Number of patterns that can be distinguished within the 8 soybean somaclones, lines, and varieties with different RAPD primers and total number of polymorphic fragments in the data set.
 (a) (b)
(a) (b)
Figure 1. RAPD banding pattern on agarose 1.5% (M: marker 1 kb, 1 = Burangrang, 2 = Dieng, 3 = B3731, 4 = 3SE4, 5 = MLG2999, 6 = 2S3, 7 = MSC8606, 8 = 8B6) (a). Band 650 bp and 450 bp amplified by OPK7 capable of distinguishing tolerance to sensitive lines and somaclones; (b). Band 2000 bp amplified by OPK12 capable of distinguishing tolerance to sensitive lines and somaclones.
variants which expressed similar levels of tolerance against drought stress did not possess similar RAPD fragment patterns. This means that the DNA sequences of those variants were different, and this difference doesn’t affect the levels of tolerance.
3.2. The Relative Distance of the Somaclones, the Initial Line and the Standard Varieties
All the RAPD fragments resulted from polymorphic primers were used for analysis of relative distance between lines using the Clad97 program. All somaclones variants were used for the construction of a dendogram. A total number of 31 polymorphic fragments was unambiguously identified and used for the computation of relative genetic similarities. A dendogram was constructed based on the RAPD fragment variations in different somaclone variants (Figure 2).
The dendogram grouped the somaclone variants into two main clusters at a similarity coefficient of 0.67. Comparing the clustering pattern with the tolerance levels, it was found that RAPDs are not capable of differentiating between levels of tolerance. The Dieng variety, which has been known as a drought-tolerant variety, clustered with MSC8606 and its somaclone 8B6, which were physiologically drought susceptible. On the other hand, the Burangrang variety, which is a standard variety for drought susceptibility, was in the same cluster with MLG2999, B3731 and 3SE4, which were mediumtolerant variants. However, in that cluster, Burangrang was positioned quite separately from the other three variants at a similarity coefficient of 0.7. Interesting data were shown by 2S3, which is the somaclone variant of MLG2999. Both variants have medium tolerance against drought. However, they were separated in two different clusters, MLG2999 positioned in a similar cluster with Dieng, which is a tolerant variety, and 2S3 was grouped with B3731, 3SE4 and Burangrang, which are mediumtolerant and susceptible varieties.
4. DISCUSSION
RAPD has been widely used as a tool to detect differences
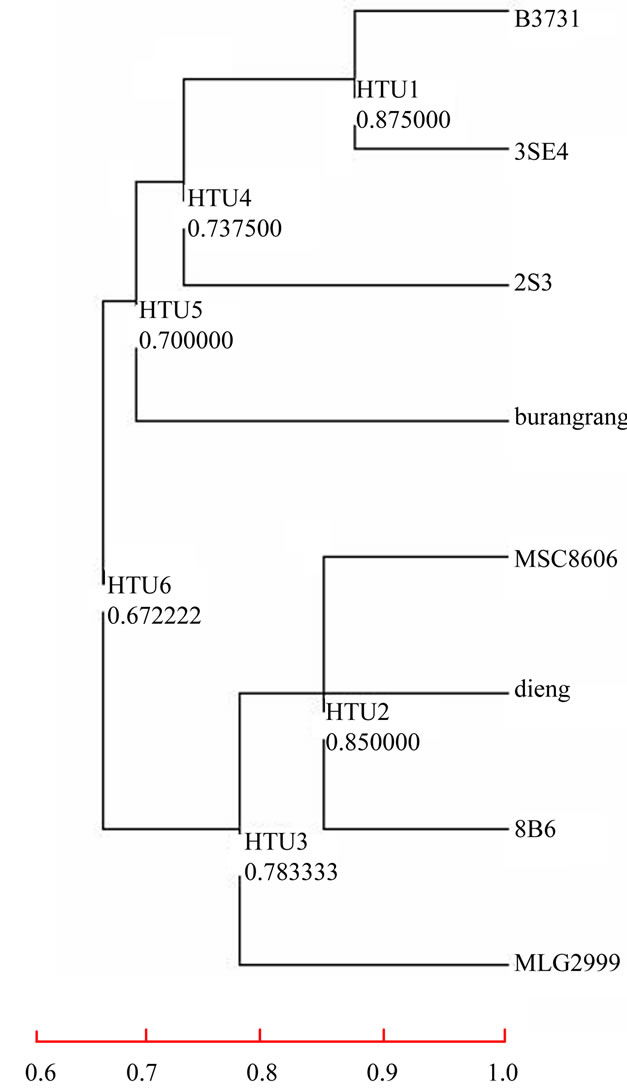
Figure 2. A dendogram showing genetic relationships among soybean lines constructed, based on the UPGMA cluster analysis of RAPDs. Drought-susceptible variety Burangrang clustered with medium-tolerant variants, MLG2999, B3731 and 3SE4. Drought-tolerant variety Dieng clustered with droughtsusceptible MSC8606 and its somaclone 8B6.
in DNA sequences of various organisms, including soybeans [27-30]. RAPD has also been used to detect genetic diversity among 18 selected soybean genotypes in breeding programs to increase the protein content [31].
The absence or presence of one specific gene or allele could cause a change in the drought resistant nature. If a sequence of a certain gene, or part of a gene, is not detected by RAPD primers, this may occur because the sequence is missing or changed into a different sequence. The amount of change will determine whether the change is up to the level of genes (different genes) or only the level of alleles (different alleles). Both these changes may cause significant changes in expression, depending on the role of the genes. In this experiment, differences in sequences were detected by the primers OPK-0, which produced 650 bp and 450 bp bands, and OPK-12 which produced a 2000 bp band. Those bands were not detected in sensitive lines. The bands may be associated with the expression of drought-tolerant genes.
A number of genes that regulate drought resistance have also been reported by many researchers. Breeding to increase resistance to stress has consistently showed that the vigor of plants in various environmental conditions is naturally regulated by multiple loci. Drought resistance is a complex phenomenon, involving the coordinated expression of genes in large numbers [32]. To get droughtresistant plants, co-expression of more than one gene is required. For example, expression of XVPer1, which allegedly protects DNA from ROS (reactive oxygen species) together with XVSAP1, which may prevent membrane leakage, and XVGols and ALDRXV4, both of which are osmoprotectans, might result in the group of proteins that are capable of producing drought resistance [33].
Plant response to drought stress can be grouped into three stages [34]. When drought stress is not too strong, the plant will try to avoid experiencing water shortages. Plants will use a little water in balance with the water escaping, such as by closing the stomata. When stress increases, the plant will try as much as possible to increase the amount of water in the cells, for example, by increasing the number of roots and reducing shoot growth. If the plant experiences prolonged drought stress which is caused by the availability of water being really low, the plant will show several responses. These include accumulation of dissolved materials or production materials or proteins, changes in metabolism, detoxification of free oxides (reactive oxygen species, ROS) to avoid cell damage due to water loss. These responses are associated with the regulation of many active genes which produces a variety of osmoprotectan compounds. One mechanism of plant tolerance to drought stress is the ability of plant cells to adjust osmotic potential by accumulating solutes, such as proline, and sugars in the cell. Proline plays a role in the adjustment of osmotic function in maintaining turgor and root growth in drought conditions, while the sugar serves to maintain the stability of cell membranes, protect proteins, and lower leaf osmotic potential so plants keep their turgidities when experiencing drought stress. Drought stress causes an increase in the proline content in all soybeans that are tolerant and drought sensitive [35]. An accumulation of proline serves as a source of cytoplasmic osmoticum, cytoplasmic enzyme protectors, and protective cellular structures. Correlation between proline accumulation and the level of plant tolerance to drought stress has been reported based on the analysis of transgenic plants. Transforming gene P5CS (delta 1-pyrroline- 5-carboxylate synthetase), which plays a role in proline accumulation in soybeans, is capable of producing plants which are more tolerant to drought [36].
The presence or absence of RAPD bands may be associated with regulatory or structural genes corresponding to drought stress. Drought stress acts as a signal which will be received by receptors (ion channel, histidine kinase, GPCR, RLK), which then activate the Ca2 ions and molecules such as inositol secondary identifiers, ROS, and ABA, to activate the transcription factor. Transcription factors (DREB, bzipped, EREB/AP2, MYC, MYB) induce genes, in response to drought stress (LEA-like, P5CS, HVA, AtABA, rd21, TPSP etc.), to produce osmoprotectan compounds to maintain cell osmotic pressure, keeping cell turgor, etc. [37].
The ability of the RAPD technique to distinguish between tolerant or sensitive varieties of the strains and somaclones in this experiment shows that this technique is capable of detecting the presence of somaclonal variations. Somaclonal diversity is derived from the genetic diversity of explant sources and mutations that occur during a tissue-culture process. The diversity of the explants was due to mutations as well as the presence of polysoms in a particular tissue. Genetic diversity in tissue cultures, among others, is caused by doubling the number of chromosomes, chromosomal structural changes, changes in genes and changes in genetic material in the cytoplasm [38].
A variation of banding patterns between the original lines and their somaclones shows that, indeed, there is a change of sequence on somaclones as a result of the simulation of drought stress with PEG-6000. However, the banding pattern that is summarized in the dendogram cannot be associated with tolerant or sensitive natures. There are several possibilities that cause this to happen. The first is a change in the DNA sequence that is not expressed into the physiological level. The second possibility is that changes in the DNA sequence occur in the genes that are not vital, so they do not alter the gene expression. The third possibility is changes in the DNA sequence which occur only in one particular gene, while producing resistance to drought takes several genes that cooperate with each other. This phenomenon seems to be the result of differential mutations on different loci which convert the level of tolerance against drought, which is possible because there is a family of genes that control drought tolerance. This finding is in accordance to previous research [39] which stated that tolerance of drought is a complex phenomenon. The tolerance changes according to drought intensity and duration, and the plant’s developmental stage during which drought occurs. It was also reported that one stress may affect the plant which lead to the activation of many genes as a response to the stress. So, plant adaptation to drought induces, more or less, some reactions, such as signaling pathways, target-gene expression, and biochemical/metabolic changes. As a result, differences in water-stress tolerance among cultivars, or within a cultivar at various developmental stages, may result from differences in the expression of genes in signal-perception and transduction mechanisms.
5. CONCLUSION
RAPD variations were detected among lines and somaclones, suggesting that drought stress simulation using PEG-6000 is capable of altering DNA sequence. However, this alteration can not be associated with drought-tolerant or sensitive natures, suggesting that the changing of DNA sequences detected in this research was not the only component to convert drought tolerance. It may occur only in one particular gene, while producing resistance to drought takes several genes that cooperate with each other.
6. ACKNOWLEDGEMENTS
The authors would like to thank Research Institute for Legumes and Tuber Crops, Kendalpayak, Malang, Indonesia for providing soybean seeds, Agustin Sri Mulyatni for technical assistance, and Dr. Larry Cothren from the Intensive English Language Institute (IELI) Flinders University, South Australia, for English language assistance with the manuscript. The research was supportedby Directorate General of Higher Education (DGHE)’s PHB project.
REFERENCES
- Taji, T., Seki, M., Satou, M., Sakurai, T. Kobayashi, M., Ishiyama, K., Narusaka, Y., Narusaka, M., Zhu, J.K. and Shinozaki, K. (2004) Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiology, 135, 1697-1709. doi:10.1104/pp.104.039909
- Stolf-Moreira, R., Medri, M.E., Neumaier, N., Lemos, N.G., Pimenta, J.A., Tobita, S., Brogin, R.L., MarcelinoGuimarães, F.C., Oliveira, M.C.N., Farias, J.R.B., Abdelnoor, R.V. and Nepomuceno, A.L. (2010) Soybean physiology and gene expression during drought. Genetics and Molecular Research, 9, 1946-1956. doi:10.4238/vol9-4gmr851
- Widoretno, W., Guhardja, E., Ilyas, S. and Sudarsono. (2002) The effectivity of Polyetylene Glycol for evaluating the response of soybeans genotype toward drought stress in the seedling phase. Hayati, 9, 33-36.
- Pastori, G.M. and Foyer, C.H. (2002) Common components networks and pathways of cross-tolerance to stress the central role of “redox” and abscisic acid-mediated. Plant Physiology, 129, 460-468. doi:10.1104/pp.011021
- Shinozaki, K., Yamaguchi-Shinozaki, K. and Seki, M. (2003) Regulatory network of gene expression in the drought and cold stress responses. Current Opinion in Plant Biology, 6, 410-417. doi:10.1016/S1369-5266(03)00092-X
- Bartels, D. and Sunkars, R. (2005) Drought and salt tolerance in plants. Critical Reviews in Plant Science, 24, 23-58. doi:10.1080/07352680590910410
- Yamaguchi-Shinozaki, K. and Shinozaki, K. (2005) Organization of cis-acting regulatory elements in osmoticand cold-stress-responsive promoters. Trends in Plant Science, 10, 88-94. doi:10.1016/j.tplants.2004.12.012
- Shinozaki, K. and Yamaguchi-Shinozaki, K. (2007) Gene networks involved in drought stress response and tolerance. Journal of Experimental Botany, 58, 221-22. doi:10.1093/jxb/erl164
- Singh, K., Foley, R.C. and Onate-Sanchez, L. (2002). Transcription factors in plant defense and stress responses. Current Opinion in Plant Biology, 5, 430-436. doi:10.1016/S1369-5266(02)00289-3
- Bocca, S.N., Magioli, C., Mangeon, A., Junqueira, R.M., Cardeal, V., Margis, R. and Sachetto-Martins, G. (2005). Survey of glycine-rich proteins (GRPs) in the Eucalyptus expressed sequence tag database (ForEST). Genetic and Molecular Biolology, 28, 608-624. doi:10.1590/S1415-47572005000400016
- Hannah, M.A., Wiese, D., Freund, S., Fiehn, O., Heyer, A.G. and Hincha, D.K. (2006) Natural genetic variation of freezing tolerance in Arabidopsis. Plant Physiology, 142, 98-112. doi:10.1104/pp.106.081141
- Aharon, R., Shahak, Y., Wininger, S., Bendov, R., Kapulnik, Y. and Galili, G. (2003) Overexpression of a plasma membrane aquaporin in transgenic tobacco improves plant vigor under favorable growth conditions but not under drought or salt stress. Plant Cell, 15, 439-447. doi:10.1105/tpc.009225
- Aroca, R., Alguacil, M. M., Vernieri, P. and Ruiz-Lozano, M. (2008) Plant responses to drought stress and exogenous aba application are modulated differently by mycorrhization in tomato and an aba-desficient mutant (Sities). Microbial Ecology, 56, 704-719. doi:10.1007/s00248-008-9390-y
- Schafleitner, R., Gaudin, A., Rosales, R.O.G., Aliaga, C.A.A. and Bonierbale, M. (2007) Proline accumulation and real time PCR expression analysis of genes encoding enzymes of proline metabolism in relation to drought tolerance in Andean potato. Acta Physiologiae Plantarum, 29, 19-26. doi:10.1007/s11738-006-0003-4
- Iuchi, S., Kobayashi, M., Yamaguchi-Shinozaki, K. and Shinozaki, K. (2000) A stress-inducible gene for 9-cisepoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiology, 123, 553-562. doi:10.1104/pp.123.2.553
- Yamaguchi-Shinozaki, K. and Shinozaki, K. (2005) Organization of cis-acting regulatory elements in osmoticand cold-stress-responsive promoters. Trends in Plant Science, 10, 88-94. doi:10.1016/j.tplants.2004.12.012
- Krishna, H. and Singh, S.K. (2007) Biotechnological advances in mango (Mangifera indica L.) and their future implication in crop improvement: A review. Biotechnology Advances, 25, 223-243. doi:10.1016/j.biotechadv.2007.01.001
- Welsh, J. and McClelland, M. (1990) Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Research, 18, 7213-7218. doi:10.1093/nar/18.24.7213
- Williams, J.G.K., Kubelik, A.K. and Livak, K.J. (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Research, 18, 6531-6535. doi:10.1093/nar/18.22.6531
- Anping, G., Jianguang, S. and Xingi, C. (1998) Genome relationship among hibiscus section furcaria species based on random amplified polymorphic DNA (RAPD). Proceedings of the Workshop on Application of Biotechnology in the Improvement of Jute, Kenaf and Allied Fibres-Phase I, Beijing, China.
- Cheng, Z., Lu, B. and Baldwin, B.S. (2002) Comparative studies of genetic diversity in kenaf (Hibiscus cannabinus L.) varieties based on analysis of agronomic and RAPD data. Hereditas, 136, 231-239. doi:10.1034/j.1601-5223.2002.1360309.x
- Widoretno, W. and Sudarsono (2004) Evaluation on some lines of somaclonal variant soybeans arised from in vitro selection against drought. Hayati, 11, 11-20.
- Widoretno, W., Guhardja, E., Ilyas, S. and Sudarsono (2003) Polyetilene glycol effectivity in evaluating response of soybeans to drought stress at germination stage. Hayati, 9, 33-36.
- Trisnawati, L.P.A. (2006) Growth response, relative water content, and chlorophyll content after drought treatment of some soybean somaclones [Glycine Max (L)] resulted from in vitro selection using Polyetilene glycol at vegetative phase. Thesis, Faculty of Science, University of Brawijaya, Malang, Indonesia (unpublished).
- Doyle J.J. and Doyle J.L. (1987) A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochemical Bulletin, 19, 11-15.
- Balitkabi (Research Institute for Legumes and Tuber Crops) (2008) Description of superior legumes and tuber crops varieties. Research Institute for Legumes and Tuber Crops, Malang, Indonesia (unpublished).
- Pakniyat, H. and Nazari L. (2008) Genetic diversity of wild and cultivated barley genotypes under drought stress using RAPD markers. Biotechnology Issue, 7, 745-750.
- Li, Z. and Nelson, R.L. (2002) RAPD Marker Diversity among Cultivated and Wild Soybean Accessions from Four Chinese Provinces. Crop Science, 42, 1737-1744. doi:10.2135/cropsci2002.1737
- Thompson, J.A. and Nelson, R.L. (1998) Core set of primers to evaluate genetic diversity in soybean. Crop Science, 38, 1356-1362. doi:10.2135/cropsci1998.0011183X003800050034x
- Thompson, J.A., Nelson, R.L. and Vodkin, L.O. (1998) Identification soybean germplasm using RAPD markers. Crop Science, 38, 1348-1355. doi:10.2135/cropsci1998.0011183X003800050033x
- Doldi, M.L., Vollmann, J. and Lelley, T. (2006) Genetic diversity in soybean as determined by RAPD and microsatellite analysis. Plant Breeding, 116, 331-335. doi:10.1111/j.1439-0523.1997.tb01007.x
- Walters, C., Farrant, J.M., Pammenter, N.W. and Berjak, P. (2002) Desiccation stress and damage. In: Black, M. and Pritchard, H.W., Eds., Desiccation and Survival in Plants: Drying without Dying. CABI Publishing, Oxford and New York, 263-293. doi:10.1079/9780851995342.0263
- Mundree, S.G., Baker, B., Mowla, S., Peters, S., Marais, S., Willigen, C.V., Govender, K, Maredza, A., Muyanga, S., Farrant, J.M. and Thomson J.A. (2002) Physiological and molecular insights into drought tolerance. African Journal of Biotechnology, 1, 28-38.
- Verslues, P.E., Agarwal, M., Katiyar-Agarwal, S., Zhu, J. and Zhu, J.K. (2006) Techniques for Molecular Analysis: Methods and concepts in quantifying resistance to drought salt and freezing abiotic stresses that affect plant water status. The Plant Journal, 45, 523-539. doi:10.1111/j.1365-313X.2005.02593.x
- Hamim, K., Ashari, Miftahudin and Triadiati (2008) Analysis of water status of prolin and the activity of antioxidant enzym on some drought tolerant, susceptible and wild type. Agrivita, 30, 201-210.
- Simon-Sarkadi, L., Kocsy, G., Várhegyi, Á., Galiba, G. and de Ronde, J.A. (2005) Genetic manipulation of proline accumulation influence the concentrations of other amino acids in soybean subjected to simultaneous drought and heat stress. Journal of Agricultural and Food Chemistry, 53, 7512-7517. doi:10.1021/jf050540l
- Xiong, L., Schumaker, K.S. and Zhu, J.K. (2002) Cell Signaling during Cold Drought and Salt Stress. The Plant Cell, 14, 165-S183.
- Hutami, S., Mariska, I. and Supriati, Y. (2006) The increase of genetic variability of plant through somaclonal variation. Jurnal AgroBiogen, 2, 81-88.
- Chinnusamy, V., Schumaker, K, and Zhu, J.K. (2004). Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. Journal of Experimental Botany, 55, 225-236. doi:10.1093/jxb/erh005

