Advances in Bioscience and Biotechnology
Vol.3 No.3(2012), Article ID:20002,7 pages DOI:10.4236/abb.2012.33032
Ribosomal proteins expression and phylogeny in alpaca (Lama pacos) skin*
![]()
College of Animal Science and Veterinary Medicine, Shanxi Agricultural University, Taigu, China
Email: #cs_dong@sxau.edu.cn
Received 3 February 2012; revised 8 March 2012; accepted 24 April 2012
Keywords: Ribosomal Protein; Expression; Phylogenetic Tree; Alpaca
ABSTRACT
Ribosomal proteins (RP) has been reported as a central player in the translation system, and are required for the growth and maintenance of all cell kinds. RP genes form a family of homologous proteins that express in the stable pattern and were used for phylogenetic analysis. Here we constructed a cDNA library of alpaca skin and 13,800 clones were sequenced. In the cDNA library, RP genes from skin cDNA library of alpaca were identified. Then 8 RP genes were selected at random and built the phylogenetic trees from the DNA sequences by using parsimony or maximum likelihood methods for molecular and evolutionary analysis of ribosomal proteins. The results showed that the 42 RP genes of alpaca have been expressed in alpaca skin. They were highly conserved. The phylogeny inferred from all these methods suggested that the evolutionary distances of alpaca RP genes were closer to rat.
1. INTRODUCTION
The ribosome has been reported as a central player in the translation system, which consists of four RNA species and 79 ribosomal proteins (RPs) in mammals. Its function is to decode the nucleotide sequence carried by the mRNA and convert it into an amino acid primary structure by the catalysis of peptide bonds [1]. In eukaryotes, ribosomes consist of two different subunits: a 40S small subunit and a 60S large subunit. In mammals, the 40S subunit contains 33 different proteins and an 18S rRNA, whereas the 60S subunit is composed of 49 unique polypeptides and three rRNAs: a 5S, a 5.8S, and a 28S [2]. Ribosomal proteins are thought to have mainly a scaffolding/chaperone role in facilitating the processing and folding of rRNA during biogenesis and stabilizing the mature particle during protein synthesis [3]. It has been well established that global regulation of protein synthesis in eukaryotes is mainly achieved by posttranslational modification (PTM) of translation factors in response to environmental cues [4].
The family of South American camelids with four members is recognized as two wild species, the guanaco (Lama guanicoe) and the vicuna (Vicugna vicugna) and two of domestic species, the alpaca (Lama pacos) and the llama [5]. Because all potential ancestral forms are extant, South American camelids domestication represents an unusual and useful opportunity to gain insight into the origin and biodiversity of domesticated animals, an issue which is of increasing interest due to the recognized potential economic benefits of indigenous genetic resources and the threats that face marginal and extensive agricultural today (Hall & Bradley, 1995). The molecular evolutionary analysis of the family Camelidae by analyzing the full DNA sequence of the mitochondrial cytochrome b gene was reported. Estimates for the time of divergence of the Old World (Camelini) and New World (Lamini) tribes obtained from sequence data are in agreement with those derived from the fossil record. The DNA sequence data were also used to test current hypotheses concerning the ancestors of the domesticated llama and alpaca. The results showed that hybridization has occurred in the ancestry of both domesticated camelids, obscuring the origin of the domestic species (Helen et al., 1994). The evolutionary origins of South America’s domestic alpaca and llama remain controversial [5] (Miranda Kadwell et al., 2001) due to hybridization, near extirpation during the Spanish conquest and difficulties in archaeological interpretation. At present, although alpaca and llama rearing is a central element of the economy in the high Ands, it is often not profitable due to the poor quality of the animals and their fibre. The reconstruction of fine-fibre breeds and the breeding strategies needed are therefore uniquely dependent upon the contributions of archacozoology and genetic analysis [5].
The relationship between domestic alpaca and other mammalian species has been reported. The complete mitochondrial (mt) genome of an odontocete and the sperm whale (Physeter macrocephalus) were sequenced and included it in phylogenetic analyses together with the previously sequenced complete mtDNAs of two mysticetes (the fin and blue whales) and a number of other mammals, including five artiodactyls (the hippo-potamus, cow, sheep, alpaca, and pig). The most strongly supported cetartiodactyl relationship was: outgroup ((pig, alpaca), ((cow, sheep), (hippopotamus, (sperm whale, (baleen whales). As in previous analyses of complete mtDNAs, the sister-group relationship between the hippopotamus and the whales received strong support, making both Artiodactyla and Suiformes (pigs, peccaries and hippopotamuses) paraphyletic. In addition, the analyses identified a sister-group relationship between Suina (the pig) and Tylopoda (the alpaca), although this relationship was not strongly supported [6]. Ribosomal RNA (rRNA) genes commonly found in eukaryotic and prokaryotes, they are more conservation and have a constant evolution rate variability during the process of evolution. So ribosomal RNA (rRNA) genes is a useful molecular marker for studying organic evolution [7]. Here, we report 8 RP genes (selected randomly) sequences and a phylogenetic analysis of alpaca to find the relationship between alpaca and other animals.
2. MATERIAL AND METHODS
2.1. Animals
The alpacas used in these experiments was maintained at the Shanxi alpaca farm of Shanxi Agricultural University. All animal experiments were performed according to the protocols approved by the institutional committee for use and care of animals.
2.2. cDNA Library Production
The total RNA is extracted by Trizol reagent (Stratagene) and mRNA is isolated by Oligotex (Qiagen). The first strand cDNA is produced at 42˚C in 10 µL reaction with 1 µL PowerScript reverse transcriptase. The second and double strands cDNA(ds cDNA) are produced by LD PCR with 5’PCR primer and CDSIII primer in 100 µL reaction for 24 cycles (95˚C 5 s; 68˚C 6 min). Following the digestion by proteinase K and Sfi, ds cDNA are isolated by CHROMA APIN-400 in the molecular weight order and collected cDNA together with the aimed size. The ds cDNA is ligated with λTriplEx2 vector in the ligation reaction at 16˚C and then the ligation production was packaged in λ-phage (Gigapack III plus packaging extract, Stratagene) at 22˚C.
2.3. Sequencing
The package production was transferred into the XL1- Blue at 37˚C for a night with X-gal and IPTG. 17,400 white clones were picked at random and then were converted from λTriplEx2 to ρTriplEx2 in E. coli BM25.8 at 31˚C for circularization of a complete plasmid from the recombinant phage. The clone of circularization production were sequenced in both directions with T7 and 5’special primer.
2.4. Assembly of the Sequences of Targeted Genes
The sequences were assembled by the DNAMAN software and the consensus sequences will be used in the next phylogentic analysis.
2.5. Sequence Analysis
Using alpaca sequences as queries, search for RP sequences was performed in database accessible with the Basic Local Alignment Search Tool (BLAST) on the server of the National Center for Biotechnology Information (NCBI). The obtained nucleotide sequences were loaded into ORF on NCBI, translated into amino-acid sequences and aligned with CLUSTALW in DNAMAN.
2.6. Computer Sequences and Phylogenetic Analysis
The determined nucleotide and amino acid sequences were analyzed using BLAST program search of GenBank for homology with known sequences. The sequence data herein have been submitted to GenBank and the accession numbers assigned in phylogenetic analysis were presented in Table 1. Phylogenetic analysis was performed using CLUSTAL X program, the transition/ transversion rates were calculated using PUZZLE 4.0.2 program. Bootstrappimg values were calculated using the modules SEQBOOT (random number seed: 13; 100 replicates). PROTDIST (distance estimation maximum likelihood; analysis of 100 data sets). NEIGHBOR (Neighbor joining and UPGMA method; random number seed: 13; analysis of 100 data sets) and CONSENSE from the PHYLIP package version 3.65. TREEVIEW version 1.6.0 was used for visualization of the trees.
3. RESULTS
3.1. Characteristerization of RP Genes
We got 7286 ESTs from 13,800 clones which have been deposited into NCBI (GenBank name: ASCD). In the cDNA library, 42 RP genes from skin cDNA library of alpaca were identified (Table 1). Animals possess three classes of acidic ribosomal P proteins: RPLP0, RPLP1


Table 1. Members of RP from the cDNA library of alpaca skin and accession number.
and RPLP2. Interestingly, RPLP1 and RPLP2 have their own specific characteristics on both expression profiling and amino acid composition by our analyses. In our expression profile, RPLP1 and RPLP2 were highly co-expressed in LND and keratinocytes, forming a sub-cluster. As only RPLP1 and RPLP2 form dimers in the silkworm, they may have gene expression machinery different from those of the other RP genes. This may indicate that RPLP0 is a specific gene not only for P proteins but also for the RP gene family. On the other hand, because all three P protein genes belonged to the main cluster in the study of synonymous codon composition, evolutionarily they might have been affected by selective pressure on codon usage along with other RP genes. From these results, we conclude that RPLP0, RPLP1, and RPLP2 are unique and specific genes compared with the major RP genes, but that these P protein genes are members of the RP gene family. Conserved regions with lengths of over 100 bp were found in regions upstream of the TSS in the following RPgenes: RPS2, RPS4X, RPS7, RPS10, RPS12, RPS14, RPS18, RPS23, RPS27A, RPS30, RPL6, RPL7, RPL10, RPL15, RPL17, RPL18, RPL19, RPL21, RPL22, RPL26, RPL27A, RPL32, RPL35, RPL35A, RPL36A, RPL40, and RPLP1. Most importantly, 14 RP genes were found to have conserved upstream regions of over 100 bp adjacent to the TSS. Conserved intronic regions with lengths of over 100 bp were found in RPS3, RPS6, RPS8, RPS19, RPS27, RPL7, RPL22, RPL23A, and RPL30.
Among those RP genes, there were 15 genes which have the complete ORF and we selected 9 members at random to be the subject for this study. All animals with those genes were searched basing on BLAST (Table 2). And we found that some RP genes have the another RP
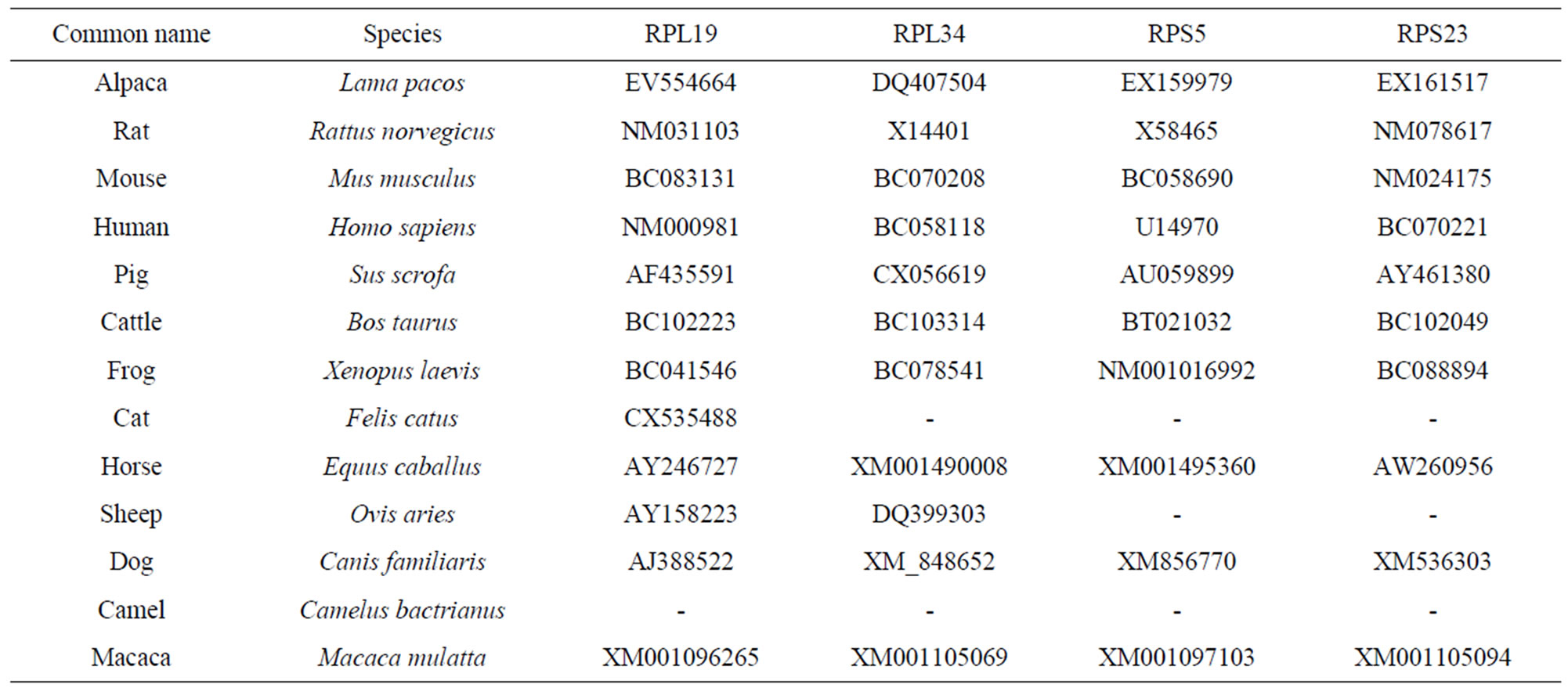
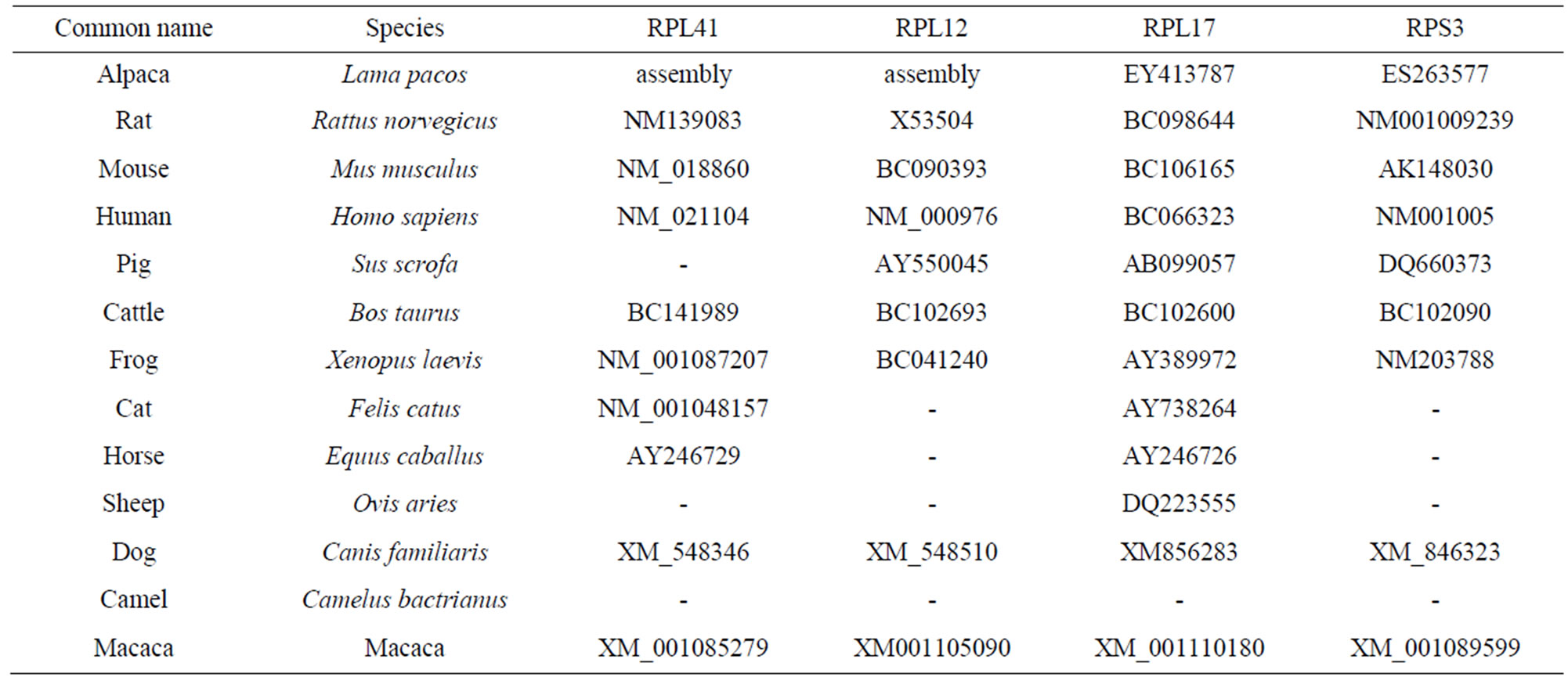
Table 2. Accession numbers of nucleotide and amino acid sequences of RP for alignment and phylogenetic analysis.
genes domain, which is RPL12 with RPL11 domain, RPS11 with RPS17, RPS5 with S7, RPS12 with RPL7, RPL23a with RPS15p, RPL17 (L23) with RP1M, RPS9 with RPS4, RPS2 with RPS5, RPS29 with RPSN, RPS14 with RPSK.
These genes formed 3 main clusters. One cluster contained RPS5, RPL34 and so on. One cluster contained RPL13 and RPS19. The another cluster contained RPL18. In the first cluster, RPP2 and RPLP2 formed a sub-cluster. These genes were named according to their similarity to mammalian RP genes.
3.2. The Homology of RP Genes of Alpaca with Other Animals
Reference gene sequences were obtained from NCBI. Alignments were made with the program DNAMAN. The homology of 9 RP between alpaca and other animals was over 82% at the lever of nucleotide (Table 3). Of particular interest was the fact that RPL12 have the conserved RPL11 domain, RPS11 have RPS17 domain, RPS5 have RPS7 domain, RPL17 have RP1M domain, which suggested that RPL12, RPS11, RPS5 and RPL17 may have the similar function to RPL11, RPS17, RPS7 and RP1M, respectively.
3.3. The Phylogenetic Analysis of Alpaca Comparing with Other Animals Collected in GenBank
Phylogenetic analysis were conducted on the basis of amino acid sequences of ribosomal protein genes collected in this study and sequences from the GenBank for several other animals which could be classified into 5 major groups of Rodentia (mouse and rat), Primate (human and macaca), Perissodactyla (horse), Artiodactyla
(cattle, sheep, pig, camel) and Carnivora (dog and cat). Additionally, we collected the sequences of frog to be aligned with these sequences. The alignments of these sequences with the homologous regions. The phylogenetic analysis of the sequences was carried out with the program package PHYLIP 3.65. The phylogenetic unrooted trees were constructed with maximum-likelihood (ML) and neighbor-joining (NJ) method. The outgroup special was not set and the multiple dataset was set 100. Because of the limited the species in GenBank, we tried to collect the representatives of every group. The trees of these RP with ML and NJ exhibited the same results which indicated that alpaca clustered together with rat and were more closely to rat on the evolutionary tree, since no RP genes were found in the camel and camelids (Figure 1).
4. DISCUSSION
RPS5 is highly conserved in all life forms [8,9]. Of particular interest was the fact that RPL12 have the conserved RPL11 domain, RPS11 have RPS17 domain, RPS5 have RPS7 domain, RPL17 have RP1M domain, which suggested that RPL12, RPS11, RPS5 and RPL17 may have the similar function to RPL11, RPS17, RPS7 and RP1M, respectively. RPL34, RPS5, RPL12, RPL17 and RPS3 of alpaca have the highly conserved regions with that of other counterpart species which may have an equally important translation role in those species and be required for the retention of activity of RP proteins. In past studies, the control mechanisms of gene expression and RP functions were believed to be identical [10]. This highly rate of sequence conservation among difference orders could be due to the highly selective pressure necessitated by the fundamental role of RP in translation function. We
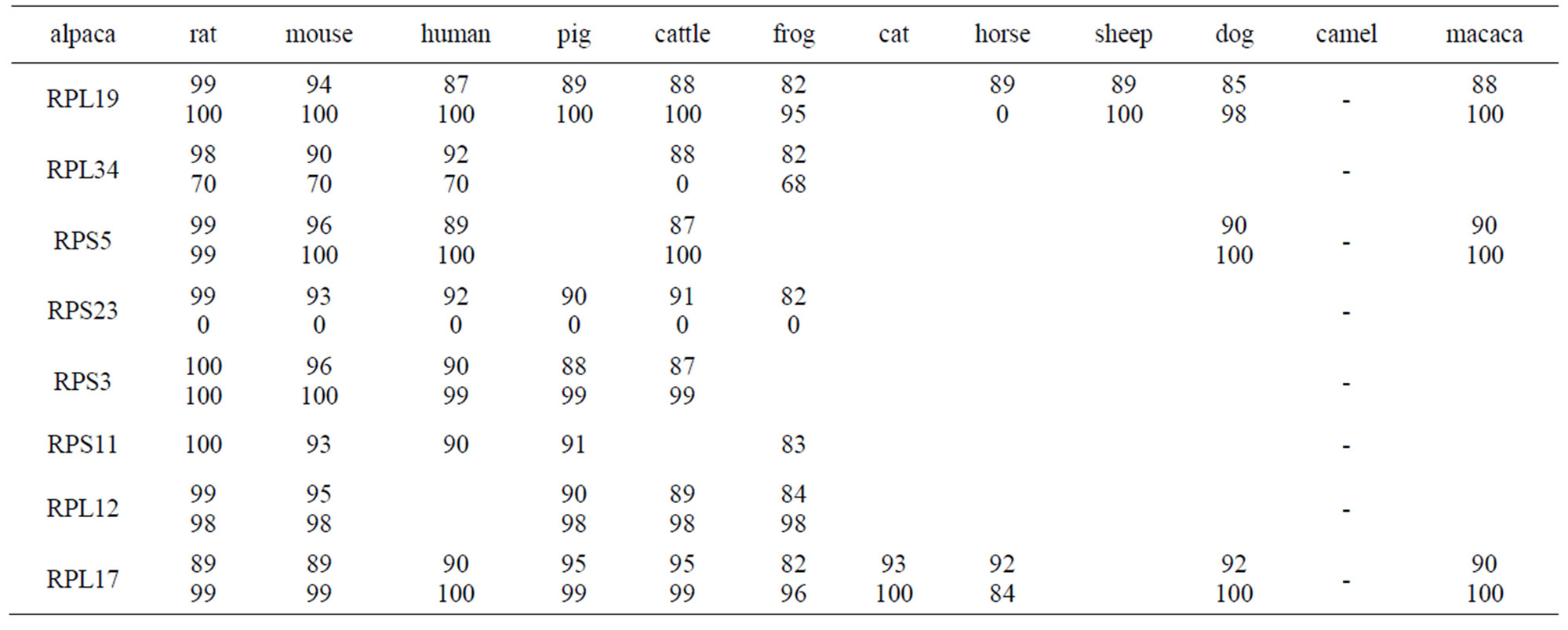
Table 3. Identity of nucleotide and amino acid sequences of RP for alignment and phylogenetic analysis % nucleotide (amino acid) homology.
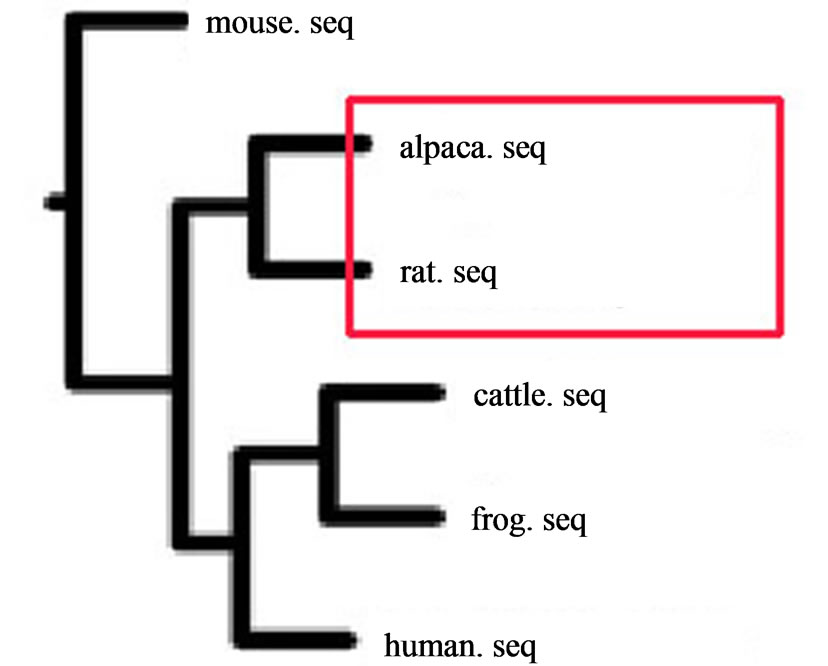
Figure 1. Phylogenetic analysis based on 8 RP amino acid sequences in Table 3 with NJ and ML produced an evolutionary tree showing the relationship between alpaca and other species. Reerence sequences are from GenBank and accession numbers are in Table 2.
found that RP genes were the highly conserved and stable characters that decides that RP can be used in the phylogenetic analysis. It has become clear that in S. cerevisiae the transcription of ribosomal protein genes, which makes up a major proportion of the total transcription by RNA polymerase II, is controlled by the interaction of three transcription factors, Rap1, Fhl1, and Ifh1 [11]. The RP genes that have characteristically poor TATA boxes [12]. The ATG initiation codon of two thirds genes is present in a C(G)ATG sequence. There also is a marked preference for a C or G before the initiation, while in chloroplasts the preference for a T before the initiation codon. We can speculate they represent an evolutionarily acquirement.
Analysis of agnathans will be necessary to determine the timing of emergence of RP relative to. The differences to those trees obtained with ribonuclease protein sequences can be explained by the influence of convergence of pancreatic RNases from hippopotamus, camel, and ruminants and by taking into account the information from third codon positions in the DNA based analyses. The evolution of sequence features of ribonucleases such as the distribution of positively charged amino acids and of potential glycosylation sites is described with regard to increased double-stranded RNA cleavage that is observed in several cetacean and artiodactyls RNases which may have no role in ruminant or ruminant-like digestion protein synthesis takes place at the ribosome, a ribonucleoprotein complex divided into two subunits that in total contains one third protein and two thirds RNA.
According to the arrangement of these genes of E. coli, RPS10 are followed by RPL3, RPL4, RPL23, RPL2, RPS19, RPL22, RPS3, RPL16, RPL29, RPS17 and these genes are separated by some bases spacer, but because of no genome of alpaca, it is a pity that we can not arrange those genes on genome. However, RP genes’ high conservation may decide these genes’ location like that of E. coli. As to the absent genes, they might not exit at the expected position on the genome, therefore, they are not always functional in alpaca skin, as some of them demonstrated in spinach. Another, they might exit and are functional, which had less copies and were not found.
The phylogenetic position of the alpaca became more interesting. Commonly used mitochondrial rDNA is one of the best molecular marker to resolve closer kinship between the species [13]. However, it is difficult to objectively phylogenetic analysis when only used a few genes as molecular marker because of the existence evolutionary rate difference between different genes. Therefore, it is necessary to find new molecular marker.
In our study, there was at least one species of ruminantia and suiform used as representatives to be aligned. Alpaca is monophyletic based on the phylogenetic tree of RPL17 and divergent closer to pig which appear to support the relationship with pig (suiform). As to the relationship of them, the consensus trees constructed with NJ and ML exhibited the same topology. Pig was the first divergent member of artiodactyla and alpaca can not really cluster into a group with pig in this tree because the divergence of the pig occurred before the gene duplication event that happened in an ancestor. In the analysis of 8 RP deduced amino acid sequences constructing phylogenetic trees, none can support the relationship with ruminantia. Therefore, using RP genes to support the close relationship between tylopoda, suiforms and ruminantia was not complete.
The placenta is essential for the success of therian mammalian reproduction. Intense selective pressure has shaped changes in placental anatomy and function during mammalian cladogenesis. Among placentals, rodents with their great fossile and extant diversities appear as a model group to understand the variance between dates derived from fossils and sequences. Molecular studies usually make the palaeontological dates for the origin of rodent clades older than about 25 - 55 million years [14]. These species were shown to have faster rates of sequence evolution [15] and it is known that contrasted substitution rates can severely affect divergence estimate. Rodents are also the most diversified mammals [16]. Using phylogenetic and statistical analyses of molecular and morphological data, it is demonstrated that the ancestral eutherian mammalian placenta had the distinctive features of 1) hemochorial placental interface, 2) a discoid shape, and 3) a labyrinthine maternofetal interdigitation. Alpaca and rat or mouse share these common features. These results reveal that the first eutherians had a deeply invasive placenta existed throughout the eutherian lineage that descended from the last common ancestor of placental mammals. The ancestral state reconstructions demonstrate both clade-specific patterns of placentation and specific cases of convergent evolution within individual eutherian clades. In our study, of interest was that alpaca and Rodent were clustered into a group in RP genes phylogenetic trees. The high conservation of 8 RP genes from prokaryotes to eukaryotes suggested that this protein might have been subjected to a strong selective pressure during evolution, and its role might have a substantial biological meaning from prokaryotes to eukaryotes [17]. It is because of gene duplication of RP nearly at the same time and both of them face the approximate same evolutionary pressures. These data, therefore, point towards a very close genetic affinity between the alpaca and rodent. The implications of these data are potentially important for the way in which these genetic resources are managed in the future.
Like other genes of a gene family at some stage of evolution, RP genes have arisen by gene duplication. The evolution of genes belonging to the same gene family does not occur at random, but is controlled by a mechanism which is termed gene conversion resulting in concerted evolution of the genes. This meant that if a mutation occurs in one of the genes, it can be repaired by homologous recombination with the other genes [18].
5. CONCLUSION
The goal of this study was to add both more molecular data and analytical rigor to the phylogenetic study of basal relationship of alpaca and ruminant and pig. To the best of our knowledge, this is the first report describing the alpaca evolution by the RP genes. While it pointed towards a very close genetic affinity between the alpaca and rodents. The implications of these data are potentially important for the way in which these genetic resources are managed in the future.
REFERENCES
- Marshall, R.A., Aitken, C.E., Dorywalska, M. and Puglisi, J.D. (2008) Translation at the single-molecule level. Annual Review of Biochemistry, 77, 177-203.
- Wool, I.G., Chan, Y.L. and Gluck, A. (1995) Structure and evolution of mammalian ribosomal proteins. Biochemistry and Cell Biology, 73, 933-947. doi:10.1139/o95-101
- Dresios, J., Panopoulos, P. and Synetos, D. (2006) Eukaryotic ribosomal proteins lacking a eubacterial counterpart: Important players in ribosomal function. Molecular Microbiology, 59, 1651-1663. doi:10.1111/j.1365-2958.2006.05054.x
- Gebauer, F. and Hentze, M.W. (2004) Molecular mechanisms of translational control. Nature Reviews Molecular Cell Biology, 5, 827-835. doi:10.1038/nrm1488
- Wheeler, J.C. (1995) Evolution and present situation of the South American Camelidae. Biological Journal of the Linnean Society, 54, 271-295. doi:10.1016/0024-4066(95)90021-7
- Björn M.U., Kerryn, E.S. and Ulfur A. (2000) Subordinal artiodactyl relationships in the light of phylogenetic analysis of 12 mitochondrial protein-coding genes. Zoologica Scripta, 29, 83-88. doi:10.1046/j.1463-6409.2000.00037.x
- Tauta, D., Hancock, J.M., Webb, D.A., et al. (1988) Complete sequences of the rRNA genes of Drosophila melanogaster. Molecular Biology and Evolution, 5, 366- 376.
- Ramakrishnan, V. and Stephen, W.W. (1992) The structure of ribosomal protein S5 reveals sites of interaction with 16S rRNA. Nature, 358, 768-771. doi:10.1038/358768a0
- Kadwell, M., Fernandez, M., Stanley, H.F., Baldi, R., Wheeler, J.C., Rosadio, R. and Bruford, M.W. (2001) Genetic analysis reveals the wild ancestors of the llama and the alpaca. Proceedings of the Royal Society B: Biological Sciences, 268, 2575-2584. doi:10.1098/rspb.2001.1774
- Xu, X., Janke, A. and Arnason, U. (1996) The complete mitochondrial DNA sequence of the Greater Indian Rhinoceros, Rhinoceros unicornis, and the phylogenetic relationship among Carnivora, Perissodactyla and Artiodactyla (+Cetacea). Molecular Biology Evolution, 13, 1167- 1173. doi:10.1093/oxfordjournals.molbev.a025681
- Rudra, D., Mallick, J., Zhao, Y. and Warner, J.R. (2007) Potential interface between ribosomal protein production and pre-rRNA. Processing of Molecular Cell Biology, 27, 4815-4824. doi:10.1128/MCB.02062-06
- Mencia, M., Moqtaderi, Z., Geisberg, J.V., Kuras, L. and Struhl, K. (2002) Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Molecular Cell, 9, 823-833. doi:10.1016/S1097-2765(02)00490-2
- Ortí, G. and Meyer, A. (1997) The radiation of characiform fishes and the limits of resolution of mitochondrial ribosomal DNA sequences. Systems Biology, 46, 75-100. doi:10.1093/sysbio/46.1.75
- Kumar, S. and Hedges, B. (1998) A molecular timescale for vertebrate evolution. Nature, 392, 917-920. doi:10.1038/31927
- Huchon, D., Catzeflis, F.M. and Douzery, E.J.P. (1999) Molecular evolution of the nuclear von willebrand factor gene in mammals and the phylogeny of rodents. Molecular Biology and Evolution, 16, 577-589. doi:10.1093/oxfordjournals.molbev.a026140
- Wheeler, J.C., Russel, A.J.F. and Redden, H. (1995) Llamas and alpacas: Pre-conquest breeds and post-conquest hybrids. Journal of Archaeological Science, 22, 833-840. doi:10.1016/0305-4403(95)90012-8
- Vaccaro, A., Patel, T., Fischgrund, J., Anderson, D.G., Truumees, E., Herkowitz, H., Phillips, F., Hilibrand, A. and Albert, T.J. (2003) A pilot safety and efficacy study of OP-1 putty (rh BMP-7) as an adjunct to iliac crest autograft in posterolateral lumbar fusions. European Spine Journal, 12, 495-500. doi:10.1007/s00586-003-0561-8
- Shmuel, R., Yogev, D. and Naot, Y. (1998) Molecular biology and pathogenicity of mycoplasmas. Microbiology and Molecular Biology Reviews, 62, 1094-1156.
NOTES
*This work was supported by a grant from the National Natural Science Foundation of China (No. 30671512), and by Shanxi Province Science Foundation for Youths (No. 2008021038-3).
#Corresponding author.

