American Journal of Plant Sciences
Vol.4 No.7(2013), Article ID:33991,6 pages DOI:10.4236/ajps.2013.47169
Inheritance of the Number of Ovules per Ovary and Selection of Cacao Genotypes
![]()
1Departamento de Ciências Biológicas, Universidade Estadual de Santa Cruz, Ilhéus, Brasil; 2Centro de Pesquisas do Cacau, Comissão Executiva do Plano da Lavoura Cacaueira, Itabuna, Brasil; 3MARS Center for Cocoa Science, Itajuípe, Brasil.
Email: cassiabahia@yahoo.com.br, ronanxc@uesc.br, ronaldocs1964@yahoo.com.br, Brazil regina.machado@effem.com, darioa@uesc.br
Copyright © 2013 Rita de Cássia Bahia et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received April 8th, 2013; revised May 9th, 2013; accepted June 17th, 2013
Keywords: Theobroma cacao; Flower Trait; Heritability; Reproductive Biology; Genetic Improvement
ABSTRACT
We have characterized the number of ovules per ovary (NoOV) in cacao plants originated from crossing the clones CCN 51 and TSH 1188 (segregating progeny) and determined the heritability of this characteristic in order to select plants with higher NoOV in this progeny. The NoOV was calculated as the average of 10 flowers per plant and ranged from 44.8 to 58.6 between the six clones (two parents and four clones belonging to their genealogy). In the progeny (n = 209 plants) the NoOV averaged 54.3 (range 44.1 to 67.8). The NoOV was distributed uniformly among the progeny indicating that this trait is conditioned by polygenes. Its heritability was estimated at 67.7%. The 32 plants with NoOV similar or superior to the genitor CCN 51 (highest amount among clones) were selected for use in the genetic improvement program. We showed that this population is suitable for genetic mapping, molecular marker identification and selection of superior cacao genotypes.
1. Introduction
Various attributes of the flower have been used for the identification of the descriptors of interest in Theobroma species. A great variability of characteristics related to the flower was verified for Theobroma cacao L. (cacao) and Theobroma grandiflorum (Willd. ex Spreng.) K. Schum. (cupuassu), which are used to describe the diversity of these species [1,2]. In addition to this, the studies on the variations of the flowers’ characteristics and production of seeds are necessary as subsidy for the cacao genetic improvement program [3] and has been in-depth studied from 221 clones considered representative of the species [4].
The number of ovules per ovary (NoOV) is one of the seven descriptors related to the flower, present on the list of 27 descriptors that involve leaf, flower, fruit and agronomic characteristics, adopted for cacao [5], as well as among the 29 descriptors proposed for cupuassu [1]. This characteristic shows itself as consistent within each cacao access, suffers low environmental variability and may be used to estimate the potential number of seeds per fruit, because NoOV is a component for cacao production [4, 6].
The NoOV ranges from 35 to 70 for cacao, and some fruits with 67 seeds were already seen [5]. Supposedly this number may reach about 80 ovules per ovary, as fruits of the cultivar P 18 with 74 seeds and IMC with 77 seeds were already reported [2]. In general, higher numbers of ovules are found in forastero cacao, whereas in trinitario type lower values are found [7]. The cultivars evaluated by CEPEC in the 1970’s were classified based on this characteristics on the following categories: materials of high NoOV, if the values are higher than or equal to 52; average NoOV, if it range from 46 and 51; and low, if these are equal to or lower than 45. Additionally, a heritability of 70% was estimated for this trait in experiments carried out in the cacao producing region of Bahia, Brazil [8], up to 91% in populations carried out in Costa Rica [6], and of 81% in a large collection of 221 cocoa clones must of them from the international germplasm [4]. So, the NoOV is considered a clonal descriptor due to the specificity of this trait in each genetic material [7]. However, NoOV is not well understood for most of the clones that are currently used in different breeding programs cocoa. Also, the analyses of this trait from a controlled cross are not available for important breeding materials.
The ovary is the main reproducing organ of the plant and, therefore, an important component for the production of fruits. Even so, few studies have been carried out as regards this characteristic in cacao. The number of seeds per fruit in cupuassu is not proportional to NoOV, because the number of seeds is a production trait that depends on the fertilization percentage [9]. There are reports of cupuassu fruits collected in the Amazon with up to 67 seeds, therefore, close to the higher NoOV found in cacao [5]. The NoOV in cacao determines the number of seeds that may be produced in each fruit [2], and agroecological factors can interfere with this potential. Based on the analysis of 31 cacao clones, it was shown the positive and significative correlation (r = 0.73) between NoOV and number of seeds per fruit [3]. However, this correlation was lower (r = 0.48) in a representative sampling of the cocoa diversity and in a different environmental conditions [4]. Those data evidence the importance of NoOV to characterize the productive potential of the cultivars in specific genetic background of interest in a breeding program.
This work aimed to characterize the NoOV in cacao genotypes originated from the crossing of the clones CCN 51 and TSH 1188 and determine the heritability of this characteristic. Additionally, we select genotypes with higher NoOV from this population.
2. Material and Methods
The sample consisted of 209 plants from a population resulting from the artificial crossing of the clones CCN 51 and TSH 1188. The procedures of crossing and cultivation, the date and local of experiment were previously described [10]. Although there are around 600 plants in the experimental field, only those that had enough number of flower buds at the 2006/2007 year were sampled. Besides of the progeny and respective genitors, we also examined the clones available at the Germplasm Bank of Mars Center for Cocoa Sciences-MCCS, Barro Preto, BA, that are part of the genealogy of these genitors: CCN 51 originated from crosses among three different clones [(ICS 95 × IMC 67) × “Canellos” nacional clone)] as well as TSH 1188 (IMC 67, ICS 1, Sca 6). Out of this, only the clones Canellos and P18 were not found in the germoplasm collection of MCCS. In previous studies based on SSR markers, it was shown that the clones TSH 1188 and CCN 51 have higher heterozygosity than clones involve in their genealogy; also, the genetic clustering are consistent with the genealogy of this material [11].
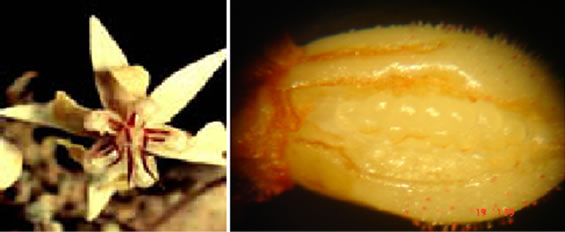
From each one of the plants and from each clone, 30 to 40 days after the appearance of the flower buds, ten recently open flowers were collected (Figure 1(a)) for the counting of ovules per ovary (Figure 1(b)). This population also shows segregation for resistance to cacao’s black pod and witch’s broom disease [10,12]. Additionally, the clones involved in the genealogy of these two genitor clones show variation in NoOV (Table 1). Therefore, these attributes were used in order to confirm this population as adequate for this study.
The flowers were collected during the first hours of the morning and transported in 15 mL test tubes with sterile distilled water to the laboratory. With the help of a 10× magnifying glass, and after a lateral incision on the ovary, the number of ovules was counted for each sampled flower per plant. The number of flowers per plant was nine per plant, the same adopted before to evaluate the
 (a) (b)
(a) (b)
Figure 1. Cacao flower and ovary. (a) General aspects, in which the petal presents average width of 2.68 mm and corolla with the width of 15.89 mm; (b) Ovary, with average dimension of 1.52 mm of length and 1.13 mm of width.
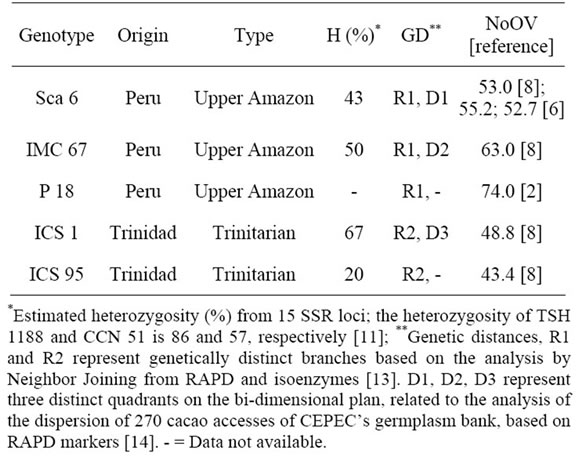
Table 1. Origin, type, Heterozygosity (H), molecular genetic diversity (GD), and number of ovules per ovary (NoOV) in cacao genotypes pertaining to the genealogy of the clones TSH 1188 and CCN 51.
number of ovules per ovary in cupuassu [1].
The descriptive statistics were estimated based on maximum, minimum and average values, standard deviation, frequency distribution and average variance of the NoOV, by means of the use of the SAS statistical package SAS [15]. For the definition of the categories of the plants according to NoOV, the definition of three categories proposed by CEPEC [8] was used, based on the analysis of 31 clones, making the adaptation to 10 categories that better held up the variation on this characteristic for the material analyzed on this work.
We calculated phenotypic variance (Vp) of each genotype involved in the genealogy of the segregating population. The environmental variance (Ve) was estimated from the Vp according to the following model: 1) the Ve of the clones is the same as Vp; 2) the Ve of the progeny is the arithmetical mean of the Vp of the genitor and progenitor clones [Ve = (VSca 6 + VIMC 67 + VICS 1 VICS 95 + VCCN 51 + VTSH 1188)/6]. The genetic variance (Vg) was calculated from environmental and phenotypic variation (Vg = Vp of Progeny − Ve). The broad sense heritability (Ha) was calculated based on the ratio between the estimates of the Vg and Vp of the progeny, assuming that the trait variances in the clones involved in the genealogy are due to the environment.
3. Results and Discussion
The NoOV averages of the plants of the progeny uniformly distributed themselves around the general average of the progeny (Figure 2). It was observed that the NoOV averages of the progeny ranged from a minimum of 44.1 to a maximum of 67.5, showing that there is variation in this progeny, confirming the segregating nature of this characteristic on the population under study. When the set of data was divided into four equal parts, the first quartile was NoOV = 51.3 and the last quartile was NoOV = 57.4, indicating that the population has a tendency to a higher NoOV.
The plants of the progeny were grouped into 10 cate-
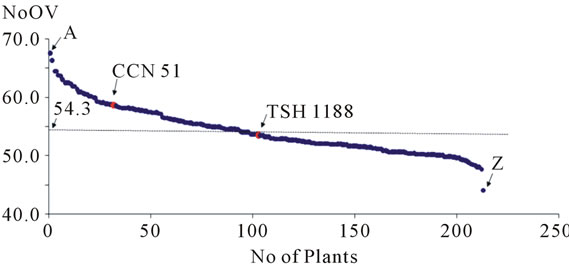
Figure 2. Distribution of the plants of the progeny in relation to the number of ovules per ovary (NoOV), evidencing the extremes of the progeny (plants A and Z) and the positioning of the genitors (TSH 1188 and CCN 51) of this cacao progeny in relation to the average (54.3).
gories, with intervals of three ovules per ovary between them, revealing a distribution of frequency of plants of this progeny by category of NoOV typical of a polygenic characteristic and with environmental interaction (Figure 3). The population has showed a positive asymmetric distribution (0.7), where the average (54.3) is higher than the median (53.2). Also low standard deviations and variation coefficients were observed, indicating that the dispersion around the average is low, and the individuals tend to concentrate close to the average. The three categories with lower NoOV (<48) were the less frequent, corresponding to six plants of the segregating population and to the clone ICS 95. On the other hand, the three categories with higher NoOV (>60) corresponded to 18 plants of the segregating population and did not include none of the clones analyzed. The intermediate categories (49 to 60 ovules) were the most frequent (185 plants, 88.5% of the progeny). The polygenic nature of this characteristic in this population is according to the data from a different genetic background of cocoa [16], which mapped eight quantitative trait loci (QTL) related to NoOV, distributed into six different chromosomes in segregating populations resulting from the crossing of the forasteiro high Amazon clone IMC 78 and two trinitario materials: DR1 and S52. Among these eight loci, four alleles were identified as favorable to the increase of NoOV, supposedly from forasteiro ancestry.
The analysis of the variance of the progeny resulting from the crossing CCN 51 × TSH 1188 revealed a significative difference between the individuals of the progeny and the genitors (Table 2). The average NoOV of the progeny differs from the average of the CCN 51. However, it does not differ from the average of the TSH 1188 that also has lowered NoOV (Table 3). This data allows the inference that a dominance effect for the genes that reduce the number of ovules for this population is
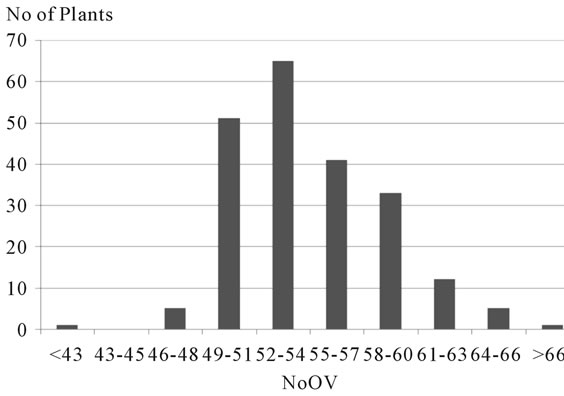
Figure 3. Categories of number of ovules per ovary (NoOV), and respective number of plants of the progeny resulting from the crossing of the cacao clones TSH 1188 and CCN 51.
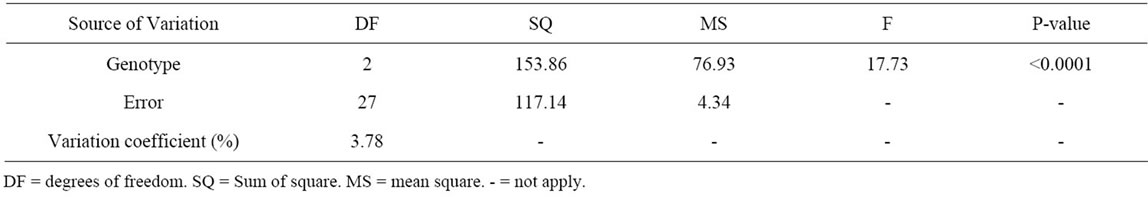
Table 2. Variance analysis of the number of ovules per ovary on the three cacao clones: clone CCN 51; clone TSH 1188; progeny CCN 51 × TSH 1188.
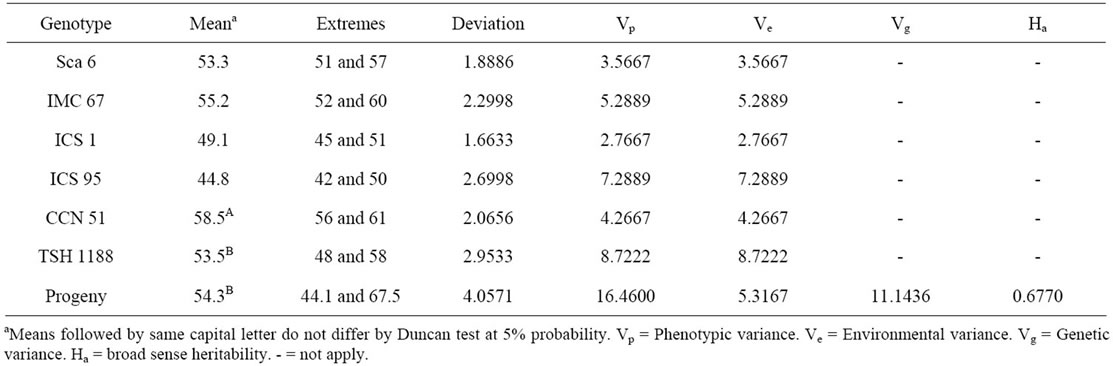
Table 3. Estimated averages and variances for the number of ovules per ovary in cacao clones pertaining to the genealogy of the progeny resulting from the crossing of CCN 51 and TSH 1188.
happening. A similar trend was reported previously [17]. However, this trend differs from other studies about the inheritance of this characteristic once that genes related to higher NoOV seem to exert dominance effect on genes related to lower NoOV [2]. And, indeed, we observe individuals in the progeny with the highest NoOV that both his parents.
Although the average of the genitor TSH 1188 has not differentiated significantly in relation to the average of the progeny, it was verified that at least 32 plants of the progeny (15% of the plants) showing number of ovules equal or higher than CCN 51 (genitor of higher NoOV) and meaningfully differ of the genitor with lower number of ovules (Figure 4). The clone P 18 has around 74 ovules per ovary, being the possible origin of genes favorable to higher NoOV on IMC clones, some of which produce fruits with up to 77 seeds [2]. P 18 is one of the clones involved in the genealogy of IMC 67, which is part of the genealogy of both genitors of the progeny used for this work. Thus, these 32 plants must have inherited many of the alleles typical to IMC 67 and P 18, once that these had NoOV values close to those described for these clones. Among these, one plant shows a NoOV that ranged from 63 to 72, with an average of 67.5 —higher than all the others—and an average deviation of 1.49. Supposedly, these 32 plants and others with NoOV close to that of the CCN 51 have combinations of favorable alleles to the increase of NoOV in amounts equal to or higher than that of this genitor of higher NoOV. Therefore, the evaluation of this population on the whole, may reveal at least 100 plants with NoOV higher than 58.5, what provides great chances of finding genotypes with this characteristic and that also have satisfactory pattern as regards the other characteristics that are under evaluation on this cacao improvement program.
The NoOV of the clones Sca 6, IMC 67, ICS 1 and ICS 95 have confirmed that this characteristic is typical of each cacao clone (Table 3). These values were similar to those obtained in previous works [6,8] (Table 1). This proves that the NoOV is a very stable trait for the different cultivars or cacao populations. Besides to that, the NoOV is very variable among the different cacao genetic groups, and may be used either for the characterization of the genetic material or to study the variability of the populations [7].
The clones TSH 1188 and CCN 51 have showed intermediate NoOV among those that are part of its genealogy (Table 3). But the averages of CCN 51 and progeny were different among themselves according to the Duncan test at 5% probability. The segregating progeny resulting from the crossing of these two clones have shown itself very variable for this characteristic and 157 plants (74% of the progeny analyzed) have flowers with more than 51 ovules. This number of ovules is considered high for cacao [8]. Additionally, a more large range of NoOV was observed for the progeny involving the
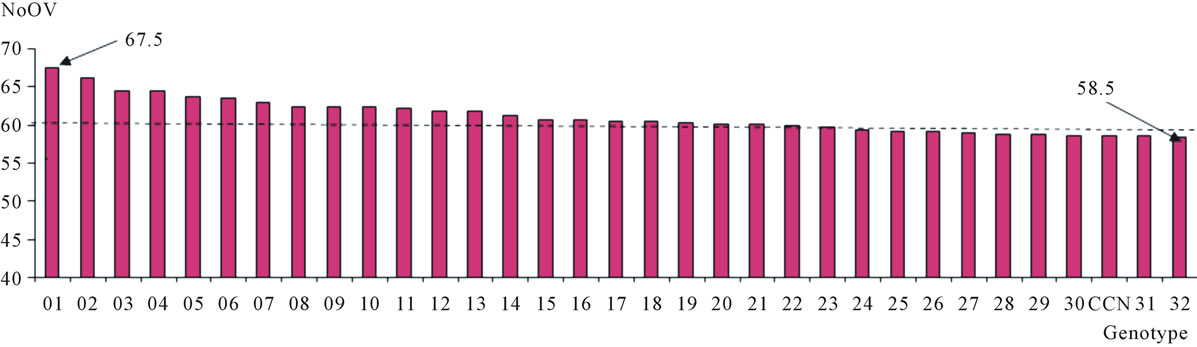
Figure 4. Distribution of the 32 plants selected from the progeny resulting from the crossing of the cacao clones TSH 1188 and CCN 51 in relation to the number of ovules per ovary (NoOV). The mean of selected plants is indicated as a horizontal line.
crossing of the clone IMC 78 with Catongo (NoOV = 58.7 ± 2.10; extremes 52.7 to 63.5) [16]. The progeny studied on this work (Table 3: NoOV = 54.3 ± 4.03; extremes 44.1 and 67.5) shows higher NoOV variation than that involving IMC 78 and Catongo (NoOV = 58.7 ± 2.10). In the analysis of 200 clones involved in cocoa breeding program, different selection strategies for seed weight and number of seeds per fruit reveal that TSH 1188 and CCN 51 are between 7.5% more productive clones, and that NoOV contributes 23% of potential yield [4]. Therefore, our data confirm the great mapping potential of genes related to this characteristic and selection of superior genotypes in this population.
The estimated Ha based on the population derived from the crossing of CCN 51 and TSH 1188 was 67.7% (Table 3). This value is close to the value found on other populations, in experiments carried out in the cacao producing region of Bahia [8], which was of 70%, but lower than those found based on experiments carried out in Costa Rica [6], that were of 79.43% in the experimental station of Limón and 91.01% in the experimental station of Turrialba. Besides of reflecting the effects of different environments, it must be considered that in each case crossings involving different clones to those used in this work were used. Therefore, the environmental effects and the different genotypes are the main sources for variation in the heritability values. These values reveal that this characteristic has a strong genetic component, and it is adequate for the selection of material with higher productive potential.
Our results have confirmed that NoOV is very stable in the same cacao population or cultivar and has high heritability. It has also shown that 15% of the progeny shows NoOV higher than that of the genitor of higher value. It was previously proved that this characteristic is determining for the potential number of seeds per fruit once that it presents a positive correlation with number of seeds per fruit [3,4]. Also in relation to production, evidences of gene grouping were detected with NoOV and other characteristics of the seed, in the form of QTL co-location for both in a same region of chromosome 4 [16], indicating a possible grouping of genes genetically linked and with effects on these characteristics. Therefore, the plants selected in the progeny resulting from the crossing of TSH 1188 × CCN 51 represent an elite in terms of potential for the seed production in this population. As this progeny is segregating for the NoOV trait, the adaptation of this population for genetic mapping and selection of superior genotypes is proved. The productive potential of the population is high, qualifying it for the development of improvement works directed to the obtaining of varieties with higher number of seeds per fruit and resistant to cacao’s black pod and witch’s broom disease [10,12].
Considering the high NoOV values found in this population, the high fertilization rate observed in cacao, and the high productivity history of the genitors of this progeny, it may be inferred that this is a promising progeny as regards the production trait. Although production is the result of a great many number of factors, the genetic factor related to NoOV is conclusive as to the productive potential in cacao.
4. Conclusions
1) The number of ovules per ovary in cacao is a polygenic trait, has high heritability and displays great variability among different cacao genotypes.
2) The progeny resulting from the crossing of the clones CCN 51 and TSH 1188 is segregating for NoOV, and it is sufficiently suitable for the genetic mapping studies and selection of superior genotypes as to production potential.
3) The 32 plants selected from the progeny carry combination of favorable alleles to higher NoOV in proportions equal to or higher than those of the genitor with higher value for this trait.
5. Acknowledgements
The authors are grateful to Lindolfo Santos Filho, Alex-Alan Furtado de Almeida and Uilson Vanderley Lopes for suggestions. Thanks to the MARS Center for Cocoa Sciences (Mars Inc.), for logistic support provided, keeping the population in the experimental station, and particularly to Marcos for cooperation in obtaining and analyzing the cocoa flowers. The first author was recipient of a fellowship from CNPq.
REFERENCES
- R. M. Alves, A. A. F. Garcia, E. D. Cruz and A. Figueira, “Seleção de Descritores Botânico-Agronômicos Para Caracterização de Germoplasma de Cupuaçuzeiro,” Pesquisa Agropecuária Brasileira, Vol. 38, No. 7, 2003, pp. 807-818. doi:10.1590/S0100-204X2003000700004
- B. G. D. Bartley, “The Genetic Diversity of Cocoa and Its Utilization,” CABI, Cambridge, 2005. doi:10.1079/9780851996196.0000
- G. C. T. Castro, T. N. S. Pereira, G. A. Carleto and B. G. D. Bartley, “Caracterização de Recursos Genéticos do Cacaueiro. III Flor Das Seleções CEPEC, EEG, SIAL, BE, MA, RB, CA e CAS,” Agrotropica, Vol. 1, No. 1, 1989, pp. 27-33.
- C. Cilas, R. Machado and J.-C. Motamayor, “Relations between Several Traits Linked to Sexual Plant Reproduction in Theobroma cacao L.: Number of Ovules per Ovary, Number of Seeds per Pod, and Seed Weight,” Tree Genetics & Genomes, Vol. 6, No. 2, 2010, pp. 219- 226. doi:10.1007/s11295-009-0242-9
- L. A. S. Dias, “Melhoramento Genético do Cacaueiro,” Editora Folha de Viçosa, Viçosa, 2001.
- O. López, G. A. Enríquez and J. Soria, “Herencia del Número de Óvulos por Ovario en Theobroma cacao L.,” Turrialba, Vol. 38, No. 3, 1988, pp.163-167.
- P. Lachenaud, F. Bonnot and G. Oliver, “Use of Floral Descriptors to Study Variability in Wild Cocoa Trees (Theobroma cacao L.) in French Guiana,” Genetic Resources and Crop Evolution, Vol. 46, No. 5, 1999, pp. 491-500. doi:10.1023/A:1008763321959
- CEPEC—Centro de Pesquisas do Cacau, “Informe Técnico,” CEPLAC, Ilhéus, 1974.
- R. M. Silva, “Estudo do Sistema Reprodutivo e Divergência Genética do Cupuaçuzeiro (Theobroma grandiflorum Willd ex Spreng Schum),” Dissertação (Mestrado em Genética e Melhoramento de Plantas)—Escola Superior de Agricultura Luiz de Queiróz, Universidade de São Paulo, Piracicaba, 1996.
- R. M. F. Santos, U. V. Lopes, R. C. Bahia, R. C. R. Machado, D. Ahnert and R. X. Corrêa, “Marcadores Microssatélites Relacionados Com a Resistência à Vassoura-de-Bruxa do Cacaueiro,” Pesquisa Agropecuária Brasileira, Vol. 42, No. 8, 2007, pp. 1137-1142. doi:10.1590/S0100-204X2007000800010
- M. M. Yamada, F. G. Faleiro, A. B. Flores, U. V. Lopes, J. L. Pires, R. X. Corrêa and R. F. Santos, “Microsatellite Diversity and Heterozygosity of Parents of a Cocoa Breeding Population,” Crop Breeding and Applied Biotechnology, Vol. 9, No. 1, 2009, pp. 17-22.
- R. C. Bahia, U. V. Lopes, E. D. M. N. Luz, R. S. Carvalho, R. C. R. Machado, D. Clement, R. M. F. Santos, D. Ahnert and R. X. Corrêa, “Caracterização Fenotípica de uma População Segregante de Cacaueiro Quanto à Resistência à Podridão-Parda,” 15th International Cocoa Research Conference, Vol. 1, San José, 2007, pp. 445- 451.
- O. Sounigo, R. Umaharan, Y. Christopher, A. Sankar and S. Ramdahin, “Assessing the Genetic Diversity in the International Cocoa Genebank, Trinidad (ICG,T) Using isozyme Electrophoresis and RAPD,” Genetic Resources and Crop Evolution, Vol. 52, No. 8, 2005, pp. 1111-1120. doi:10.1007/s10722-004-6110-4
- J. M. Marita, J. M. Rodriguez and J. Nienhuis, “Development of an Algorithm Identifying Maximally Diverse Core Collections,” Genetic Resources and Crop Evolution, Vol. 47, No. 5, 2000, pp. 515-526. doi:10.1023/A:1008784610962
- SAS Institute, “SAS/STAT User’S Guide: Version 6,” 4th Edition, SAS Institute, Cary, 1989.
- D. Clement, A. M. Risterucci, J. C. Motamayor, J. A. K. N’Goran and C. Lanaud, “Mapping Quantitative Trait Loci for Bean Traits and Ovule Number in Theobroma cacao L.,” Genome, Vol. 46, No. 1, 2003, pp. 103-111. doi:10.1139/g02-118
- J. V. Soria and O. Esquivel, “Alguns Resultados del Programa de Mejoramiento Genético de Cacao en el IICA-Turrialba,” Internacional Cocoa Research Conference, 2nd Edition, Bahia, 1969, pp. 35-42.
NOTES
*These authors contributed equally to this work.

