Advances in Bioscience and Biotechnology
Vol.2 No.4(2011), Article ID:6861,4 pages DOI:10.4236/abb.2011.24035
Regulation of glucose and protein syntheses by Micrococcus luteus during the fermentation of a Nigerian rice, Oryza sativa variety “Igbimo”
![]()
Department of Microbiology, Federal University of Technology, Akure, Nigeria.
Email: *boboye_b@yahoo.com
Received 28 April 2011; revised 17 June 2011; accepted 20 July 2011.
Keywords: Regulation; Glucose; Protein; Micrococcus luteus; Rice Fermentation
ABSTRACT
Micrococcus luteus synthesises glucose and protein during the fermentation of a Nigerian rice. To regulate the formation of these substances, mutation was carried out with an alkylating agent: ethylmethyl sulphonate (EMS). Screening the mutants generated for the levels of the traits expressed, four major groups were obtained. These are poor, moderate, good and super producers of either glucose or protein. They produced the properties at 0 - 1.00 (poor), 1.01 to 1.99 (moderate) in 2.0 to 2.99 (good) and 3.0 and above (super) mg·mL–1 of each substance. The classes were made up of 37, 40, 20 and 3 mutants for glucose production and 13, 35, 40 and 12 mutants for protein synthesis. The wild strain bacterium made 0.86 mg·mL–1 glucose and 1.2 mg·mL–1 protein describing the M. luteus as poor glucose maker and moderate protein producer. It was also noticed that the mutation caused some variants (25%) to form more glucose than protein; the remaining 75% of the population are made up of two sets viz: mutants having better ability to synthesise protein at higher concentrations than glucose and those that formed about the same amounts of the substances. It thus follows that the glucose and protein productions in M. luteus are genetically based and can be regulated by genetic manipulation.
1. INTRODUCTION
Rice belonging to the genus Oryza has more than 20 different species out of which very few are cultivated. The cultivated ones include O. sativa and O. glaborrima. China, India and Indonesia are the most important rice producing countries in the world [1]. In the sub-Saharan Africa, West Africa leads the producers and consumers of rice. West Africa accounts for 64.2% and 61.9% of total rice production and consumption respectively in sub-Saharan Africa [2,3]. Nigeria ranked highest as both producer and consumer of rice in the sub-Saharan Africa. More than 50% of rice produced in this region is from Nigeria [2].
Rice is one of the most staple foods consumed by many people in the world. Except for Burkina Faso and Niger, rice is a staple crop throughout West Africa especially in Côte d’Ivoire, Gambia, Guinea Bissau, Liberia, Senegal and Sierra Leone [4]. In Nigeria, the rice ranks first socially among the food crops (sorghum, millet, cowpea, cassava and yam) consumed [3]. Since the mid- 1970s, rice consumption in the country has risen tremendously (10.3% and above per annum) due to increase per capita consumption (+7.3% per annum) [5].
Rice is an energy source food containing 80% carbohydrate, little protein, fat, vitamins (B1, B3, B6, B12, E, and folic acid) and minerals (phosphorus, calcium, potassium, sodium and iron). More than 90% of the energy in rice comes from the carbohydrates. These carbohydrates are made up of both complex type (starch-amylose and fibres) and simple form comprising of glucose, fructose, lactose and sucrose [6,7]. Rice is useful in treating hypertension because it is free from cholesterol. It is also used in the preparation of baby foods being free from allergens [8]. Rice starch serves as a source of glucose in oral rehydration solution for diarrhoeal infants [9].
Rice fermentation is carried out commonly in the Asia to improve its nutrient quality. Fermentation of foods increases food security, preserves it from spoilage, salvages wastes, removes antinutrients, improves nutrition, increase digestibility, increase vitamins and flavour of the rice [10]. Fermentation converts rice to various products including wine (sake), vinegar, rice-coconut mix, oats, crispies, gruel, beer, pudding, ragi, tapai, candy and sap [11,12]. Natural fermentation using microbes associated with surrounding environment is used in batch fermentation to produce Philippino rice. In the more advanced system, “starters” are added which are remnants of a previously successful batch fermentation. These are added to the new one [13]. Many microorganisms are used as “starters” particularly the fungi, yeasts (Saccharomyces, Amylomyces, Endomycopsis, Hansenula anomala), molds (Rhizopus, Aspergillus, Mucor) and bacteria (Lactic acid bacteria such as Lactobacillus lactis, L. acidophilus, Leuconostoc mesenteroides, Leuconostoc dextranicum and Pediococcus cerevisiae, acetic acid bacteria (Acetobacter aceti) [14].
In Nigeria, rice is not fermented for consumption and when natural fermentation sets into left-over cooked rice, it is considered spoilt and thrown away. In order to employ nutrient enrichment of fermentation in rice consumption, Nigerian indigenous rice variety “Igbimo” was fermented [15]. Chemical analysis of the fermented rice showed that there was improvement in the nutrient contents of the rice. Boboye and Alabi [16] showed that individual microorganisms isolated during the natural fermentation of the rice carried out specific functions in the synthesis and catabolism of nutrients. Micrococcus luteus was identified as the best carbohydrate fermenter.
Mutation is important for the study of traits in living systems. Ethylmethyl sulphonate (EMS) has been widely used to induce mutations both in lower and higher organisms [17]. In order to investigate the genetic basis of synthesizing glucose and protein in rice during natural fermentation, Micrococcus luteus was mutated with EMS. The mutants were used to ferment rice individually and amounts of glucose and protein produced were assayed. This is important to show whether glucose and protein anabolisms in the bacterium are directly controlled by specific gene(s) or not.
2. MATERIALS AND METHODS
2.1. Mutation Experiment
Micrococcus luteus was grown in 5 mL nutrient broth at 37˚C for 18 hours with agitation at 80 rpm. Using ethylmethyl sulphonate (EMS), the culture was mutated at 28˚C according to the method of Parkinson [18] modified as described by Boboye and Alao [19]. Mutational rate was estimated. Mutants were screened for the synthesis of glucose and protein during the fermentation of rice.
2.2. Fermentation of Rice with Mutants and Wild-Type Yeast
Each mutant was inoculated into 5 mL of nutrient broth and incubated at 37˚C for 24 hours after which cells were spun down at 12,168 × 103 g for 15 min (MSE Minor 35 Centrifuge). Cells were washed and resuspended in sterile distilled water. Equal cell load (0.867 at 670 nm) of each mutant was prepared and used to inoculate 4 g cooled rice previously sterilized for 15 min at 121˚C. The wild-type strain was used as a control. Fermentation was carried out for 7 days at 27˚C after which glucose and protein contents were determined.
2.3. Determination of Glucose and Protein Contents
Amount of glucose released by each mutant and wildtype was determined by using the standard dinitrosalicylic acid (DNSA) method slightly modified as described by Boboye and Alao [19]. DNSA reagent (0.5 mL) was added to 1.0 mL of fermented rice liquor, boiled for five minutes after which 4.5 mL of distilled water was added. It was cooled and the absorbance was read at 540 nm. Glucose standard curve was used to estimate the amount of glucose released during fermentation. Glucose content was defined as the amount of glucose (mg/mL) released during fermentation. Protein concentration in the rice sample fermented with each mutant and wild-type was estimated in milligram by standard Biuret method [20] as reported by Boboye and Alao [19].
3. RESULTS AND DISCUSSION
The mutational rate of the mutagen (EMS) was 99.9%. This is a very high rate implying a very high chance of hitting every part of the genome. Different types of mutant were obtained. None among these variants formed 0 mg/mL of glucose. All of them have ability to form the glucose ranging in content from 0.12 glucose mgmL–1 (mutant 23) to 4.15 glucose mgmL–1 (mutant 62) (Figure 1). Some mutants (32%) produced the carbohydrate at lower level and many (66%) at higher level than the wild-type Micrococcus which formed 0.86 mgmL–1 glucose. Generally, the poor glucose producers made the carbohydrate at ≤1.00 mgmL–1 (Table 1). These are 37 mutants and they span across low and high levels glucose producers. Only two mutants (No. 19 and 48) (Figure 1) synthesised the same amount of glucose as the unmutated bacterium and these are named normal glucose producers. Thus, the mother strain is also a poor glucose former. The higher level glucose synthesisers are made up of three major groups: 1.01 to 1.99 (40 mutants) and 2.0 to 2.99 (20 mutants) and 3.0 and above (3 mutants) mgmL–1 glucose makers. They are considered as moderate, good and super glucose producers.
Mutants 23 and 5 respectively made the lowest (0.09 mgmL–1) and highest (5.0 mgmL–1) concentrations of
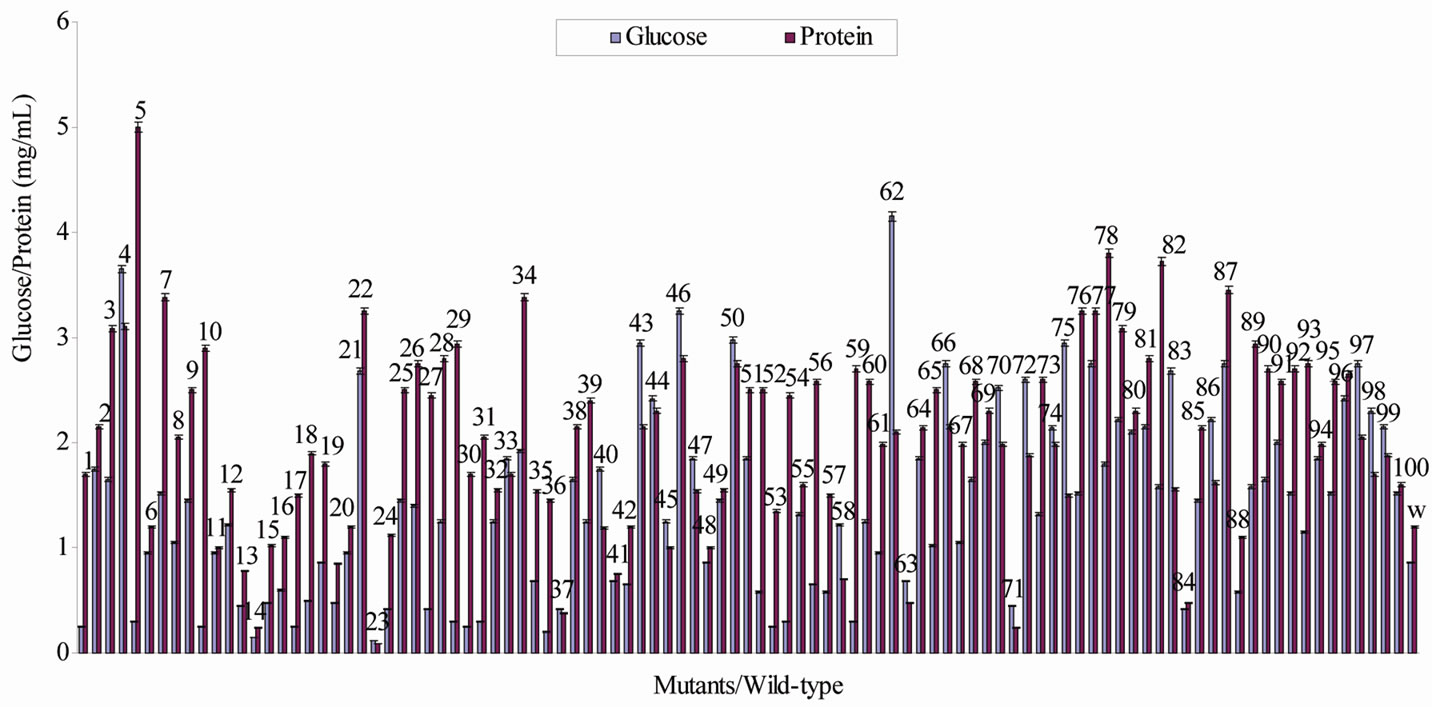
Figure 1. Concentrations of glucose and protein produced by the mutants and wild strain of Micrococcus luteus during the fermentation of a Nigerian rice, Oryza sativa var. “Igbimo”.
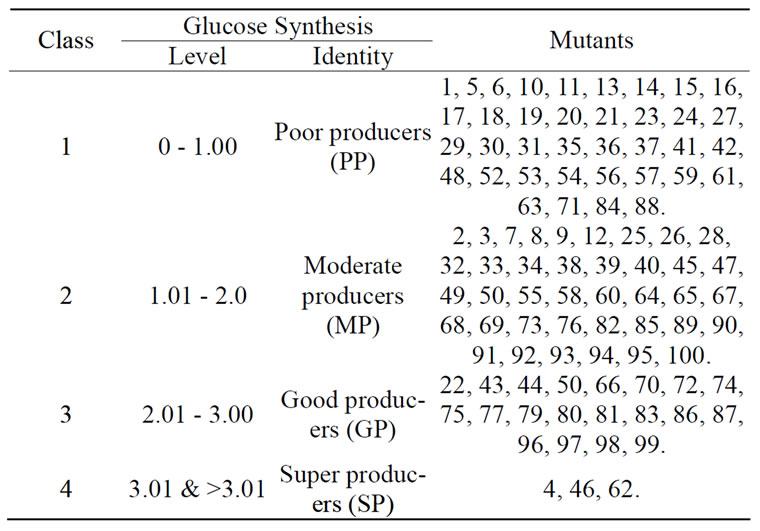
Table 1. Groups of Micrococcus luteus variants involved in glucose synthesis during the fermentation of a Nigerian rice, Oryza sativa var. “Igbimo”.
protein (Figure 1). Using the same classification scale as for the glucose, the variants were grouped into classes namely: poor, moderate, good and super protein producers (Table 2). They are composed of 13, 35, 40 and 12 mutants respectively. The wild strain bacterium made 1.2 mgmL–1 protein which lies in the range of 1.01 to 1.99 mgmL–1 protein; indicating that the original bacterium is naturally a moderate protein producer as determined under the growth conditions used in this experiment.
Some mutants formed more glucose than protein; these are mutants 4, 23, 33, 37, 40, 43, 44, 45, 46, 47, 50, 58, 62, 63, 66, 70, 71, 72,74, 75, 83, 86, 97, 98 and 99 (Figure 1). Other variants were better in the making of protein than glucose. The mother microbe synthesised less glucose (0.86 mgmL–1) than protein (1.2 mgmL–1).

Table 2. Mutants classes of Micrococcus luteus involved in the total protein production during the fermentation of a Nigerian rice, Oryza sativa var. “Igbimo”.
This data means that the genes encoding the formation of the two substances were affected by EMS to cause higher production of glucose than protein and vice versa.
Ethylmethyl sulphonate is known to alkylate nucleotides [17]. It binds between purines on opposite strands of DNA causing the removal of the base(s) affected. The action results into mispairing. At times, the EMS can cause baseless site [17,21] The mutants that produced glucose or protein lower than the wild strain have the genes coding for the synthesis of the products deleted such that the bacterium could not form the macromolecules appropriately; hence, they were repressed. The mutation appeared to have induced the relevant gene(s) producing glucose and protein above the normal levels. This underlies the good and super production of glucose and protein. Variation in the concentrations of the products obtained in this work is an indication of different levels of hit impacted by the mutagen on the concerned genes; thus concluding that glucose and protein anabolisms in the bacterium are directly controlled by specific gene(s).
4. ACKNOWLEDGEMENTS
We appreciate Federal University of Technology Akure, Ondo State, Nigeria for provision of facilities, materials and reagents used for this study.
REFERENCES
- Yong, Y., Tao, W.-Y., Liu, Ye-Jia and Feng, Z. (2007) Inhibition of Bacillus cereus by lactic acid bacteria starter cultures in rice fermentation. Food Control, 19, 159-161. doi:10.1016/j.foodcont.2007.03.002
- Imolehin, E.D. and Wada, A.C. (2000) Meeting the rice production and consumption demands of Nigeria with improved technologies. International Rice Commission Newsletters, 49, 23-41.
- Longtau, S. (2003) Multi-agency partnerships in West African agriculture: A review and description of rice production systems in Jos, Nigeria. EDO-DFID, 50.
- Akande, T. (2001) An overview of the Nigerian rice economy. Monograph of the Nigerian Institute of Social and Economic Research (NISER), Ibadan, 11.
- Akpokoje, G., Loncon, F. and Erenstein, O. (2001) Nigerian’s rice economy: State of the art. Draft Report, WARDA-NISER Collaborative Study, 21.
- Steinkraus, K.H. (1994) Nutritional significance of fermented foods. Marcel Dekker, New York, 671.
- Primrose, S.B. (1995) Molecular biotechnology. 2nd Edition, Blackwell Scientific Publishing Co. Inc., Westport, 251-252.
- James, S.A. and McCaskill, D. (1993) Handbook of energy crops, 5.
- Juliano, B.O. (1995) Protein and energy utilization of rice milling fractions by rats. Qualitas Plantarum-Plant Foods for Human Nutrition, 31, 371-376.
- Anon (2010) Fermented foods. www.sakthifoundation.org.
- Batra, L.R. and Millner, P.D. (1974) Some Asian fermented foods, beverages and associated fungi and bacteria. Mycobacteria, 66, 942-950.
- Steinkraus, K.H. (1989) Industrialization of indigenous fermented Foods. Marcel Dekker, New York, 140-169.
- Steinkraus, K.H. (1987) Indonesian tape ketan fermentation. Applied Environmental Microbiology, 1, 1067-1073.
- Yokotsuka, T. (1985) Fermented protein foods in the Orient with emphasis on Koji and Miso in Japan, Nuruk and Jeji in Korea. In: Wood, B.B. Ed., Microbiology of Fermented Foods, Elsevier, London, 1, 197-247.
- Boboye, B. and Terwase, D.E. (2004) Bacteria and nutritional changes associated with natural fermentation of a Nigerian indigenous rice (Oryza sativa “IGBIMO”). Oriental Journal of Chemistry, 20, 235-237.
- Boboye, B. and Alabi, O.J. (2011) Effect of monoculture fermentation on the proximate composition and sensory properties of a Nigerian indigenous rice (Oryza sativa “IGBIMO”). Food and Nutrition Sciences, submitted.
- David, T., Anthony, J.F., Jeffrey, H. and Richard, C. (2000) An introduction to genetic analysis. 3rd Edition, Freeman, W.F. and Company, New York, 127-143.
- Parkinson, J.S. (1976) Che A, Che B and Che C genes of Escherichia coli and their role in chemotaxis. Journal of Bacteriology, 126, 758-770.
- Boboye, B. and Alao, A. (2008) Effect of mutation on Trehalose-catabolic-enzyme synthesized by a tropical Rhizobium species F1. Research Journal of Microbiology, 3, 269-275. doi:10.3923/jm.2008.269.275
- Gornall, A.J., Bardwill, C.S. and David, M.M. (1949) Quantitative determination of protein. Journal of Biological Chemistry, 177, 751.
- Taggart, R. and Starr, C.B. (2006) The unity of life: Mutated genes and their protein products, 227.

