Advances in Bioscience and Biotechnology
Vol.4 No.3A(2013), Article ID:29295,14 pages DOI:10.4236/abb.2013.43A061
Metabolic regulation of Escherichia coli cultivated under anaerobic and aerobic conditions in response to the specific pathway gene knockouts
![]()
1Department of Bioscience and Bioinformatics, Kyushu Institute of Technology, Fukuoka, Japan
2Institute of Advanced Bioscience, Keio University, Tokyo, Japan
Email: *shimi@bio.kyutech.ac.jp
Received 5 January 2013; revised 16 February 2013; accepted 25 February 2013
Keywords: Metabolic Regulation; Single-Gene Knockout Mutant; Escherichia coli
ABSTRACT
Effect of the specific gene knockout on the main metabolism in Escherichia coli was reviewed, and the regulation mechanisms were clarified based on different levels of information such as gene expressions, enzyme activities, intracellular metabolite concentrations, and metabolic fluxes together with fermentation data. The effects of the knockout of such genes as pflA, pta, ppc, pykF, adhE, and ldhA on the metabolic changes were analyzed for the case under anaerobic condition. The effects of the knockout of such genes as pgi, zwf, gnd, ppc pck, pyk, and lpdA on the metabolic changes were also analyzed for the case under aerobic condition. The metabolic regulation analysis was made focusing on the roles of transcription factors.
1. INTRODUCTION
Escherichia coli is a Gram-negative, facultative anaerobic and non-sporulating bacterium. The E. coli cells are typically rod-shaped with length of about 1 - 2 μm having a cell volume of 0.6 - 0.7 μm3. This organism has been identified by Theodor Escherich in 1885, and is now classified as part of Enterobacteriaceae family of gamma-proteobacteria [1]. This bacterium grows optimally at 37˚C and can survive even at higher temperature up to about 50˚C. E. coli is commonly found in the intestine of warm-blooded organisms. Although most E. coli strains are harmless to host organisms, and those are part of normal flora of the gut, and can benefit the host organisms by providing some vitamins, and by preventing the establishment of pathogenic bacteria within the intestine, some strains such as O157:H7 cause serious poisoning to human beings [2,3]. Escherichia coli is now the most popular bacterial model organism due to its well-known characteristics and easy for gene manipulation, and the research on such organism is useful for medical, pharmaceutical, and fermentation applications.
The most typical wild type strain E. coli K-12 has about 4605 open reading frame (ORF) in a 4.6 Mbp long [1]. The genes are located in 3386 transcription units, where it is operative unit for gene expression based on promoter and terminator regions, which correspond to the start and terminus of transcription, respectively. Of the total genes, about 300 are considered to encode transcription factors (TFs) and 7 for σ factors [4,5]. The detailed information is updated as documented in RegulonDB [6] and EcoCyc [7]. The hierarchical structure of such TFs and the regulated genes have been identified to some extent [8], and some of the metabolic regulation mechanism has been investigated [9,10].
As mentioned above, E. coli has been often utilized to produce useful metabolites by metabolic engineering. However, the success of pathway engineering is limited, where the expected result is rarely obtained for strain improvement. The main reason is due to limited knowledge on the metabolic changes caused by gene level and enzyme level regulation. Here, an attempt is made to clarify the regulation mechanisms for the several specific gene knockout mutants under both anaerobic and aerobic conditions based on different levels of information focusing on the roles of transcription factors.
2. REGULATION OF THE METABOLISM BETWEEN ANAEROBIC AND AEROBIC CONDITIONS
Global regulators such as Fnr and ArcAB are mainly responsible for the regulation of the metabolism under oxygen limited condition, where Fnr regulates the expressions of metabolic pathway genes under anaerobic condition [11], while ArcAB regulates under both anaerobic and microaerobic conditions [12,13]. It has been shown that ArcA/B system exerts more significant effect on the cell metabolism under microaerobic condition than under aerobic or anaerobic condition. The effect of ArcAB system on the flux distribution at pyruvate node has been investigated based on the extracellular metabolite concentrations [14-16]. It was shown that lactate can be overproduced by arcA/fnr double mutant [14] in a similar way as pfl gene knockout [17,18].
Reoxidation of the reducing equivalents such as NADH generated by the oxidation of the energy source occurs in the respiratory chain under aerobic or microaerobioc condition. In E. coli, NADH is oxidized in the respiratory chain via a coupled NADH dehydrogenage NDH-1 encoded by nuo or an uncoupled dehydrogenase NDH-2 encoded by ndh, and the electron flows into quinone and quinol pool. Quinol is then oxidized by either the cytochrome bo or the cytochrome bd terminal oxidase complex, which in turn passes the electrons to oxygen with concomitant production of water. The cyoABCDE operon is represses by both ArcA and Fnr, while cydAB operon is activated by ArcA and repressed by Fnr [19].
The reducing equivalents such as NADH are reoxidized in the respiratory chain, where oxygen, nitrate, fumarate, and dimethyl sulfoxide etc. are the electron acceptors. This process is coupled to the formation of a proton mortive force (PMF), which is utilized for ATP generation from ADP and Pi. In the absence of oxygen, or other electron accepters, ATP is generated via substrate level phosphorylation through the process of degradation of carbon source in the metabolic pathways. The expressions of pfl genesis activated by ArcA and Fnr, and it becomes higher at lower oxygen concentrations, whereas aceE,F which encode α and β subunits of PDHc is repressed by ArcA under oxygen limited condition. At the branch point of AcCoA, the product of both Pfl and PDHc reactions, is converted to either acetate and ethanol or subsequently undergo further oxidation in the TCA cycle.
The Arc (anoxic respiration control) system, composed of ArcA, the cytosolic response regulator, and ArcB, the membrane bound sensor kinase, regulates the TCA cycle genes depending on the oxygen level or redox state. Upon stimulation by the redox state, ArcB undergoes autophosphorylation, and the phosphoryl group is transferred to ArcA. The phosphorylated ArcA binds to the promoter regions of the TCA cycle and other genes. It has been reported that ArcA, when phosphorylated, represses the expressions of the genes involved in the TCA cycle and the glyoxylate shunt genes such as gltA, acnAB, icdA, sucABCD, sdhCDAB, fumA, mdh, and aceA,B (Appendix A). Moreover, the genes which encode the primary dehydrogenases such as glpD, lctPRD, aceE,F and lpdA are also repressed by ArcA (Appendix A).
Alexeeva et al. [13] investigated the effects of different oxygen supply rates on the catabolism in arcA mutant. A strong link is demonstrated between redox ratio (NADH/NAD+) and acetate overflow in E. coli [20]. It was shown that the commencement of acetate overflow occurred above the critical NADH/NAD+ ratio of 0.06 [20]. Moreover, acetate overflow is delayed by the expression of heterologous NADH oxidase (NOX), an enzyme that serves to reduce the NADH/NAD+ ratio [20]. The redox state has been reported to trigger the Arc regulon [21,22].
Since phosphorylated ArcA represses TCA cycle genes, the arcA gene deletion activates the TCA cycle, resulting in the reduction in the acetate formation [20]. The NADH oxidation by the expression of NOX in the arcA gene knockout mutant further reduced the acetate formation, resulting in the increased recombinant protein production [20]. Since TCA cycle is the main source of energy generation and provides important precursors for amino acids such as glutamate, lysine etc., it is of practical interest to enhance the TCA cycle activity. As stated above, the arcA/B genes knockout in E. coli transcriptionally activates the TCA cycle and overproduces NADH, which may in turn repress the TCA cycle by its allosteric regulation. Moreover, it has been reported that ArcAB does not control the TCA cycle under aerobic condition due to the fact that oxidized quinone electron carriers inhibit autophosphorylation of ArcB, and it cannot transphosphorylate ArcA [21]. As expected from the above mentioned regulation, the TCA cycle is activated by arcA/B gene knockout, which then causes higher NADH/ NAD ratio, which in turn represses TCA cycle activity [23]. Vemuri et al. [24,25] considered to express heterologous nox gene to oxidize NADH, and in turn activate TCA cycle, while nicotinic acid and Na nitrate may also activate TCA cycle [26].
Since the TCA cycle is the source of energy generation and provide some of the precursors for the cell synthesis, the activation of the TCA cycle may lead to the improvement of ATP production for the cell growth and/or the TCA cycle-related metabolite productions in practice. Moreover, the activation of TCA cycle reduces the acetate production rate, which is the common obstacle for the metabolite production using E. coli. However, the activation of the TCA cycle causes the decrease in the cell yield due to higher production of CO2 in the TCA cycle.
Respiration is a fundamental cellular process utilizing different terminal electron acceptors such as oxygen and nitrate etc. The ability to sense these electron acceptors is a key for the cells to survive. Escherichia coli is a metabolically versatile chemoheterotroph grown on a variety of substrate under various oxygen concentrations with fumarate or nitrate, replacing oxygen as terminal electron acceptor under anaerobic condition [27]. Many bacteria utilize oxygen as the terminal electron acceptor, but they can switch to other acceptors such as nitrate under oxygen limitation. In E. coli, this switch from aerobic to anaerobic respiration is controlled by Fnr (fumarate and nitrate reduction). Under oxygen limitation, Fnr binds a [4Fe-4S]2+ cluster, and becomes a transcriptionally active dimeric form.
The metabolic regulation is made by the binding of dimeric Fnr to the promoter regions of the relevant genes with affinities depending on the redox state [28]. The ability of Fnr to bind DNA is regulated by the change in equilibrium between monomeric apo Fnr (inactive) and dimeric Fnr (active) in vivo. The active form of Fnr binds to DNA to regulate the corresponding genes under anaerobic condition. Molecular oxygen can oxidize the ionsulfur cluster of the corresponding region, resulting in monomerization of the protein and subsequent loss of its ability to bind DNA [29].
Since Fnr is known to activate frd and pfl genes, the fnr mutant produced less succinate and formate [30]. Although arcA is known to be activated by Fnr, the regulation mechanism is somewhat complicated. Namely, cyo and cyd genes are repressed by Fnr, while cyo is repressed and cyd is activated by ArcA. The fnr mutant shows decreased gene expression of arcA, and increased gene expressions of both cyoA and cydB. This implies that the activated cytochrome oxydase increases quinone pool, which inhibits ArcB phophorylation, and in turn decreases phosphorylation of ArcA, where arcA gene expression also decreases due to fnr gene knockout.
Under anaerobic condition, TCA cycle is the branched pathway patterns, while arcA mutant or nitrate was used as electron accepter, TCA cycle functions as the case under aerobic conditions [31].
3. EFFECT OF A SPECIFIC GENE KNOCKOUT ON THE METABOLISM UNDER ANAEROBIC CONDITION
Here, we consider the metabolic regulation of E. coli cultivated under anaerobic condition in response to the specific pathway gene knockout on the metabolic fluxes as well as other levels of information (Figure 1). Let us first consider the fermentation characteristics and some enzyme activities for pflA, pta, ppc, adhE, and pykF mutants, as compared to the wild type BW25113 [18]. It was shown that the pflA mutant gives much higher activities of GAPDH and LDH, which imply the coupling between the NADH production and consumption between the two corresponding reactions. Similarly to the pflA mutant, the simultaneous up-regulations of GAPDH and LDH are also observed in the pta mutant. Interestingly, Pfl activity is repressed in this mutant, which implies a common regulatory mechanism for these two strains. The significant difference between pta and pflA mutants is the specific activity of Ppc. In the pta mutant, this enzyme activity changed little compared to the parent strain, but thismutant produced more succinate than the pflA mutant, despite lower Ppc activity. This implies that the intracellular pool size of PEP plays another important role in the metabolic regulation, where PEP concentration is higher for ppc mutant, while it is low for pflA mutant [18].
The enzyme activities of Ack and ADH, both of which are the AcCoA assimilation pathway enzymes, increased in the ppc mutant. However, the enzyme activity of Pfl, which supplies AcCoA for these two reactions, is much lower than that of the parent strain, and less acetate and ethanol are formed in the ppc mutant than those of the parent strain. Although the activity of LDH is lower, lactate production is higher in the ppc mutant compared to that of the parent strain. The activity of Pyk in the ppc mutant is about one-quarter of that in the parent strain. Pyk activity in the adhE mutant is also down-regulated. The activities of LDH and Pfl are both down-regulated in the adhE mutant, which is consistent with relatively lower lactate and formate yield in thisstrain. The pykF mutant shows higher Ppc activity, while the activities of LDH and ADH are both down-regulated as compared to the parent strain [18].
The pool sizes of G6P, FDP, and PYR increase, while PEP concentration is lower in the pflA mutant than the wild type strain. The low PEP concentration causes less succinate production, even with high Ppc activity, where this may be caused by the allosteric activation by the increased FDP. The increased FDP concentration also allosterically activated Pyk. The metabolite concentrations in the pta mutant are similar to those in the pflA mutant, except for significantly accumulated PEP. In the case of pykF mutant, all intracellular metabolites in the glycolysis are significantly accumulated, as compared to the parent strain and other mutants. Note that the value of ATP/AXP (ATP + ADP + AMP) is less in the mutants as compared to the parent strain [18].
Among the different levels of information, the metabolic flux distribution is the most important, where it is located on top of the hierarchy of informations [32]. Metabolic flux is defined as the intracellular reaction rate per cell (weight) and per time (mmol∙gDCW−1∙h−1). The information on the metabolic flux distribution is particularly useful in metabolic engineering. Although one of the catabolic pathways is blocked by the specific gene knockout, the higher glucose uptake rate, followed by higher glycolytic flux, may be seen in such strains as pflA, pta, and pykF mutants, as compared to the parent
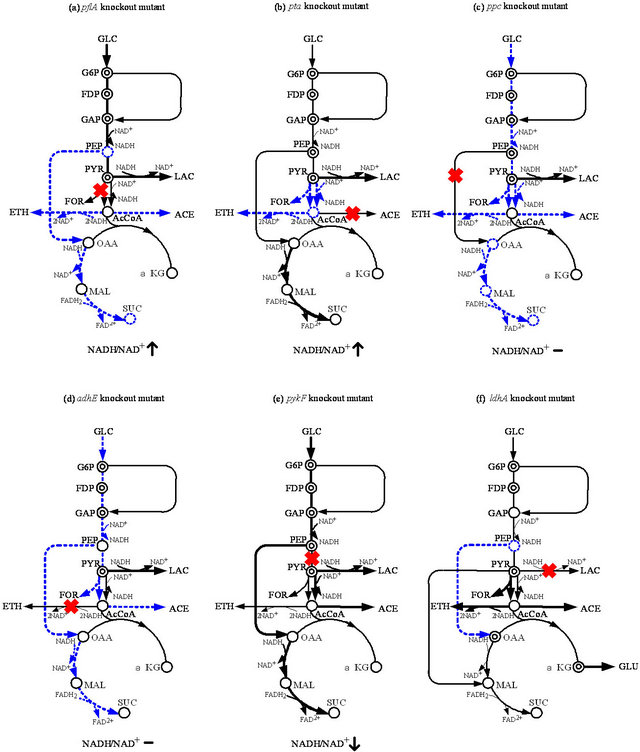
Figure 1. Schematic illustration of the specific pathway gene knockout on the metabolism under anaerobic condition: (a) pflA; (b) pta; (c) ppc; (d) adhE; (e) pykF; and (f) ldhA knockout mutants.
strain [18]. The flux through GAPDH is shown to be regulated by the intracellular NADH/NAD+ ratio [33,34]. It has been shown that NADH competitively combines with GAPDH and inhibits the reaction through this enzyme. The GAPDH activities in the pflA, pta, and pykF mutants were all higher than that of the parent strain. The intracellular NADH/NAD+ ratio was significantly lower in the pykF mutant, and therefore, the higher glycolysis flux is not restricted by the competitive inhibition of NADH on the reaction through relatively high levels of GAPDH. The high glycolytic flux of pflA and pta mutants results in a high intracellular NADH/NAD+ ratio, and subsequently lactate is produced to regenerate NAD+ so that the glycolysis continues to work [18].
The lactate producing flux in the ppc mutant is lower than those of the pflA and pta mutants. However, as compared to the parent strain, the lactate production rate in the ppc mutant is significantly higher. The glycolytic flux in the ppc mutant is about 17% of that in the wild type strain. The flux through Ppc in the pykF mutant was higher than that of the parent strain. From enzyme activity data, Ppc activity in the pykF mutant is up-regulated as compared to the parent strain. The reason for the high Ppc flux may be due to high PEP concentration and synergistic activation by FDP [35,36]. Since the product of the reaction through Ppc is OAA, which is the precursor for biosynthesis, the ppc gene knockout causes low biomass synthesis flux [18].
Pyruvate is competed by the reactions through Pfl and LDH under anaerobic condition. In wild-type E. coli, the LDH reaction is not as competitive as the reaction through Pfl, and therefore, acetate and formate are the main metabolites instead of lactate. The knockout of the pflA gene blocks the pyruvate assimilation through the Pta-Ack and ADH pathways, which are commonly used for ATP production and NADH re-oxidation, respectively, in the wild-type strain. Since the glycolytic flux is promoted by anaerobiosis with a higher ATP requirement, lactate is produced to satisfy both the stoichiometric and intracellular redox balances [17].
Note the higher acetate production in the pykF mutant as compared to the parent strain. The reaction from PEP to OAA through Ppc is the main pathway to replenish OAA in E. coli under oxygen-limited conditions. Succinate is derived from OAA, and two equivalents of NADH are required for 1 mole of succinate production. In PTS (phosphotransferase system), PEP is used to transport phosphate for glucose utilization. The reaction through Pyk also uses PEP to produce PYR and ATP. When the pykF mutant was cultivated under anaerobic condition, the specific glucose uptake rate increased as compared to the parent strain. Flux through glycolysis is reported to be controlled by the ATP requirement in E. coli. By optimizing additional ATP hydrolysis, the glycolytic flux increases significantly [37].
Consider next the effect of the ldhA gene knockout on the metabolism of E. coli under anaerobic conditions. It was shown that that the extracellular formate, acetate, ethanol, pyruvate, and glutamate production rates increased in the ldhA mutant as compared to the parent strain [38]. Intracellular metabolite concentrations of G6P, F6P, 6PG, FBP, PYR, OAA, and α-KG are increased in the ldhA mutant, as compared to those of the parent strain. The enzyme activities of Pfk, GAPDH, Pgk, Pyk, and PDHc were up-regulated significantly in the ldhA mutant as compared to the parent strain. Citrate synthase (CS) was up-regulated in the ldhA mutant compared to the parent strain. This up-regulation caused an increase in the carbon flux from citrate to glutamate via α-KG, as is evidenced by the up-regulation of ICDH, and high concentration of intracellular α-KG in response to ldhA gene deletion [38]. The anaplerotic enzyme Ppc is down-regulated in the ldhA mutant compared to the parent strain. This is mainly due to the shortage of PEP in the ldhA mutant, as evidenced by the measurement of intracellular PEP concentration [38]. The reason for the decrease in PEP concentration is mainly due to the significant up-regulation of Pyk and PDH activities that channel more carbon flux toward the formate, acetate, and ethanol pathways, from the PYR and AcCoA pool, as well as increased NAD+-Mez (Sfc) activity. The activity of NADP+-specific malic enzyme (Mez) remains unchanged, whereas the NAD+-specific malic enzyme (Sfc) is up-regulated significantly in the ldhA mutant compared to that in the parent strain, implying that the inactivation of the ldhA gene leads to the production of malate from pyruvate.
Deletion of the LDH pathway blocks carbon flow from pyruvate and results in a higher pyruvate excretion rate. Metabolic flux analysis shows that most of the carbon flux in ldhA mutant E. coli is forced through formate, acetate, and ethanol production pathways, resulting in a concomitant increase in these fluxes. When PEP is converted to MAL via OAA by Ppc and MDH, the free energy is wasted. In contrast, one ATP is produced when PEP is directly converted to pyruvate by Pyk. Therefore, a large fraction of PEP is channeled through the Pyk pathway and this might be why the ldhA mutant induces the Sfc pathway to supply the C4 intermediates in the TCA cycle for conserving the free energy of PEP, as evidenced by the significant up-regulation of Sfc.
The product of the global regulatory gene cra (fruR) is known to control the transcriptional expressions of numerous genes concerned with carbon and energy metabolism. The genes ptsHI, pfkA, adhE, and pykF are regulated negatively, whereas the genes ppsA, pckA, icdA, and cydAB are regulated positively by Cra (Appendix A). Gene expression analysis shows a slight up-regulations of ptsH, pfkAB, pykF, and adhE genes in the ldhA mutant, compared to those in the parent strain. The reason is due to the higher concentration of FDP in the ldhA mutant, since the effect of the Cra protein on the transcription is counteracted by a high concentration of FDP (Appendix A). The deletion of the ldhA gene may result in a reducing power imbalance in E. coli. However, the overall NADH balance calculated, based on the metabolic flux distribution, indicates that NADH production and consumption are almost the same in both the ldhA mutant and the parent strain. The reason is mainly due to the increased carbon flux through the ethanol pathway in the ldhA mutant from pyruvate node via AcCoA.
Based on the study of gene expressions, enzyme activities, and intracellular metabolite concentrations, together with metabolic flux analysis, the overall regulation mechanisms for E. coli under anaerobic conditions in response to the specific gene deletion are shown in Figure 1.
4. EFFECT OF DIFFERENT CARBON SOURCES ON THE METABOLISM OF PFL MUTANT
Consider next the effects of other carbon sources such as gluconate, pyruvate, fructose, and glycerol on the metabolism of pfl mutant under anaerobic conditions, where pflA,B mutants are useful for the lactate production in practice [17]. A large amount of acetate is produced together with lactate, when gluconate or pyruvate is used as a carbon source. In particular, acetate is the major product when pyruvate is used as a carbon source. The fermentation patterns, when using fructose or glycerol as a carbon source, are similar to those when using glucose. However, fructose and glycerol uptake rates are significantly lower than glucose uptake rates. Compared to the case of using glucose as a carbon source, Ack activity is significantly higher for the case of using gluconate or pyruvate as a carbon source, while this enzyme activity is reduced when using glycerol or fructose as a carbon source. Ppc activity is the highest when using glucose as a carbon source, but it is significantly lower when using fructose or glycerol as a carbon source. It was shown that NADH/NAD+ratio was highest when using glycerol as a carbon source, and this ratio was relatively low when using gluconate or pyruvate as a carbon source [17]. In E. coli, the Pta-Ack pathway is related to the AcCoA pool. Since AcCoA formation through Pfl is deficient in the two pfl mutants, Pta-Ack reactions may occur in the direction of forming AcCoA using acetate and ATP. The significant induction of Ack in the pflA mutant may be related to the activation of PoxB, which produces acetate, and the cell utilizes the Ack-Pta and ACS pathway to supplement the AcCoA pool.
The cell will produce less NADH when using gluconate or pyruvate as a carbon source. This is confirmed by the lower NADH/NAD+ ratio when using gluconate or pyruvate as a carbon source than when using glucose. The activity of GAPDH is significantly higher when using gluconate as a carbon source, which indicates the partial release of NADH inhibition on this enzyme. Correspondingly, the lactate yields, when using gluconate or pyruvate, are lower. The results using gluconate or pyruvate as a carbon source may also be distinguished from those using other carbon sources, because of the significantly lower ATP/AMP ratio in the pflA mutant, which is significantly less than that when using glucose as a carbon source. One reason may be due to the difference in the metabolic networks; when using gluconate as a carbon source, some of the carbon atoms will go directly to PYR through the Entner-Doudoroff pathway, which reduces the flux through glycolysis where ATP is generated.
5. CATABOLITE REGULATION OF WILD TYPE E.COLI UNDER AEROBIC CONDITION
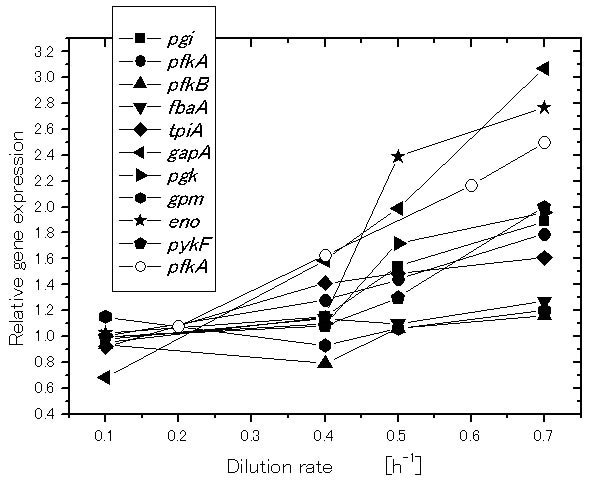
Let us consider how the specific growth rate (dilution rate in the chemostat culture) affects the global regulators and metabolic pathway genes of wild type E.coli (BW25113) [39]. It was shown that the specific acetate production rate, and the specific CO2 evolution rate (CER) increase as the dilution rate was increased [39]. Figure 2 shows the effect of the dilution rate (the specific growth rate) on gene transcript levels, where it indicates that in accordance with the increased specific glucose consumption rate, the transcript levels of ptsG, ptsH, and pfkA are increased as the dilution rate increased, where cra transcript level decreased and crp as well as mlc decreased accordingly. The transcript levels of zwf, gnd, edd, and eda increased as the dilution rate increased. The transcript level of ppc increased while pckA decreased as the dilution rate was increased. Moreover, the transcript levels of fadR and iclR increased, and aceA and aceB decreased as the dilution rate increased [39].
The central players in carbon catabolite regulation in E. coli are the transcriptional activator Crp (cyclic AMP (cAMP) receptor protein; also called as catabolite geneactivator protein (CAP)), the signal metabolite cAMP, adenylate cyclase (Cya), and the PTSs, where these systems are involved in both transport and phosphorylation of carbohydrates. The PTS in E. coli consists of two common cytoplasmic proteins, EI (enzyme I) encoded by ptsI and HPr (histidine-phosphorylatable protein) encoded by ptsH, as well as carbohydrate-specific EII (enzyme II) complexes. The glucose-specific PTS in E. coli consists of the cytoplasmic protein EIIAGlc encoded by crr and the membrane-bound protein EIICBGlc encoded by ptsG, which transport and concomitantly phosphorylate glucose as explained before. The phosphoryl groups are transferred from PEP via successive phosphorelay reactions in turn by EI, HPr, EIIAGlc and EIICBGlc to glucose. The cAMP-Crp complex and the repressor Mlc are involved in the regulation of ptsG gene and pts operon expressions. It has been demonstrated that unphosphorylated EIICBGlc can relieve the expression of ptsG
 (a)
(a)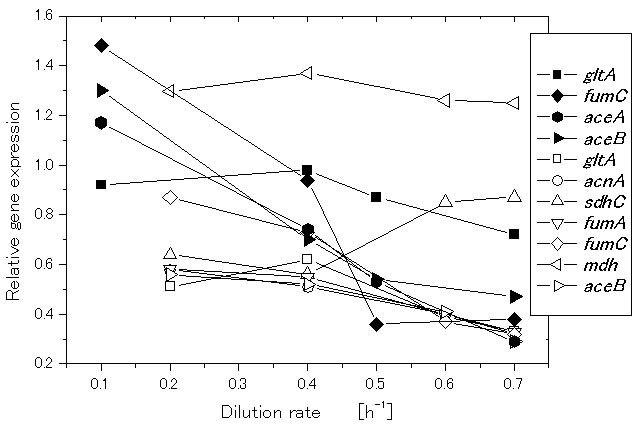 (b)
(b)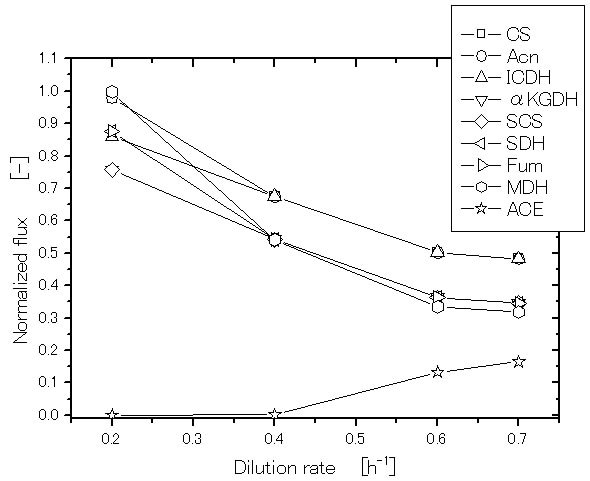 (c)
(c)
Figure 2. Effect of the growth rate (dilution rate) on the glycolysis and TCA cycle gene expressions and fluxes: (a) glycolysis genes; (b) TCA cycle genes; (c) TCA cycle fluxes and acetate formation rate.
gene expression by sequestering Mlc from its binding sites through a direct protein-protein interaction in response to glucose concentration. In contrast to Mlc, where it represses the expressions of ptsG, ptsHI and crr (Appendix A), cAMP-Crp complex activates ptsG gene expression (Appendix A).
Since intracellular cAMP levels are low during growth on glucose, these two antagonistic regulatory mechanisms guarantee a precise adjustments of ptsG expression levels under various conditions [40]. It should be noted that unphosphorylated EIIAGlc inhibits the uptake of other non-PTS carbohydrates by the so-called inducer exclusion [41], while phosphorylated EIIAGlc (EIIAGlc-P) activates adenylate cyclase (Cya), which generates cAMP from ATP and leads to an increase in the intracellular cAMP level [42]. Note that if the concentration ratio between PEP and PYR(PEP/PYR) is high, EIIAGlc is predominantly phosphorylated,whereas if this ratio islow, then EIIAGlc is predominantly dephosphorylated [43,44]. EIIAGlc is preferentially dephosphorylated when E. coli cells grow rapidly with glucose as a carbon source [43,44]. Note also that cAMP levels are low during growth with non-PTS carbohydrates, where PEP/PYR ratio is the key factor that controls phosphorylation of EIIAGlc, which explains dephosphorylation of EIIAGlc, resulting in low cAMP pool [43,44].
In addition to cAMP-Crp, which acts depending on the level of glucose concentration, the catabolite repressor/ activator protein (Cra) originally characterized as the fructose repressor (FruR) plays an important role in the control of carbon flow in E. coli [45-47]. The carbon uptake and glycolysis genes such as ptsHI, pfkA, pykF, zwf and edd-eda are reported to be repressed, while ppsA, fbp, pckA, icdA, aceA and aceB are activated by Cra (Appendix A). The gluconeogenic pathway is deactivated by the knockout of cra gene, and the carbon flow toward catabolism and the glucose consumption rate are expected to increase, since glycolysis pathway genes such as ptsHI, pfkA and pykF are activated by the cra gene knockout. It has been shown that cra gene knockout enables the increase of the glucose consumption rate and thus improve the rate of metabolite production under certain culture conditions [48]. However, the regulation mechanism is complex and it must be careful since icdA, aceA, B, and cydB genes are repressed, while zwf and edd gene expressions are activated, and thus ED pathway is activated by cra gene knockout [48].
6. EFFECT OF THE SPECIFIC PATHWAY GENE KNOCKOUT ON THE METABOLISM UNDER AEROBIC CONDITION
6.1. Effect of pgi Gene Knockout on the Metabolism
If pgi gene was knocked out, the primary route for glucose catabolism is the pentose phosphate (PP) pathway together with Entner Doudoroff (ED) pathway [49-52].
The NADPH is over-produced, which inhibits the activity of G6PDH, and thus the glucose consumption rate becomes significantly low in this mutant [52]. This in turn causes glycolysis fluxes as well as intermediate concentrations of the glycolysis to be decreased [52]. The decreases in FDP and PEP concentrations cause Ppc activity to become lower by allosteric regulation, which causes OAA concentrations to be significantly decreased, which further activated glyoxylate pathway [51-53]. The acetate formation becomes significantly low due to the activation of glyoxylate pathway [52]. The activation of glyoxylate pathway reduces NADPH production at ICDH, where some of the NADPH overproduced may be converted to NADH by transhydrogenase such as Udh [51].
Referring to Figure 3, note that the decreased glucose consumption rate may cause cAMP-Crp to be increasedwhich in turn activates TCA cycle genes (Appendix A). Moreover, the decrease in FDP concentration may cause Pyk and Ppc activities to be decreased by allosteric regulation, and the decreased FDP concentration activates Cra, which in turn represses glycolysis gene expressions, while activates gluconeogenetic pathway gene expressions as well as icdA and aceA/B genes. Those caused the acetate production to be decreased.
6.2. Effect of zwf and gnd Genes Knockout on the Metabolism
In the case of zwf gene knockout, the cell growth phenotype is little affected, where the non-oxidative PP pathway flux is reversed [51,54]. This mutant shows significant overflow metabolism, and some of the NADPH, which cannot be produced at the oxidative PP pathway,
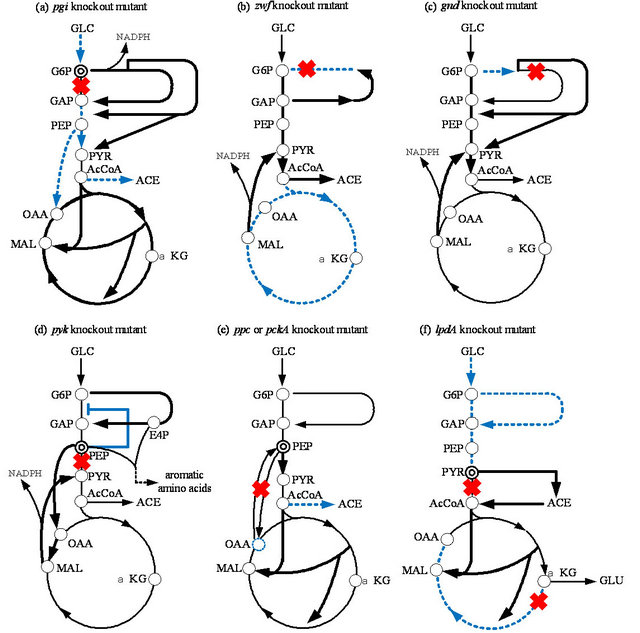
Figure 3. Schematic illustration of the specific pathway gene knockout on the metabolism under aerobic condition: (a) pgi; (b) zwf; (c) gnd; (d) pyk; (e) ppc or pckA; (f) lpdA knockout mutants.
is backed up by activating Mez (Figure 3), and by the transhydrogenase Pnt from NADH [51]. Note that zwf gene knockout causes the increase in glycolysis flux, and thus EIIA-P decreases and in turn cAMP-Crp decreases. This causes TCA cycle to be repressed. Moreover, as glycolysis flux increases, FDP concentration also increases, and in turn represses Cra activity, and thus TCA cycle is repressed by the down regulation of icdA and aceA/B genes. This is the reason why more acetate overflow metabolism was observed as compared to wild type strain [51,54].
The effect of gnd gene knockout on the metabolism is different from zwf gene knockout mutation [55]. The gnd gene knockout activates the ED pathway and causes a decrease in the flux through G6PDH, which reduces NADPH production throughthe oxidative PP pathway. This decreased NADPH production is backedup by activating Mez using MAL together with transhydrogenase Pnt. The conversion of carbon skeletons from the TCA cycle through Mez enables the cells to respond to TCA carbon depletion by regulating the carbon flux through Ppc. This can be shown in the positive correlation between the Mez and Ppc pathways.
Growth characteristics indicate that neither zwf nor gnd is essential. However, further inspection of the metabolic parameters indicates a unique alteration in the utilization of the metabolic pathways for optimal growth of the mutants under different culture conditions. The carbon can be fed into the ED pathway through two routes in E. coli grown on glucose. It is clear that the ED pathway, which is not possessed by S. cerevisiae serves as the route to relieve the toxic level of 6PG in the E. coli mutant grown on glucose. Because of the activation of this potential bypass reaction, the flux via G6PDH is reduced but not blocked. The activity of the oxidative PP pathway is therefore partly maintained. ICDH is important in producing NADPH [56]. Although the shortage of NADPH due to disruption of the oxidative PP pathway can be partially compensated for by an increased flux through ICDH, it appears that the flux via ICDH alone cannot enable the apparent shortage of NADPH to be met under certain circumstances. For this, malate is deviated out of the TCA cycle through Mez to function as the route in which an adequate supply of NADPH is generated to meet the biosynthesis requirements. The drain of carbon skeletons from the TCA cycle through Mez enables the cells to replenish the OAA pool by regulating the carbon flux through Ppc. To increase the synthesis of OAA from PEP, Ppc is therefore up-regulated in accordance with the activity of the Mez pathway. In zwf and gnd mutants, all directions of the fluxes through on oxidative PP pathways are reversed, indicating that the mutant tries to compensate for the lack of E4P and R5P through the glycolytic metabolites GAP and F6P. In thisway, the non-oxidative branch can function as an important metabolic route without the participation of the oxidative PP pathway.
6.3. Effect of ppc and pck Gene Knockout on the Metabolism
The decrease in OAA concentrations due to the pckA gene knockout causes the phosphorylation/inactivation of ICDH, which results in an increase in isocitrate concentration, and the flux through Icl also significantly increases [53]. Moreover, the pckA gene knockout causes accumulation of PEP, which in turn inhibits Pfk activity, and thus reduces the glucose consumption rate. This mutant produces less acetate and CO2, resulting in higher cell yield with less growth rate than the wild type [53]. It is shown that in vivo regulation of the Pck flux occurs mainly by modulation of enzyme activity and by the changes in PEP and OAA concentrations rather than by the ATP/ADP ratio under aerobic condition. This indicates that the reaction catalyzed by Pck can respond flexibly to the availability of PEP and OAA, which may form the metabolic cycle at low glucose concentrations [50]. PEP is an important intermediate in the metabolism, since it alone directly regulates the phosphotransferase (PTS) system but also affects Pfk and Pyk activities. Since PEP can be formed gluconeogenetically through Pck from the TCA cycle, Pck serves to maintain the relative balance between OAA and PEP pools and drain off excess carbon of the TCA cycle to supply PEP for cellular requirements [53].
In relation to the pckA gene knockout, consider the effect of the ppc gene knockout on the metabolism. Among the central metabolic pathway enzymes of E. coli, Ppc plays an anaplerotic role in replenishing OAA consumed in biosynthetic reactions and keeping the TCA cycle intermediates from starvation. Farmer and Liao [57] overexpressed Ppc and/or induced a glyoxylate shunt by the fadR gene knockout [58], to elevate TCA cycle and glyoxylate pathway activities for the reduction of acetate excretion under aerobic conditions, since acetate excretion is a major obstacle in recombinant protein production. The specific cell growth rate and specific glucose consumption rate of the ppc mutant are lower than those of the wild type [59]. The CO2 evolution rate (CER) is also reduced in the ppc mutant. Note that little acetate is excreted in the ppc mutant during cultivation as compared to the wild-type E. coli. As a consequence, an improvement of biomass yield on glucose can be observed in the ppc mutant. The activity of Pck, which catalyzes the reverse reaction to Ppc, is considerably lower in the ppc mutant. The activities of glycolytic enzymes, such as Pgi, Pfk, Fba, and GAPDH and the pentose phosphate (PP) pathway enzymes, such as G6PDH, 6PGDH, and Tal, are all significantly decreased in the ppc mutant [59]. These data correspond to the slower growth and lower glucose utilization rates of the ppc mutant.
The up-regulation of Pyk is expected to channel more carbon flux from PEP to pyruvate due to the blockage of Ppc in the ppc mutant. Accordingly CS, the first enzyme of the TCA cycle, is up-regulated in the ppc mutant [59]. This up-regulation is expected to increase the carbon flux from pyruvate into the TCA cycle for the replenishment of OAA in response to Ppc deficiency. The activities of Acn and MDH, but not ICDH, in the TCA cycle, increase in a co-ordinated manner. Note that Icl, encoded by aceA involved in the glyoxylate shunt, is significantly induced in the ppc mutant [59]. This regulation pattern clearly demonstrates that the ppc mutant utilizes an alternative anaplerotic pathway, the glyoxylate shunt, to replenish OAA in response to the blockage through Ppc. It was shown that the glycolytic intermediates, such as G6P, F6P, F1,6BP (FDP), and PEP, and the PP pathway intermediate, such as 6PG, accumulate in the ppc mutant. In contrast, the intracellular concentration of AcCoA decreases in the ppc mutant, implying higher activity of the TCA cycle (or glyoxylate pathway) relative to glycolysis.
PEP accumulates in the ppc mutant and allosterically inhibits some of the glycolytic enzymes, such as Pgi and Pfk [60]. Inhibition of these enzymes leads to higher intracellular concentrations of their intermediates, such as F6P and F1,6BP (FDP), which in turn affect some other enzymes. For instance, G6PDH is allosterically inhibited by FDP and PRPP and induced by glucose, while 6PGDH is inhibited by FDP, PRPP, GAP, Ru5P, E4P, and NADPH, and induced by gluconate [61]. Apparently, the higher concentrations of intracellular F1,6BP (FDP) in the ppc mutant partially cause down-regulation of both enzymes. Although Cra activity may be repressed by the increased FDP concentration, and icdA and aceA gene expressions may be repressed, PEP/PYR ratio decreased, and the phosphorylated EIIA increased, and in turn cAMP-Crp increased, which may have caused TCA cycle to be activated. The higher flux through the TCA cycle produces more NADPH in the ppc mutant, which may also be considered the reason for the down-regulation of 6PGDH. The transcript of the glucose transport gene, ptsG, is also associated with accumulation of the glycolytic intermediates, such as G6P and F6P, which degrade the mRNA of ptsG by activating the RNaseP enzyme [62]. Both down-regulation of the glycolytic and PP pathway enzymes results in slower growth rates and lower glucose uptake rates in the ppc mutant. The remarkably reduced Pck activity in the ppc mutant may be caused by the higher intracellular concentration of PEP, since Pck is allosterically inhibited by nucleotides ATP and PEP [63]. Note that the activation of the glyoxylate shunt contributes to the reduction in CO2 production in the ppc mutant.
6.4. Effect of pyk Gene Knockout on the Metabolism
Let us consider how the cell regulates upon pyk gene knockout [52,64,65]. The flux through the anaplerotic reaction catalyzed by Ppc in the pykF mutant is high in the pykF mutant as compared to the wild-type strain. It should be noted that the flux through Pck is also high. It can also be seen that the flux through Mez is high for the pykF mutant. Moreover, the glycolytic flux from G6P to F6P is low for the pykF mutant as compared to the wildtype strain, while the flux through the oxidative PP pathway is higher for the pyk mutant as compared to the wild type strain.
In short, the blockage of PEP to the PYR pathway activitivates both Ppc and Mez to supply PYR by forming an alternative pathway, and increases the PEP pool in the mutant, which might inhibit Pfk enzyme activity in the glycolytic pathway. The accumulation of PEP and the activation of the PP pathway (or increased E4P concentration) may activate the aromatic amino acids synthetic pathways [66].
6.5. Effect of lpdA Gene Knockout on the Metabolism
The lipoamide dehydrogenase (LPD) encoded by the lpdA gene is a component of PDHc, KGDH, and glicine cleavage multi-enzyme (GCV) system. This lpdA gene knockout produces more pyruvate and L-glutamate under aerobic conditions than the wild-type strain [67]. It is shown that AcCoA is considered to be formed by the combined rerouting pathways through PoxB, Acs, Ack, and Pta in the lpdA mutant. Metabolic flux analysis of the lpdA gene knockout mutant indicates that the ED pathway and glyoxylate pathway are activated, while the glycolysis and the oxidative PP pathway, as well as the TCA cycle, are down-regulated [67]. If only KGDH was blocked by the sucA gene knockout, metabolism is a little different from the lpdA gene knockout mutant [68]. The regulation of the anaplerotic pathway, such as the glyoxylate shunt, plays a role in these mutants. It was shown that the cell growth of the lpdA mutant is slow compared to its parent strain, and that a fair amount of pyruvate is produced during exponential growth phase of lpdA mutant. The accumulated pyruvate begins to decrease rapidly after glucose exhaustion. Then the cell starts to grow again using pyruvate as another carbon source, and produces acetate, which is subsequently utilized after the pyruvate is consumed. Although cell growth is depressed in the mutant, the final cell concentrations were higher than that in the parent strain.
The activity of the oxidative PP pathway enzyme, such as G6PDH, is up-regulated in the mutant and also the activity of the ED pathway enzymes and the glyoxylate shunt enzyme. It was shown that intracellular PYR concentration was higher in the lpdA mutant as compared to that in the parent strain. In addition, accumulation of L-Glu was higher than in the parent strain. It was shown by the continuous culture that the biomass yield of the lpdA mutant is higher than that of the parent strain, while glucose uptake rates are lower for the mutant and the specific CO2 evolution rate is much reduced in the mutant.
To maintain growth, the cell can generate AcCoA from PYR using another less efficient route by first converting PYR to acetate using PoxB, and then convert acetate to AcCoA via Acs and Ack-Pta. The activation of PoxB can be detected in the PDHc-deficient mutant by in vitro PoxB enzyme assay [69]. It has been shown that poxB gene transcription depends on the rpoS gene product, σ38 (Appendix A). It is assumed that the PYR assimilation in the lpdA mutant might be done via PoxB and Ppc. It has been reported that the double mutants lacking the PDHc and Pps, or the PDHc and Ppc, could not metabolize PYR accumulated during growth on glucose [70]. Thus, the direct utilization of PYR is made by the combining reactions catalyzed by PoxB and Pps-Ppc. The inactivation of KGDH in the lpdA mutant causes the TCA cycle to be interrupted, which may activate the glyoxylate shunt to supply the precursor for cell growth. The low concentration of AcCoA causes down-regulation of CS in the TCA cycle. The synthesis of Glu needs NADPH, which may be supplied by the overproduction of NADPH in the PP pathway due to the upregulated activity of the oxidative PP pathway enzymes.
7. CONCLUSION
In the present article, an attempt was made to clarify the metabolic regulation mechanisms in response to the specific pathway gene knockout. It is shown to be quite important to focus on the roles of transcription factors, which gives insight into the essential metabolic regulation.
REFERENCES
- Blattner, F.R., Plunkett 3rd, G., Bloch, C.A., Perna, N.T. Burland, V., Riley, M., Collado-Vides, J., Glasner, J.D., Rode, C.K., Mayhew, G.F., Gregor, J., Davis, N.W., Kirkpatrick, H.A., Goeden, M.A., Rose, D.J., Mau, B. and Shao, Y. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453-1462. doi:10.1126/science.277.5331.1453
- Ohnishi, M., Tanaka, C., Kuhara, S., Ishii, K., Hattori, M., Kurokawa, K., Yasunaga, T., Makino, K., Shinagawa, H., Murata, T., Nakayama, K., Terawaki, Y. and Hayashi, T. (1999) Chromosome of the enterohemorrhagic Escherichia coli O157:H7; comparative analysis with K-12 MG 1655 revealed the acquisition of a large amount of foreign DNAs. DNA Research, 6, 361-368. doi:10.1093/dnares/6.6.361
- Steed, H., Macfarlane, G.T. and Macfarlane, S. (2008) Prebiotics, synbiotics and inflammatory bowel disease. Molecular Nutrtion and Food Research, 52, 898-905. doi:10.1002/mnfr.200700139
- Madan Babu, M. and Teichmann, S.A. (2003) Evolution of transcription factors and the gene regulatory network in Escherichia coli. Nucleic Acids Research, 31, 1234- 1244. doi:10.1093/nar/gkg210
- Pérez-Rueda, E. and Collado-Vides, J. (2000) The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Research, 28, 1838- 1847. doi:10.1093/nar/28.8.1838
- Gama-Costello, S., Salgado, H., Peralta-Gil, M., SantosZavaleta, A., Muniz-Rascado, L., Solano-Lira, H., JimenezJacinto, V., Weiss, V., Garcia-Sotelo, J.S., Lopez-Fuentes, A., Porron-Sotelo, L., Alquicira-Hernandez, S., MedinaRivera, A., Martinez-Flores, I., Alquicira-Hernandez, K., Martinez-Adame, R., Bonavides-Martinez, C., MirandaRios, J., Huerta, A.M., Mendoza-Vargas, A., ColladoTorres, L., Taboada, B., Vega-Alvarado, L., Olvera, M., Olvera, L., Grande, R., Morett, E. and Collado-Vides, J. (2011) RegulonDB version 7.0: Transcriptional regulation of Escherichia coli K-12 integrated within genetic sensory response units (Gensor Units). Nucleic Acids Research, 39, D98-105. doi:10.1093/nar/gkq1110
- Keseler I.M., Collado-Vides J., Santos-Zavaleta A., PeraltaGil, M., Gama-Castro, S., Muniz-Rascado, L., BonavidesMartinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A.G., Mackie, A., Paulsen, I., Gunsalus, R.P. and Karp, P.D. (2011) EcoCyc: A comprehensive database of Escherichia coli biology. Nucleic Acids Research, 39, D583-D590. doi:10.1093/nar/gkq1143
- Martínez-Antonio, A. (2011) Escherichia coli transcriptional regulatory network. Network Biology, 1, 21-33.
- Shimizu, K. (2012) Bacterial cellular metabolic systems. Woodhead Publishing Ltd., Cambridge.
- Shimizu, K. (2013) Metabolic regulation of a bacterial cell system with emphasis on Escherichia coli metabolism. International Scholarly Research Network Biochemistry, in Press.
- Kang, Y., Weber, K.D., Qiu, Y., Kiley, P.J. and Blattner, F.R. (2005) Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. Journal of Bacteriology, 187, 1135-1160. doi:10.1128/JB.187.3.1135-1160.2005
- Gunsalus, R.P. (1992) Control of electron flow in Escherichia coli: Coordinated transcription of respiratory pathway genes. Journal of Bacteriology, 174, 7069-7074.
- Alexeeva, S., Hellingwerf, K.J. and Teixeira de Mattos, M.J. (2003) Requirement of ArcA for redox regulation in Escherichia coli under microaerobic but not anaerobic or aerobic conditions. Journal of Bacteriology, 185, 204-209. doi:10.1128/JB.185.1.204-209.2003
- Zhu, J., Shalel-Levanon, S., Bennett, G. and San, K.Y. (2006) Effect of the global redox sensing/regulation networks on Escherichia coli and metabolic flux distribution based on C-13 labeling experiments. Metabolic Engineering, 8, 619- 627. doi:10.1016/j.ymben.2006.07.002
- Levanon, S.S., San, K.Y. and Bennett, G.N. (2005) Effect of oxygen on the Escherichia coli ArcA and FNR regulation systems and metabolic responses. Biotechnology and Bioengineering, 89, 556-564. doi:10.1002/bit.20381
- Shalel-Levanon, S., San, K.Y. and Bennett, G.N. (2005) Effect of oxygen, and ArcA and FNR regulators on the expression of genes related to the electron transfer chain and the TCA cycle in Escherichia coli. Metabolic Engineering, 7, 364-374. doi:10.1016/j.ymben.2005.07.001
- Zhu, J. and Shimizu, K. (2004) The effect of pfl genes knockout on the metabolism for optically pure D-lactate production by Escherichia coli. Applied Microbiology and Biotechnology, 64, 367-375. doi:10.1007/s00253-003-1499-9
- Zhu, J. and Shimizu, K. (2005) Effect of a single-gene knockout on the metabolic regulation in E. coli for Dlactate production under microaerobic condition. Metabolic Engineering, 7, 104-115. doi:10.1016/j.ymben.2004.10.004
- Alexeeva, S., de Kort, B., Sawers, G., Hellingwerf, K.J. and de Mattos, M.J. (2000) Effects of limited aeration and of the ArcAB system on intermediary pyruvate catabolism in Escherichia coli. Journal of Bacteriology, 182, 4934-4940. doi:10.1128/JB.182.17.4934-4940.2000
- Vemuri, G.N., Minning, T.A., Altman, E. and Eiteman, M.A. (2005) Physiological response of central metabolism in Escherichia coli to deletion of pyruvate oxidase and introduction of heterologous pyruvate carboxylase. Biotechnology and Bioengineering, 90, 64-76. doi:10.1002/bit.20418
- Georgellis, D., Kwon, O. and Lin, E.C. (2001) Quinones as the redox signal for the arc two-component system of bacteria. Science, 292, 2314-2316. doi:10.1126/science.1059361
- Malpica, R., Franco, B., Rodriquez, C., Kwon, O. and Georgellis, D. (2004) Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proceedings of the National Academy of Sciences of the United States of America, 101, 13318-13323. doi:10.1073/pnas.0403064101
- Nizam, S.A., Zhu, J.F., Ho, P.Y. and Shimizu, K. (2009) Effects of arcA and arcB genes knockout on the metabolism in Escherichia coli under aerobic condition. Biochemical Engineering Journal, 44, 240-250. doi:10.1016/j.bej.2008.12.017
- Vemuri, G.N., Altman, E., Sangurdekar, D.P., Khodursky, A.B. and Eiteman, M.A. (2006) Overflow metabolism in Escherichia coli during steady-state growth: Transcriptional regulation and effect of the redox ratio. Applied and Environmental Microbiology, 72, 3653-3661. doi:10.1128/AEM.72.5.3653-3661.2006
- Vemuri, G.N., Eiteman, M.A. and Altman, E. (2006) Increased recombinant protein production in Escherichia coli strains with overexpressed water-forming NADH oxidase and a deleted ArcA regulatory protein. Biotechnology and Bioengineering, 94, 538-542. doi:10.1002/bit.20853
- Nizam, S.A. and Shimizu, K. (2008) Effects of arcA and arcB genes knockout on the metabolism in Escherichia coli under anaerobic and microaerobic conditions. Biochemical Engineering Journal, 42, 229-236. doi:10.1016/j.bej.2008.06.021
- Crack, J.C., Le Brun, N.E., Thomson, A.J., Green, J. and Jervis, A.J. (2008) Reactions of nitric oxide and oxygen with the regulator of fumarate and nitrate reduction, a global transcriptional regulator, during anaerobic growth of Escherichia coli. Methods in Enzymology, 437, 191- 209. doi:10.1016/S0076-6879(07)37011-0
- Green, J. and Guest, J.R. (1993) Activation of FNR-dependent transcription by iron: An in vitro switch for FNR. FEMS Microbiology Letters, 113, 219-222. doi:10.1111/j.1574-6968.1993.tb06517.x
- Kiley, P.J. and Reznikoff, W.S. (1991) Fnr mutants that activate gene expression in the presence of oxygen. Journal of Bacteriology, 173, 16-22.
- Marzan, L.W., Siddiquee, K.A. and Shimizu, K. (2011) Metabolic regulation of an fnr gene knockout Escherichia coli under oxygen limitation. Bioengineered Bugs, 2, 331-337. doi:10.4161/bbug.2.6.16350
- Toya, Y., Nakahigashi, K., Tomita, M. and Shimizu, K. (2012) Metabolic regulation analysis of wild-type and arcA mutant Escherichia coli under nitrate conditions using different levels of omics data. Molecular BioSystems, 8, 2593-2604. doi:10.1039/c2mb25069a
- Sauer, U. (2006) Metabolic networks in motion: 13Cbased flux analysis. Molecular Systems Biology, 2, 62. doi:10.1038/msb4100109
- Garrigues, C. Loubiere, P., Lindley, N.D. and CocaignBousquet, M. (1997) Control of the shift from homolactic acids to mixed-acid fermentation in Lactococcus lactis: Predominant role of the NADH/NAD+ ratio. Journal of Bacteriology, 179, 5282-5287.
- De Graef, M.R., Alexeeva, S., Snoep, J.L. and Teixeira de Mattos, M.J. (1999) The steady-state internal redox state (NADH/NAD) reflects the external redox state and is correlated with catabolic adaptation in Escherichia coli. Journal of Bacteriology, 181, 2351-2357.
- Smith, T.E., Balasubramanian, K.A. and Beezley, A. (1980) Escherichia coli phosphoenolpyruvate carboxylase. Studies on the mechanism of synergistic activation by nucleotides. The Journal of Biological Chemistry, 255, 1635-1642.
- McAlister, L.E., Evans, E.L. and Smith, T.E. (1981) Properties of a mutant Escherichia coli phosphoenolpyruvate carboxylase deficient in coregulation by intermediary metabolites. Journal of Bacteriology, 146, 200-208.
- Koebmann, B.J., Westerhoff, H.V., Snoep, J.L., Nilsson, D. and Jensen, P.R. (2002) The glycolytic flux in Escherichia coli is controlled by the demand for ATP. Journal of Bacteriology, 184, 3909-3916. doi:10.1128/JB.184.14.3909-3916.2002
- Kabir, M.M., Ho, P.Y. and Shimizu, K. (2005) Effect of ldhA gene deletion on the metabolism of Escherichia coli based on gene expression, enzyme activities intracellular metabolite concentrations, and metabolic flux distribution. Biochemical Engineering Journal, 26, 1-11. doi:10.1016/j.bej.2005.05.010
- Yao, R., Hirose, Y., Sarkar, D., Nakahigashi, K., Ye, Q. and Shimizu, K. (2011) Catabolic regulation analysis of Escherichia coli and its crp, mlc, mgsA, pgi and ptsG mutants. Microbial Cell Factories, 10, 67. doi:10.1186/1475-2859-10-67
- Bettenbrock, K., Fischer, S., Klemling, A., Sauter, F.T. and Gilles, E.D. (2006) A quantitative approach to catabolite repression in Escherichia coli. The Journal of Biological Chemistry, 281, 2578-2584. doi:10.1074/jbc.M508090200
- Gorke, B. and Stulke, J. (2008) Carbon catabolite repression in bacteria: Many ways to make the most out of nutrients. Nature Reviews Microbiology, 6, 613-624. doi:10.1038/nrmicro1932
- Park, Y.H., Lee, B.R., Seok, Y.J. and Peterkofsky, A. (2006) In vitro reconstruction of catabolite repression in Escherichia coli. The Journal of Biological Chemistry, 281, 6448-6454. doi:10.1074/jbc.M512672200
- Bettenbrock, K., Sauter, T., Jahreis, K., Klemling, A., Lengeler, J.W. and Gilles, E.D. (2007) Correlation between growth rates, EIIACrr phosphorylation, and intracellular cyclic AMP levels in Escherichia coli K-12. Journal of Bacteriology, 189, 6891-6900. doi:10.1128/JB.00819-07
- Hogema, B.M., Arents, J.C., Bader, R., Eijkemans, K., Yoshida, H., Takahashi, H., Aiba, H. and Postma, P.W. (1998) Inducer exclusion in Escherichia coli by non-PTS substrates: the role of the PEP to pyruvate ratio in determining the phosphorylation state of enzyme IIAGlc. Molecular Microbiology, 30, 487-498. doi:10.1046/j.1365-2958.1998.01053.x
- Moat, A.G., Foster, J.W. and Spector, M.P. (2002) Microbial physiology. 4th Edition, Wiley-Liss Inc., New York. doi:10.1002/0471223867
- Saier Jr., M.H., Ramseier, T.M. and Reizer, J. (1996) Regulation of carbon utilization. In: Neidhardt, F.C., Curtiss III, R., Ingraham, J.L., Lin, E.C.C., Low, K.B., Magasanik, B., Reznikoff, W.S., Riley, M., Schaechter, M. and Umbarger, H.E. Ed., Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd Edition, ASM Press, Washington DC, 1325-1343.
- Saier Jr., M.H. and Ramseier, T.M. (1996) The catabolite repressor/activator (Cra) protein of enteric bacteria. Journal of Bacteriology, 178, 3411-3417.
- Sarkar, D. and Shimizu, K. (2008) Effect of cra gene knockout together with other genes knockouts on the improvement of substrate consumption rate in Escherichia coli under microaerobic conditions. Biochemical Engineering Journal, 42, 224-228. doi:10.1016/j.bej.2008.06.019
- Canonaco, F., Hess, T.A., Heri, S., Wang, T., Szyperski, T. and Sauer, U. (2001) Metabolic flux response to phosphoglucose isomerase knock-out in Escherichia coli and impact of overexpression of the soluble transhydrogenase UdhA. FEMS Microbiology Letters, 204, 247-252. doi:10.1111/j.1574-6968.2001.tb10892.x
- Fischer, E. and Sauer, U. (2003) Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC-MS. European Journal of Biochemistry, 270, 880-891. doi:10.1046/j.1432-1033.2003.03448.x
- Hua, Q., Yang, C., Baba, T., Mori, H. and Shimizu, K. (2003) Responses of the central carbon metabolism in Escherichia coli to phosphoglucose isomerase and glucose- 6-phosphate dehydrogenase knockouts. Journal of Bacteriology, 185, 7053-7067. doi:10.1128/JB.185.24.7053-7067.2003
- Toya, Y., Ishii, N., Nakahigashi, K., Hirasawa, T., Soga, T., Tomita, M. and Shimizu, K. (2010)13C-metabolic flux analysis for batch culture of Escherichia coli and its pyk and pgi gene knockout mutants based on mass isotopomer distribution of intracellular metabolites. Biotechnology Progress, 26, 975-992. doi:10.1002/btpr.420
- Yang, C., Hua, Q., Baba, T., Mori, H. and Shimizu, K. (2003) Analysis of Escherichia coli anaprelotic metabolism and its regulation mechanisms from the metabolic responses to altered dilution rates and phosphoenolpyruvate carboxykinase knockout. Biotechnology and Bioengineering, 84, 129-144. doi:10.1002/bit.10692
- Zhao, J., Baba, T., Mori, H. and Shimizu, K. (2004) Effect of zwf gene knockout on the metabolism of Escherichia coli grown on glucose or acetate. Metabolic Engineering, 6, 164-174. doi:10.1016/j.ymben.2004.02.004
- Jiao, Z., Baba, T., Mori, H. and Shimizu, K. (2003) Analysis of metabolic and physiological responses to gnd knockout in E. coli by using C-13 tracer experiment and enzyme activity measurement. FEMS Microbiology Letters, 220, 295-301. doi:10.1016/S0378-1097(03)00133-2
- Choi, I.Y., Sup, K.I., Kim, H.J. and Park, J.W. (2003) Thermosensitive phenotype of Escherichia colimutant lacking NADP+-dependent isocitrate dehydrogenase. Redox Report, 8, 51-56. doi:10.1179/135100003125001251
- Farmer, W.R. and Liao, J.C. (1997) Reduction of aerobic acetate production by Escherichia coli. Applied and Environmental Microbiology, 63, 3205-3210.
- Peng, L. and Shimizu, K. (2006) Effect of fadR gene knockout on the metabolism of Escherichia coli based on analyses of protein expressions, enzyme activities and intracellular metabolite concentrations. Enzyme and Microbial Technology, 38, 512-520. doi:10.1016/j.enzmictec.2005.07.019
- Peng, L., Arauzo, M. and Shimizu, K. (2004) Metabolic flux analysis for a ppc mutant Escherichia coli based on 13C-labelling experiments together with enzyme activity assays and intracellular metabolite measurements. FEMS Microbiology Letters, 235, 17-23. doi:10.1111/j.1574-6968.2004.tb09562.x
- [61] Fraenkel, D.G. and Neidhardt, F. (1999) Escherichia coli and Salmonella: Cellular and molecular biology. American Society for Microbiology Press, Washington DC.
- [62] Sugimoto, S. and Shiio, I. (1987) Regulation of 6-phosphogluconate dehydrogenase in Brevibacterium flavum. Agriculture and Biological Chemistry, 51, 1257-1263. doi:10.1271/bbb1961.51.1257
- [63] Morita, T., EI-Kazzaz, W., Tanaka, Y., Inada, T. and Aiba, H. (2003) Accumulation of glucose 6-phosphate or fructose 6-phosphate is responsible for destabilization of glucose transporter mRNA in Escherichia coli. The Journal of Biological Chemistry, 278, 15608-15614. doi:10.1074/jbc.M300177200
- [64] Krebs, A. and Bridger, W.A. (1980) The kinetic properties of phosphoenolpyruvate carboxykinase of Escherichia coli. Canadian Journal of Biochemistry, 58, 309- 318. doi:10.1139/o80-041
- [65] Al Zaid Siddiquee, K., Arauzo-Bravo, M.J. and Shimizu, K. (2004) Metabolic flux analysis of pykF gene knockout Escherichia coli based on 13C-labeled experiment together with measurements of enzyme activities and intracellular metabolite concentrations. Applied Microbiology and Biotechnology, 63, 407-417. doi:10.1007/s00253-003-1357-9
- [66] Siddiquee, K.A.Z., Arauzo-Bravo, M. and Shimizu, K. (2004) Effect of pyruvate kinase (pykF gene) knockout mutation on the control of gene expression and metabolic fluxes in Escherichia coli. FEMS Microbiology Letters, 235, 25-33. doi:10.1016/j.femsle.2004.04.004
- [67] Kedar, P., Colah, R. and Shimizu, K. (2007) Proteomic investigation on the pyk-F gene knockout Escherichia coli for aromatic amino acid production. Enzyme and Microbial Technology, 41, 455-465. doi:10.1016/j.enzmictec.2007.03.018
- [68] Li, M., Yao, S. and Shimizu, K. (2006) Effect of lpdA gene knockout on the metabolism in Escherichia coli based on enzyme activities, intracellular metabolite concentrations and metabolic flux analysis by 13C-labeling experiments. Journal of Biotechnology, 122, 254-266. doi:10.1016/j.jbiotec.2005.09.016
- [69] Li, M., Ho, P.Y., Yao, S. and Shimizu, K. (2006) Effect of sucA or sucC gene knockout on the metabolism in Escherichia coli based on gene expressions, enzyme activities, intracellular metabolite concentrations and metabolic fluxes by 13C-labeling experiments. Biochemical Engineering Journal, 30, 286-296. doi:10.1016/j.bej.2006.05.011
- [70] Abdel-Hamid, A.M., Attwood, M.M. and Guest, J.R. (2001) Pyruvate oxidase contributes to the aerobic growth efficiency of Escherichia coli. Microbiology, 147, 1483- 1498.
- [71] Dietrich, J. and Henning, U. (1970) Regulation of pyruvate dehydrogenase complex synthesis in Escherichia coli K 12. Identification of the inducing metabolite. European Journal of Biochemistry, 14, 258-269. doi:10.1111/j.1432-1033.1970.tb00285.x
APPENDIX A
Regulation of global regulators on the metabolic pathway gene.
| Global regulator | Metabolic pathway gene |
| Cra | +:aceBAK, cydB, fbp, icdA, pckA, pgk, ppsA -:acnB, adhE, eda, edd, pfkA, pykF, zwf |
| Crp | +:acnAB, aceEF, focA, fumA, gltA, malT, manXYZ, mdh, mlc, pckA, pdhR, pflB, pgk, ptsG, sdhCDAB, sucABCD, ugpABCEQ -:cyaA, lpdA, rpoS |
| ArcA/B | +:cydAB, focA, pflB -:aceBAK, aceEF, acnAB, cyoABCDE, fumAC, glpD, gltA, icdA, lctPRD, lpdA, mdh, nuoABCDEFGHIJKLMN, pdhR, sdhCDAB, sodA, sucABCD |
| IclR | -:aceBAK, acs |
| FadR | +:iclR |
| Mlc | -:crr, manXYZ, malT, ptsG, ptsHI |
| PdhR | -:aceEF, lpdA |
| Fnr | +:acs, focA, frdABCD, pflB, yfiD -:acnA, cyoABCDE, cydAB, fumA, fnr, icdA, ndh, nuoA-N, sdhCDAB, sucABCD |
| RpoS | +:acnA, acs, adhE, fumC, gadAB, talA, tktB, poxB, asmC -:ompF |
NOTES
*Corresponding author.

