Pharmacology & Pharmacy
Vol.3 No.2(2012), Article ID:18720,17 pages DOI:10.4236/pp.2012.32025
NPS31807, a Standardized Extract from Sphaeranthus indicus, Inhibits Inflammatory, Migratory and Proliferative Activity in Keratinocytes and Immune Cells
![]()
1Department of Biomarker Discovery, Piramal Life Sciences India Limited, Mumbai, India; 2Department of Herbal Development, Piramal Life Sciences India Limited, Mumbai, India; 3School of Biosciences and Technology, VIT University, Vellore, India.
Email: {*muralidhara.padigaru, *somesh.sharma}@piramal.com
Received January 19th, 2012; revised February 24th, 2012; accepted March 7th, 2012
Keywords: Herbal Extract; Cytokine; Psoriasis; NPS31807; Keratinocyte; Sphaeranthus; Phytopharmaceutical
ABSTRACT
Chronic inflammation induced hyper-proliferation, and migration of keratinocytes are pathological hallmarks of psoriasis. Extracts from Sphaeranthus spp. demonstrate pharmacological activity in-vitro and in-vivo. However, the activity in modulating disease relevant pathways in psoriasis has not been reported. In the current study, a standardized herbal extract from Sphaeranthus indicus (NPS31807) was used to study the mechanistic activity under conditions of inflammation, keratinocyte proliferation and migration using cell based and gene expression assays. NPS31807 treatment reduced levels of pro-inflammatory cytokines from human macrophages and activated epidermal keratinocytes in a dose dependent manner. Treatment with NPS31807 diminished NFκB and AP-1 transcription activity in human macrophages. Lowered nuclear translocation of p65 sub-unit in macrophages by treatment confirmed reduced activity of NFκB. Gene expression profiling showed attenuated expression of genes involved with inflammation such as TNF signaling, and angiogenesis by NPS31807. Inhibition of angiogenesis and matrix metalloproteinase production in keratinocytes was confirmed using RTq-PCR assays. Pretreatment with NPS31807 led to significant reduction of STAT3 phosphorylation and mitogen induced cellular migration. NPS31807 induced inhibition of proliferative genes and BrdU uptake in epidermal keratinocytes. In summary, our study provides novel molecular insights into the anti-inflammatory, anti-migratory and anti-proliferative properties of NPS31807. In summary, NPS31807, an extract from Sphaeranthus indicus can be used as therapeutic option in inflammatory and auto-immune conditions such as psoriasis.
1. Introduction
Standardized herbal formulations represent an attractive therapeutic option due to their potential for synergistic action on multiple targets in disease conditions [1]. This warrants mechanistic studies aimed at rational development of these novel formulations in various disease settings. Sphaeranthus indicus Linn (Family: Astracea) is a popular plant widely described in traditional medicine and has been known to contain numerous pharmacological compounds such as eudesmanoids, sesquiterpene lactones, glycosides, flavanoids, and essential oil [2]. The extracts from Sphaeranthus spp. mediate anti-oxidant activities through free radical scavenging [3] and inhibit TNF-α and IL-8 release in P. acne stimulated leukocytes [4]. The herb has also shown protective activity in wound healing [5]. However, studies describing the mechanistic action of this plant in psoriasis have not been reported till date. In this report, we attempted to evaluate the mechanistic activity of NPS31807 using cell based functional assays and gene expression profiling. Psoriasis is a complex auto-immune disorder characterized by keratinocyte hyper-proliferation, abnormal differentiation, leukocyte infiltration and inflammation [6]. Macrophages and dendritic cells play a pivotal role by releasing pro-inflammatory cytokines, which in turn activate keratinocytes to undergo aberrant hyper-proliferation and differentiation [7]. These inflammatory and hyper-proliferative responses by immune cells and keratinocytes in psoriatic skin are largely mediated through the activation of numerous transcription (e.g. NFκB, AP-1 and CEBP-β) and growth factors (e.g. EGF, KGF) [8]. Current therapeutics such as Infliximab and Etanercept cause abrogation of cytokine mediated signaling in immune cells and keratinocytes [9, 10]. The clinical efficacy induced by anti-inflammatory therapies are associated with lowered infiltration of immune cells, reduced levels of pro-inflammatory cytokines and normalized proliferative responses in keratinocytes [11]. However, current therapies for psoriasis are associated with long term adverse effect [12]. In contrast, standardized herbal formulations demonstrate negligible adverse events associated with long term use in humans. NPS31807 is a standardized, orally available, methanolic extract from the fruiting and flowering heads of S. indicus plant undergoing randomized clinical testing [13]. NPS31807 has been standardized for two known marker compounds—7-hydroxylfrullanolide (5% - 9%) and Sphaeranthanolide (2%). The present study was undertaken to examine whether extracts from Sphaeranthus indicus could modulate disease relevant phenotypes in human epidermal keratinocytes and macrophages in-vitro.
2. Material and Methods
2.1. Extraction and Preparation of NPS31807
The extraction and preparation of NPS31807 and 7-HF was performed as described previously [14]. Briefly, the flowering and fruiting heads of S. indicus were dried and pulverized. The pulverized material (50 gms) was soaked in 300 ml of methanol and subsequently heated for 3 hours at 40˚C. The extract was filtered using Whatman filter paper and the filtrate was collected. The methanol extraction was repeated thrice and the filtrates were pooled and concentrated under line vacuum to obtain a thick sticky mass of crude extract (NPS31807). Further, bioactivity guided fractionation yielded 49 fractions from the crude extract and was subjected for in-vitro TNF-α activity screening in human peripheral blood monocyte cells (hPBMC). One of the active fractions from the anti-TNF-α screens was selected to isolate 7-HF and NPS31807 was standardized for the content of 2 marker compounds. The detailed phytochemical composition is stated as Table 1 and the standardized HPLC fingerprint for the extract and marker compounds (7-HF; Sphaeranthanolide) are shown in Figure 1.
2.2. Chemicals and Reagents
Recombinant tumor necrosis factor-α (TNF-α), epidermal growth factor (EGF), Oncostatin M (OSM) and Hygromycin B were procured from Sigma Aldrich (St Louis, MO, USA). Dexamethasone (Dex), BAY11-7082 (BAY), and FK-506 were procured from Sigma Aldrich (St Louis, MO, USA). Monoclonal primary antibody for p65/RELA (Clone: E379) was procured from Epitomics, Inc (Burlingame, CA, USA). Hoechst 33342 (nuclear stain) and antirabbit secondary antibody conjugated with DyLight® 549 were purchased from Thermo Fisher Scientific (Thermo Scientific, Waltham, MA). All other reagents were purchased from Sigma Aldrich (St. Louis, MO, USA) unless otherwise specified.
2.3. Human Macrophage (THP-1) Assays for Cytokine Estimation, Microarray and Viability Studies
THP-1 cells (ATCC, Rockville, MD) were seeded in 96- well flat bottom sterile culture-plates (1 × 104 cells/well) and 25 cm2 flasks (1 × 105 cells/well) for cytokine estimations and microarray assays respectively. For ELISA assays, cells were differentiated using phorbol myristate acetate (PMA, 10 ng/mL) for 2 hours and treated with varying concentrations of NPS31807 for 15 minutes, followed by stimulation with lipopolysaccharide (LPS, 1 μg/mL) for 24 hours. For microarray assays, the treatment concentrations were 10 μg/ml for NPS31807, 7-HF (3 μM), BAY11-7082 (3 μM), Dexamethasone (3 μM), and FK-506 (3 μM). The cells and supernatants at the end of the assay were collected, centrifuged, and stored at −80˚C for total RNA and cytokine estimation respectively. The concentrations of human cytokines (TNF-α, IL-1β, IL-6) in culture supernatants were evaluated by enzyme-linked immunosorbent assay (Duosets®, R & D Systems, Wiesbaden, Germany). Cell viability measure-
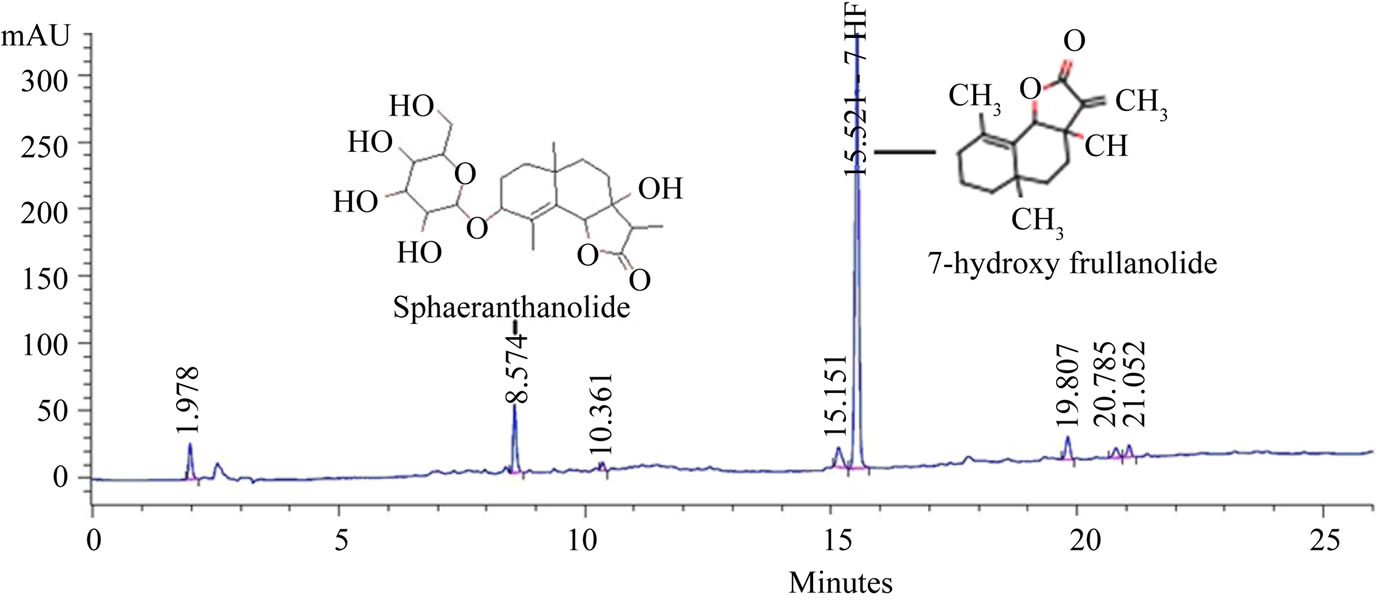
Figure 1. HPLC finger-print of NPS31807.
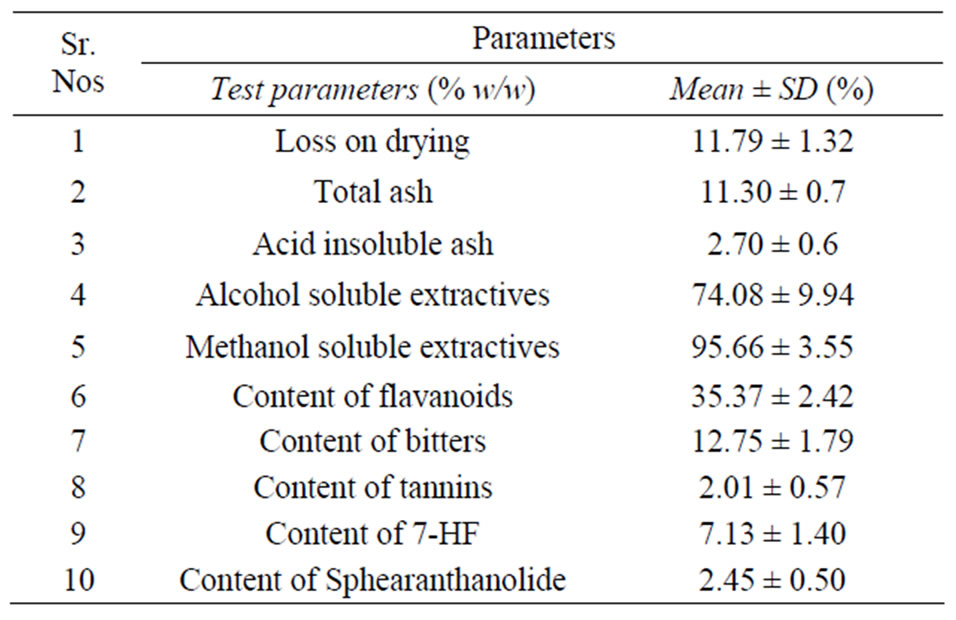
Table 1. Physico-chemical composition of NPS31807.
ments were carried out in THP-1 cells using the Cell Counting Kit-8 (CCK-8) as per the manufacturer’s instructions (Dojindo Molecular Technologies, Gaithersburg, MD).
2.4. Primary Epidermal Keratinocyte Assays for BrdU Uptake and RTq-PCR Studies
Normal human epidermal keratinocytes (NHEK) were procured from Lonza (Allendale, NJ, USA) and cultured in complete keratinocyte growth medium, KGM®-2 supplemented with Bullet Kit® (Lonza, Allendale, USA). Cells were serially passaged and assays were conducted with sub-confluent cells until passage three. For gene expression assays, NHEK cells were seeded in a 6-well tissue culture dish and treated with varying concentrations of NPS31807 (0.1 μg/mL, 1 μg/mL) for 2 hours followed by stimulation with respective stimulants. Cells were stimulated with EGF (10 ng/mL) or Oncostatin M (100 ng/mL) & TNF-α (10 ng/mL) for 24 hours and subsequently harvested for total RNA extraction. For BrdU uptake assays, NHEK cells were plated in a 96-well tissue culture plates, treated for 2 hours with varying concentrations of NPS31807 followed by stimulation with 10 ng/mL of EGF for 24 hours. Labeling and detection of BrdU was performed using the Roche BrdU ELISA kit (Roche Applied Sciences, Indianapolis, USA) as per the manufacturer’s instructions.
2.5. Total RNA Extraction, cDNA Synthesis and RT-qPCR Studies
For RNA isolation, THP-1 and NHEK cells from respective assays were harvested with chilled Trizol reagent (Invitrogen Corp, Carlsbad, USA). Total RNA was extracted and purified using the chloroform-ethanol phase separation as described earlier [15]. Two micrograms of total RNA was reverse transcribed in a 20 µL set-up using the Superscript III cDNA conversion kit as per the manufacturer’s protocol (Invitrogen Corp, Carlsbad, USA). RTq-PCR studies were carried out using Quantifast® SYBR green master-mix (Qiagen Inc., Valencia, USA) as described earlier [16]. Primers were designed using the Primer 3 software [17] and are listed in Supplementary Table 1. The cycling threshold (Ct) values for target genes in all samples were normalized using GAPDH transcript and relative fold change values were computed using the 2−∆∆Ct method as described earlier [18].
2.6. Microarray Study and Data Analysis
70-mer oligonucleotides (Operon Biotechnologies, USA) representing 267 transcripts known to play a major role in cellular signaling pathways were printed on aminosilane coated glass slides (Array IT, Sunnyvale, CA) using an arrayer (Genomic Solutions, Ann Arbor, USA) For gene expression studies, twenty micrograms of total RNA were reverse transcribed and labeled with Cy3® (Amersham Pharmacia Biotech, New Jersey, USA) using Chipshot® in-direct labeling kit (Promega, Madison, WI, USA). The microarray chip was hybridized with labeled cDNA for 18 hours at 42˚C and scanned with a laser scanner (Gene TAC, Genomic solutions, USA). Image analysis and data normalization was done using GeneTac Integrator, (Genomic Solutions, USA) followed by data analysis using Genespring 10 (Agilent Technologies, Santa Clara, USA). Welch test was used to determine differentially expressed genes [19] (F.C > 2.0, p < 0.05) affected due to NPS31807 treatment compared to LPS treatment in THP-1 cells. The differentially expressed genes were queried for functional enrichment analysis using GSEA and PANTHER database [20,21].
2.7. Reporter Expression Assays
The detailed protocol was performed as described earlier [22]. Activation of key transcription factor mediated inflammatory signaling was evaluated using the Cignal FinderTM assay kit (Qiagen Corp, Valencia, USA). Briefly, transcription factor containing specific tandem representative elements (TRE) were cloned into dual luciferase plasmid constructs encoding the mammalian firefly luciferase gene and transiently transfected into THP-1 cells using the Surefect® transfection reagent (Qiagen Corp, Valencia, USA). Cells were differentiated using PMA and subsequently treated with NPS31807 (10 µg/mL) for 15 minutes and stimulated with LPS (1 µg/mL) for 24 hours. The assay was terminated and quantified using DualGlo® luciferase reagent system [Promega, Madison, USA] as per the manufacturer’s instructions.
2.8. STAT3 Luciferase and in-Vitro Wound Assays
HeLa/STAT3-luc reporter cells (Panomics Inc., Redwood City, CA) were seeded in a white opaque 96-well plates (Nunc, Rochester, NY ) at a density of 1 × 104 cells per well and cultured for 24 hours in complete growth medium comprising of DMEM, 10% FBS and 100 ng/mL of Hygromycin B. The cells were serum starved, treated with multiple concentrations of NPS31807 for 15 minutes, followed by stimulation with Oncostatin M (100 ng/mL) and TNF-α (10 ng/mL) for 3 hours. The assays were terminated using the Promega® luciferase assay reagent system (Promega, Madison, WI) as per the manufacturer’s instructions. In-vitro wound scratch assays were performed in HeLa/STAT3-luc cells as described previously [23]. In brief, cells were pre-treated with NPS31807 (1 µg/mL) and stimulated with recombinant EGF (10 ng/mL) for 24 hours followed by fixation with 3.5% formaldehyde for 15 minutes, washed and visualized under microscope (Nikon, Tokyo, Japan). The gap width (µM) was measured using Image Pro® Plus (Media Cybernetics, Silver Spring, USA) and % inhibition as compared to basal condition was calculated.
2.9. Quantitative High Content Cell Based Imaging
Nuclear translocation of p65/RELA was quantified using cell based imaging studies in macrophage cells as described previously [24]. THP-1 cells were seeded into 96- well MTPs (Nunc, Roskilde, Denmark) at a density of 1 × 104 cells/well, treated with varying concentrations of NPS31807 for 15 minutes preceding stimulation with LPS (1 μg/mL) for 30 minutes (p65/REL). The plates were processed and quantitative imaging was performed using the high content imaging systems from Cellomics® (Thermo Scientific, USA) as described previously in detail [24]. The intensity values for treatment groups were normalized to values from LPS stimulation which was considered as 100%.
2.10. Statistical Analysis
All assays were performed in triplicates unless otherwise stated. Statistical comparison between multiple groups was evaluated using 1-way ANOVA with Dunnet’s post hoc testing in GraphPad Prism 4 (GraphPad Software, San Diego, CA). Statistical comparisons among two groups were evaluated using student’s t-test. Results are represented as mean ± SD unless otherwise stated. Statistical significance was associated with p < 0.05 (*, #).
3. Results
3.1. Effect of NPS31807 on the Production of Pro-Inflammatory Cytokines from Keratinocytes and Macrophage Cells in Vitro
We evaluated the effect of NPS31807 on pro-inflammatory cytokine production from human monocytic cell line (THP-1) and normal human epidermal keratinocytes (NHEK), since these cells are known to release significant amount of pro-inflammatory cytokines in lesional skin. THP-1 cells were seeded at a density of 1 × 103 cells/well, differentiated using PMA (10 ng/ml) for 2 hours, pre-treated with multiple concentrations of NPS- 31807 and ![]() stimulated with 1.0 μg/mL LPS for 24 hours. Addition of NPS31807 induced a dose dependent inhibition in the levels of TNF-α, IL-6 and IL-1β as measured by ELISA (Figure 2(a)). NPS31807 induced potent abrogation of cytokine levels at concentrations of 1 μg/ml, although the inhibitions at lower concentrations were more pronounced for IL-6 as compared to TNF-α and IL- 1β (Figure 2(a)). In parallel, we assessed the changes in cell viability induced by NPS31807 in THP-1 cells using the CCK-8 incorporation assay. NPS31807 treatment did not significantly decrease cell viability at concentrations up-to 10 μg/ml (Figure 2(a)). In addition, treatment maintained cell viability in epidermal keratinocytes for concentrations up-to 30 μg/ml (data not shown). Next, we evaluated the effect of NPS31807 on TNF-α and IL-8 expression in keratinocyte cells. NHEK cells were stimulated with Oncostatin M (100 ng/ml) and TNF-α (10 ng/ml) for 24 hours since TNF-α is known to induce expression of these genes [25]. We observed a robust upregulation of TNF-α levels (FC: 11.0 ± 0.66) with stimulation after 24 hours (Figure 2(b)). Pre-treatment with NPS31807 led to highly significant dose dependent decrease in TNF-α expression at 1 μg/ml (FC: 0.41 ± 0.19, Figure 2(b)). IL-8 expression was up-regulated by 13(±0.46)-fold in stimulated keratinocytes as compared to un-stimulated cells (Figure 2(c)). NPS31807 (1 μg/ml) treatment reduced expression of IL-8 to 3.2(±1.2)-fold (Figure 2(c)). This potent inhibition in levels of proinflammatory cytokine levels showed that NPS31807 mediates anti-inflammatory activity in macrophages and keratinocytes.
stimulated with 1.0 μg/mL LPS for 24 hours. Addition of NPS31807 induced a dose dependent inhibition in the levels of TNF-α, IL-6 and IL-1β as measured by ELISA (Figure 2(a)). NPS31807 induced potent abrogation of cytokine levels at concentrations of 1 μg/ml, although the inhibitions at lower concentrations were more pronounced for IL-6 as compared to TNF-α and IL- 1β (Figure 2(a)). In parallel, we assessed the changes in cell viability induced by NPS31807 in THP-1 cells using the CCK-8 incorporation assay. NPS31807 treatment did not significantly decrease cell viability at concentrations up-to 10 μg/ml (Figure 2(a)). In addition, treatment maintained cell viability in epidermal keratinocytes for concentrations up-to 30 μg/ml (data not shown). Next, we evaluated the effect of NPS31807 on TNF-α and IL-8 expression in keratinocyte cells. NHEK cells were stimulated with Oncostatin M (100 ng/ml) and TNF-α (10 ng/ml) for 24 hours since TNF-α is known to induce expression of these genes [25]. We observed a robust upregulation of TNF-α levels (FC: 11.0 ± 0.66) with stimulation after 24 hours (Figure 2(b)). Pre-treatment with NPS31807 led to highly significant dose dependent decrease in TNF-α expression at 1 μg/ml (FC: 0.41 ± 0.19, Figure 2(b)). IL-8 expression was up-regulated by 13(±0.46)-fold in stimulated keratinocytes as compared to un-stimulated cells (Figure 2(c)). NPS31807 (1 μg/ml) treatment reduced expression of IL-8 to 3.2(±1.2)-fold (Figure 2(c)). This potent inhibition in levels of proinflammatory cytokine levels showed that NPS31807 mediates anti-inflammatory activity in macrophages and keratinocytes.
3.2. Anti-Inflammatory Activity of NPS31807 Is Mediated through Decreased Activation of NFκB and AP-1 Signaling
We next evaluated whether, NPS31807 could modulate signaling pathways that regulate inflammation in immune cells. We used luciferase reporter assays (CignalfinderTM) to evaluate regulatory pathway modulation in THP-1 cells. The CignalfinderTM assays consist of a pathway focused (e.g. NFκB, CEBPβ, AP-1, etc.) inducible transcription factor responsive reporter (firefly luciferase) and a constitutive Renilla construct. Undifferentiated THP-1 cells were transfected with individual plasmid reporters, differentiated using PMA and treated with NPS- 31807 for 2 hours preceding LPS stimulation. The assay was controlled using positive and negative controls as
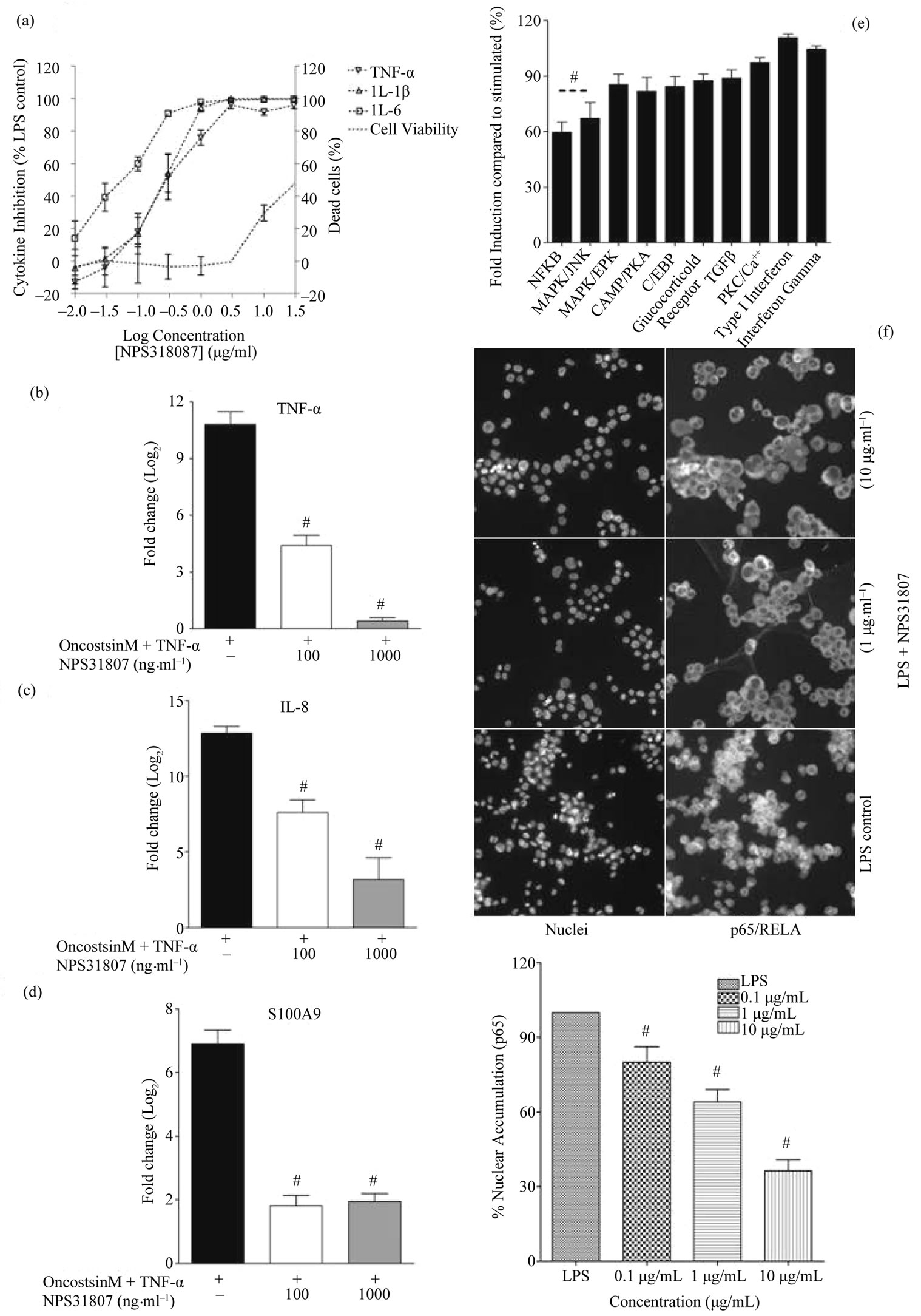
Figure 2. NPS31807 potently modulates pro-inflammatory cytokine levels via NFκB and AP-1 mediated signaling pathways. (a) Pretreatment with NPS31807 induced a concentration dependent reduction in levels of TNF-α, IL-6 and IL-1β in LPS stimulated macrophages. The raw data presents the mean ± SD for cytokine levels from three independent experiments. The right axis demonstrates the % viability associated with treatment; (b) & (c) NHEK cells were pre-treated with NPS31807 (100, 1000 ng/ml) and subsequently stimulated with OSM and TNF-α for 24 hours in triplicate assays. RTq-PCR results are represented as mean ± SD based on Log2 transformation of cycling threshold (Ct) values of TNF-α; (b) IL-8; (c) S100A9; (d) (e) The results represent the reporter expression (mean ± SD) for respective transcription factors compared to LPS control (100%); (f) NPS31807 induced a concentration dependent inhibition of nuclear translocation of p65/RELA in LPS stimulated THP-1 cells (n = 3). The LPS control classically demonstrates extensive nuclear accumulation of p65 in THP-1 nuclei. The bar graph indicates the percent (mean ± SD) nuclear levels of p65 for respective treatment conditions. Statistical significance was associated with p < 0.05.
described by the manufacturer. The relative fluorescence values were recorded for firefly and Renilla luciferase. The signal intensities of the firefly luciferase were normalized using the Renilla luciferase values and percent reporter expression for every reporter was calculated as compared to stimulated (LPS) control. Pre-treatment with NPS31807 (10 μg/ml) reduced significant activation of NFκB (60% ± 7.8%, p < 0.05) and AP-1 (62% ± 4.9%, p < 0.05) reporter activity (Figure 2(d)). Further, to confirm the NFκB activity, the nuclear translocation of p65/RELA was studied using immuno-fluorescence in THP-1 cells. THP-1 cells were treated with multiple concentration of the extract and stimulated with LPS for 15 minutes. LPS induced intense translocation of p65 subunits as shown in Figure 2(e) (LPS control). Treatment with NPS31807 significantly prevented nuclear translocation of p65 in a dose dependent manner (32% ± 2.5%, 10 μg/ml, Figure 2(e)). This substantiated that inhibition in NFκB activity was in part modulated by reduced translocation of p65 sub-unit in THP-1 cells.
3.3. NPS31807 Modulates TNF and Growth Factor Signaling in Human Macrophages
Given the fact that NPS31807 modulates pro-inflammatory cytokine production through NFκB and AP-1 signaling pathways, we further investigated the upstream signaling changes mediated by NPS31807 through the use of gene expression profiling. We used microarrays comprising of 267 genes that play a pivotal role in regulating signaling pathways to understand the mechanistic activity of NPS31807. Differential expression analysis revealed NPS31807 treatment significantly deregulated 66 transcripts (F.C > 2.0, p < 0.05) as compared to stimulated (LPS) control in human macrophages (Supplementary Table 2). Representative transcripts were verified by confirmatory assays such as RTq-PCR and the expression was found to be concordant with microarray data (Figure 3(b)). We observed a decrease in expression of TNF-α and IL-6 transcripts with NPS31807 treatment (Supplementary Table 2) correlating the ELISA results (Figure 2(a)). NPS31807 reduced expression of genes (PLAU, MMP-1, MMP-9, IL-6, CTSD, and IRF1) known to be involved with angiogenesis and matrix metalloproteinase (Supplementary Table 2). Also, NPS31807 treatment induced robust abrogation of VEGF, MMP-9 and MMP-1 expression in cytokine stimulated keratinocytes (Figures 4(f)-(h)). It was also observed that NPS31807 induced potent inhibition of TNFR pathway transcripts such as TNFAIP3, TRAF1, TRADD (Table 1) with expression of TNFSF10 (TRAIL) showing maximum deregulation in our data (16-fold reduction, Supp. Table 2). Interestingly, we found that NPS31807 modulated genes involved with the cellular apoptotic machin ery (BCL2, BID, CASP3, and AKT1) and cell cycle control (CDKN1A, EREG, BTG1). A striking observation from our data revealed decreased expression of epiregulin in THP-1 cells (Supplementary Table 2). The expression of epiregulin was confirmed by RTq-PCR and found to be concordant with the microarray readings (Figure 3(b)). Further, interpretation of the annotations associated with differentially expressed genes; gene set enrichment analysis (GSEA) and PANTHER database were used. The differentially expressed genes (n = 66) were queried against the PANTHER and GSEA databases. PANTHER annotations revealed significant treatment effects on inflammatory, proliferative and angiogenesis pathways as summarized in Supplementary Table 2. On the other hand, gene set enrichment (GSEA) allows the identification of co-expressed genes that are responsible for driving particular cellular/disease phenotypes [21]. Of note, GSEA enrichment analysis revealed a significant overlap of NPS31807 signatures (n = 9) with genes found to be over-expressed by NFκB/RELA expression in keratinocytes (Supplementary Table 3).
3.4. Effect of NPS31807 Treatment on STAT3 Phosphorylation and Cellular Migration
We next evaluated the effect of NPS31807 treatment on STAT3 phosphorylation and cellular migration upon stimulation with EGF. We employed a STAT3 reporter cell line (HeLa/STAT3-luc; Panomics) and used Oncostatin and TNF-α to activate STAT3 phosphorylation. EGF was used to activate STAT3 mediated migration in these cells using wound scratch assays. Stimulation of HeLa/STAT3-luc cells with OSM and TNF-α potently enhanced phosphorylation of STAT3 (F.C = 9.4) and significant dose dependent inhibition was achieved by pre-treatment with NPS31807 (Figure 4(a)). Further, to assess the effect of NPS31807 on mitogen induced migration; HeLa/STAT3-luc cells were plated in a 6-well tissue culture dish and assayed using the scratch wound method as described previously [23]. At the end of 24 hours, EGF (10 ng/mL) enhanced migration across the wound area (Figure 4(c)) compared to basal cells (Figure 4(b)). Pre-treatment with NPS31807 (1 μg/mL) significantly reduced migration compared to EGF alone (12% vs 60%, p < 0.05) (Figures 4(d) and (e)).
3.5. Effect of NPS31807 Treatment on Keratinocyte Proliferation
Changes in cell proliferation were assessed using BrdU uptake in NHEK cells in presence of epidermal growth factor (EGF). We observed a significant increase in BrdU uptake with basal cells (0.3 OD) compared with EGF stimulation (1.9 OD) at the end of 24 hours (Figure 5(a)) in growth factor deprived keratinocytes. NPS31807 treat-

Figure 3. Determination of mechanistic activity of NPS31807 using gene expression profiling (a); Unsupervised, un-centered, Pearson clustering of the normalized expression values from total RNA of LPS stimulated THP-1 cells with or without NPS31807, Dexamethasone, 7-HF, BAY11-7082 and FK-506. The rows represent the mean expression (n = 2) for every gene spotted in the array and columns represent the mean expression changes for individual treatments (n = 2). The list of genes represent differentially expressed genes (n = 66) deregulated by NPS31807 treatment. The color bar indicates the numerical scale for up-regulation (red) and down-regulation (green) for representative expression values (b); RTq-PCR confirmation of key transcripts. The results indicate the Log2 changes in expression values (Mean ± SD) for LPS control (Blank bars) and NPS31807 treatment (Solid bars). Statistical significance (p < 0.05) was assessed by student’s t-test.
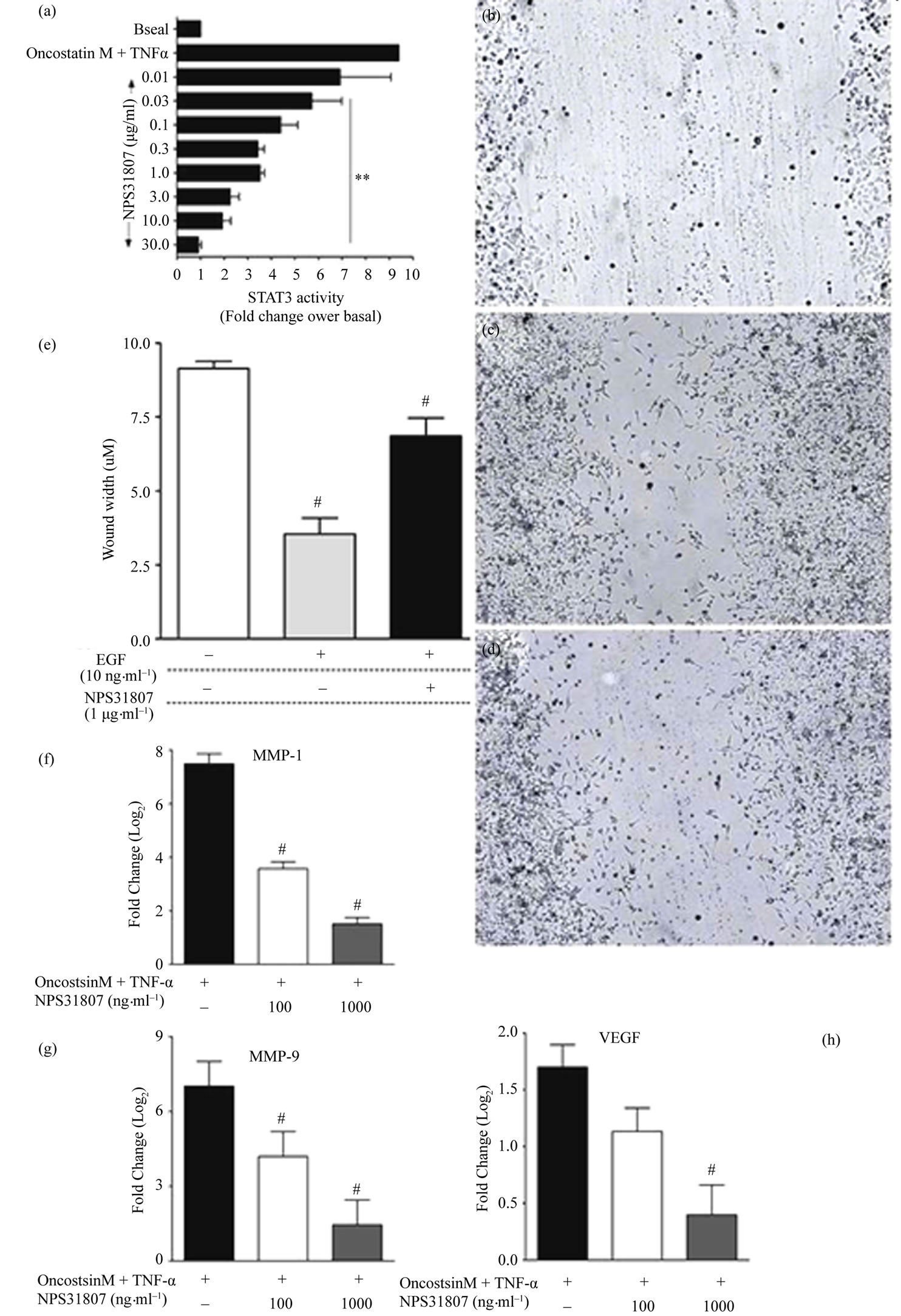
Figure 4. NPS31807 modulates expression of key genes involved with angiogenesis and EGF induced migration through inhibition of STAT3 phosphorylation. (a) NPS31807 induced dose dependent decrease in STAT3 phosphorylation. The results represent the relative fold change in STAT3 phosphorylation for respective treatment conditions with basal as 1-fold. Statistical significance (p < 0.05) was evaluated using 1-way ANOVA with Dunnet’s post hoc testing. Scratch wound induced migration was performed in Hela/STAT3-luc cells and images represent basal condition; (b) Closure of wound area by EGF (10 ng/mL) alone; (c) And wound closure in presence of NPS31807 + EGF; (d) Images represented are raw images without any image enhancements. Bar; 20 μM; (e) Wound distance (μM) was determined from the images and represents the unclosed wound area for respective treatment conditions. Statistical significance (p < 0.05) was evaluated using 1-way ANOVA and Dunnet’s post hoc testing; (f)-(h) NHEK cells were pre-treated with NPS31807 (100, 1000 ng/ml) and subsequently stimulated with OSM and TNF-α for 24 hours in triplicate. RTq-PCR results are represented as Log2 (mean ± SD) of cycling threshold (Ct) values of MMP-1 (f); MMP-9 (g); VEGF (h).
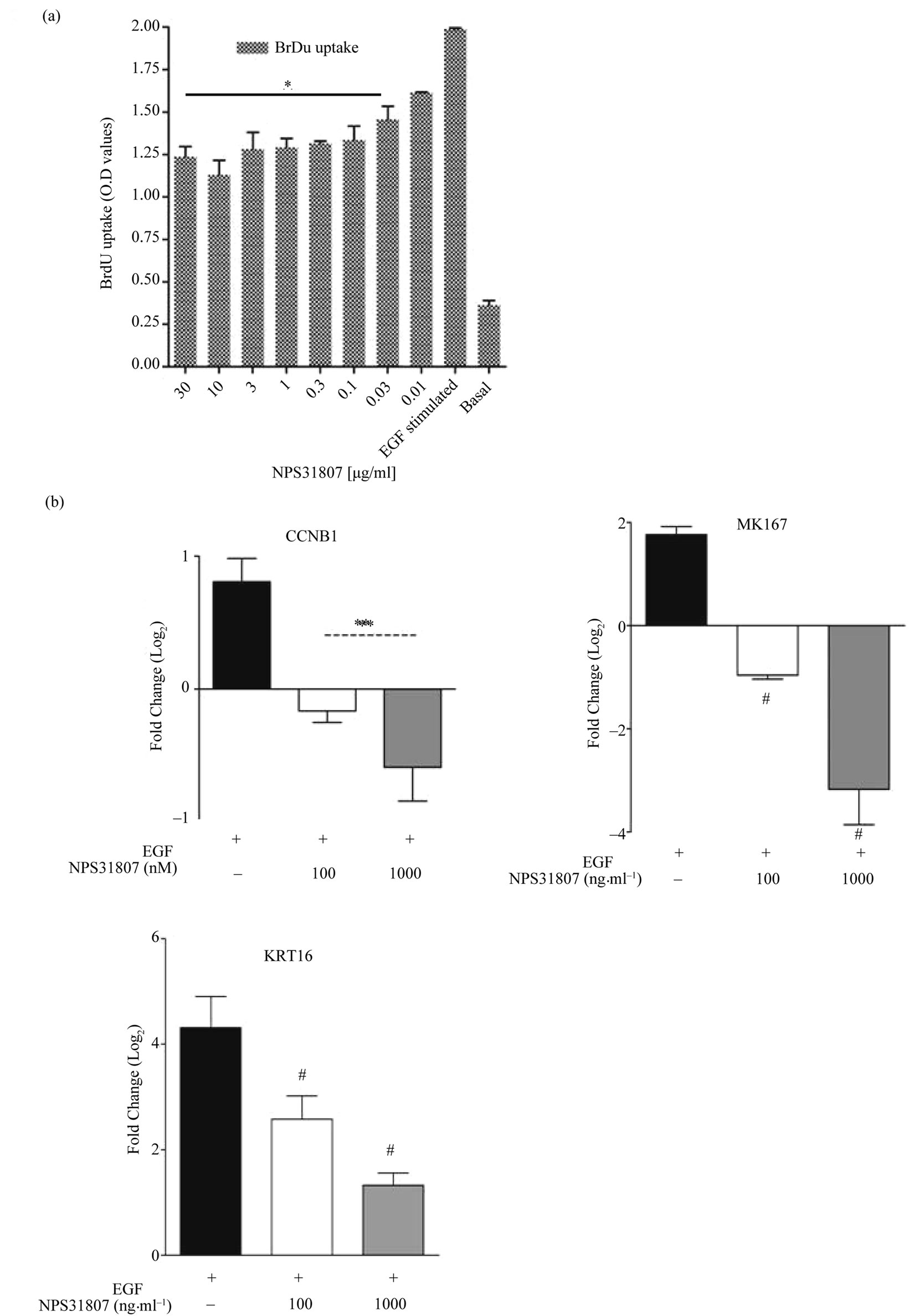
Figure 5. NPS31807 effectively abrogates keratinocyte mitotic index and hyper proliferative transcripts. (a) NHEK cells were treated with NPS31807 and subsequently stimulated with EGF (10 ng/ml) for 24 hours. (a) Pre-treatment with NPS31807 significantly abrogates BrdU uptake in keratinocytes. The results represent the O.D values for respective treatment conditions; (b) RTq-PCR results are represented as mean ± SD based on Log2 transformation of cycling threshold (Ct) values of MKI67; KRT16 and CCNB1. Normalization was carried out using Ct values of GAPDH gene as control. Statistical significance is associated with p < 0.05.
ment led to a substantial decrease in BrdU uptake in EGF stimulated keratinocytes (1.1 O.D, 10 μg/ml). Also, NPS- 31807 treatment reduced expression of MKI67 and CCNB1 (Figure 5(b)), markers associated with proliferative phenotype in epidermal keratinocytes. We further tested the expression of a keratin 16, which is over expressed in proliferative keratinocytes of psoriatic lesional skin. We observed a robust increase in expression of KRT16 upon stimulation with EGF (4.3 ± 0.59 fold increase) and subsequently, pre-treatment with NPS31807 resulted in decrease in KRT16 expression (1.3 ± 0.23-fold) after 24 hours (Figure 5(b)). In summary, these results show that extracts from Sphaeranthus indicus mediate anti-proliferative properties in proliferative keratinocytes.
4. Discussion
The objective of this study was to evaluate the mechanistic activity of extracts from Sphaeranthus indicus under relevant in-vitro models of psoriasis. We employed cell based and gene expression assays for comparative evaluation of the activity of NPS31807. In our experiments, we observed that the moisture content in NPS31807 is in the range of 10% - 12% (w/w), which is higher due to entrapment of moisture in extract and is difficult to evaporate during the drying process. High levels of total ash 11.30% (±0.7%) in the extract indicate increased content of minerals. The extract contained 2.70% (±0.6%) of acid-insoluble ash, which confirms the presence of minerals. The alcohol and methanol extractive value of this plant extract was 74.08 (±9.94%), and 95.66 (±3.55%) respectively. The higher alcoholic extractive value indicates the presence of non-polar and polar phyto-constituents in the extract. High abundance of flavanoids (35%) and bitter content (12%) in the S. indicus extract confirms the diversity of small molecules present in the extract. The extract was standardized using two known marker compounds—7-hydroxylfrullanolide (retention time; 15.52 minutes) and Sphaeranthanolide (retention time; 8.57 minutes) as shown in Figure 1. Pre-treatment with NPS31807 resulted in potent dose dependent inhibition of pro-inflammatory cytokine (e.g., TNF-α, IL-6, IL- 1β) from THP-1 cells (Figure 2(a)). This was corroborated with reduced expression of TNF-α and IL-8 in cytokine stimulated keratinocytes (Figure 2). Interleukin 8 (IL-8) is induced by TNF-α in keratinocytes [26] and is over-expressed in psoriatic lesional skin [27]. Also, TNF- α, IL-6 and IL-1β are over-expressed in patients, correlates with disease severity and subsequent treatment outcomes in psoriasis [28]. The inhibition in cytokine levels was devoid of any decrease in cell viability at the concentrations used in the study (Figure 2). We used OSM and TNF-α to stimulate keratinocytes since both these cytokines are over expressed in psoriatic skin and reproduce human disease conditions in-vitro [25,29]. S100A9, a key mediator of keratinocyte growth [30] was found to be over-expressed in keratinocytes upon stimulation with OncostatinM and TNF-α (Figure 2(d)) in accordance with previously published results [29] and pretreatment with NPS31807 resulted in reduced expression of S100- A9 (Figure 2(d)). Plants of Sphaeranthus spp. exert antiinflammatory effects due to the presence of lactone, flavones and tannins [31]. NPS31807 was found to have appreciable levels of tannin (12%) and flavones (34%) (Table 1) which could explain the anti-inflammatory properties observed in the extract. Also, the novel sesquiterpene lactone (7-HF), isolated from Sphaeranthus indicus [32,33] demonstrates anti-inflammatory activity under in-vitro and in-vivo conditions [14]. This could mean that the anti-inflammatory activity of NPS31807 could be in-part due to the content of 7-HF. Indeed, the primary mechanism of 7-HF was found to be mediated through NFκB [24] in human peripheral blood monocytes. Similarly, NPS31807 reduced nuclear translocation of p65/RELA in THP-1 cells (Figure 2(f)) and reporter expression of NFκB and AP-1 as revealed by plasmid transfection assays (Figure 2(e)). RELA (p65) translocation is established as one of the key steps in the canonical activation of NFκB [34]. Although, interestingly gene expression profiling showed that the expression changes with 7-HF treatment correlated with BAY- 11-7082 (NFκB inhibitor) whereas NPS31807 treatment correlated to that of Dexamethasone (Figure 3(a)). These results prove that although 7-HF is one of the active components of NPS31807, other phyto-constituents synergistically enhance the activities of the extract as observed in our study. NFκB and Rel proteins are expressed in psoriatic skin as compared to normal skin and demonstrate inhibition by anti-TNF-α therapies [35]. Moreover, IL-8 is a target gene of NFκB transcription and modulates key disease activity in psoriasis [36]. The inhibition in reporter expression of NFκB and AP-1 with concomitant inhibition of its target genes; PTGS2 (Figure 3(b)) and IL-8 (Figure 2(c)) effectively confirmed the antiinflammatory activity of NPS31807.Thus, our results show that the phytochemical composition of NPS31807 exerts potent anti-inflammatory properties in immune and keratinocyte cells. A unique observation from microarray data revealed that NPS31807 modulates TNFSF10 (TRAIL) levels in THP-1 cells. TNFSF10 plays a key role in mediating inflammatory and apoptotic responses in variety of cell types [37]. Additionally, TRAIL modulates NFκB mediated cytokine release in macrophages (RAW264) through toll like receptor engagement (TLR) and was found to be regulated by RELA [38]. NPS- 31807 treatment resulted in the inhibition of TRAIL and RELA expression at the end of 24 hours when stimulated with LPS (Supplementary Table 2). This could indicate that NPS-31807 interrupts pro-inflammatory cytokine release through NFκB via abrogation of RELA transcript levels. Further, recent reports have revealed that CD11c+ dendritic cells in lesional skin have higher expression of TNFSF10 in lesional skin and inhibition of TNFSF10 could lead to disease remission in psoriasis [39,40]. Hence, the potent inhibition of TRAIL and RELA transcripts by NPS31807 in macrophages could be considered as an efficacy mechanism of NPS31807 in psoriasis. Inflammation induces angiogenesis, cellular migration of leukocytes in psoriasis. We, first, evaluated the expression of angiogenic mediators in cytokine (TNF-α) stimulated keratinocytes since TNF-α induces angiogenesis in keratinocytes [25]. The distinct down-regulation of VEGF, MMP-1 and MMP-9 in keratinocytes (Figures 4(f)-(h)) confirmed the results observed in THP-1 cells (Supplementary Table 2). Angiogenesis is regulated by STAT3 activity [41] that allows interaction between keratinocytes and immune cells in psoriasis [42]. The potent ihibition of STAT3 activity (Figure 4(a) and EGF induced migration (Figure 4(e)) comprehensively proved that NPS31807 could reduce STAT3 mediated angiogenesis and migration [43].
We also observed that NPS31807 modulated expression of epiregulin—a potent EGFR ligand in THP-1 cells (Supplementary Table 2). The expression of epiregulin was confirmed by RTq-PCR assays (Figure 3(b)). Epiregulin is a classical EGF ligand released through proteolysis by EGFR auto-phosphorylation, expressed in lesional skin and macrophages [44] and induces PTGS2 expression [45,46]. All treatments abolished epiregulin levels except Dexamethasone (Figure 3(a)). Surprisingly, the highest inhibition of epiregulin was achieved by BAY11-7082, a potent NFκB inhibitor [47]. Our results confirm that expression of growth factors such as epiregulin could be mediated by NFκB in inflammatory conditions. Moreover, previous reports have hypothesized that the EGF ligands could be a crucial link connecting immune cell infiltration and epidermal hyperproliferation [48]. Our observation supports this hypothesis and demonstrates that levels of EGF ligands such as epiregulin are modulated in immune cells by cytokine antagonists and could eventually reduce epidermal hyper-proliferation. Immune cell infiltration and cytokine release leads to abnormal hyper-proliferation, differentiation and increased migration in epidermal keratinocytes [40], which finally culminates in epidermal thickening. BrdU is incorporated in the DNA during S-phase, whereas markers of proliferation like MKI67 and CCNB1 express across different phases of cell cycles in keratinocytes [49]. The inhibition in proliferation observed with NPS31807 (Figure 5(a)) is relatively modest as compared to systemic therapies such as methotrexate that induce potent S-phase arrest and concomitant apoptosis [50] in keratinocytes. Keratin 16 (KRT16) is pre-dominantly expressed in hyper-proliferative keratinocytes [51], known to contain EGF responsive elements in its distal promoters [52] and found to be over-expressed in lesional skin. NPS31807 inhibited expression of KRT16, CCNB1 and MKI67 and thus proved that extracts of Sphaeranthus indicus could modulate anti-proliferative effects at the molecular level. This anti-proliferative effect could be explained due to the tannin content found in the extract (Table 1). In summary, our report provides novel fact finding insights into the mechanistic activity of NPS31807 in macrophages and keratinocytes. Our report indicates that NPS31807 induces transcriptional abrogation of NFκB/REL and AP-1 driven pathways in human macrophages and keratinocytes as an effector mechanism for its anti-inflammatory activity. The extract reduced expression of genes involved with angiogenesis in cytokine stimulated keratinocytes and macrophages. Treatment induced inhibition of cellular migration was found to be mediated via ablation in STAT3 activity. NPS31807 revealed anti-proliferative effects in epidermal keratinocytes through reduced BrdU uptake and expression of proliferative genes. The fact that NPS31807 inhibited expression of pathological markers of psoriasis such as keratin 16 (KRT16) and Calgranulin B (S100A9) further substantiates the anti-psoriatic activity of NPS31807. Thus, our results confirm the potential use of NPS31807, an extract from Sphaeranthus indicus as a therapeutic option for psoriasis treatment.
5. Acknowledgements
We acknowledge Dr. Periyasamy Giridharan, Nilesh Dagia, Asha Almeida and Lyle Fonseca for critical discussions and reagents. We wholly acknowledge Sandip Kedar, Monica Mendon and Bindu Hegde for providing valuable technical assistance. Dr. Jayaraman and Dr. Bhaskaran Muthuvelan do not declare any conflict of interest with this work.
REFERENCES
- H. Wagner, “Synergy Research: Approaching a New Generation of Phytopharmaceuticals,” Fitoterapia, Vol. 82, No. 4, 2011, pp. 34-37. doi:10.1016/j.fitote.2010.11.016
- V. J. Galani, B. G. Patel and D. G. Rana, “Sphaeranthus Indicus Linn: A Phytopharmacological Review,” International Journal of Ayurveda Research, Vol. 1, No. 4, 2009, pp. 247-253. doi:10.4103/0974-7788.76790
- A. Shirwaikar, K. S. Prabhu and I. S. Punitha, “In Vitro Antioxidant Studies of Sphaeranthus indicus (Linn),” Indian Journal of Experimental Biology, Vol. 44, No. 12, 2006, pp. 993-996.
- A. Jain and E. Basal, “Inhibition of Propionibacterium Acnes-Induced Mediators of Inflammation by Indian Herbs,” Phytomedicine, Vol. 10, No. 1, 2003, pp. 34-38. doi:10.1078/094471103321648638
- F. Sadaf, R. Saleem, M. Ahmed, S. I. Ahmad and Z. Navaid-Ul, “Healing Potential of Cream Containing Extract of Sphaeranthus indicus on Dermal Wounds in Guinea Pigs,” Journal of Ethnopharmacology, Vol. 107, No. 2, 2006, pp. 161-163. doi:10.1016/j.jep.2006.02.022
- M. P. Schon and W. H. Boehncke, “Psoriasis,” New England Journal of Medicine, Vol. 352, 2005, pp. 1899-1912. doi:10.1056/NEJMra041320
- D. J. Marble, K. B. Gordon and B. J. Nickoloff, “Targeting TNFα Rapidly Reduces Density of Dendritic Cells and Macrophages in Psoriatic Plaques with Restoration of Epidermal Keratinocyte Differentiation,” Journal of Dermatological Science, Vol. 48, No. 2, 2007, pp. 87-101. doi:10.1016/j.jdermsci.2007.06.006
- M. Tomic-Canic, M. Komine, I. M. Freedberg and M. Blumenberg, “Epidermal Signal Transduction and Transcription Factor Activation in Activated Keratinocytes,” Journal of Dermatological Science, Vol. 17, No. 3, 1998, pp. 167-181. doi:0.1016/S0923-1811(98)00016-4
- U. Chaudhari, P. Romano, L. D. Mulcahy, L. T. Dooley, D. G. Baker and A. B. Gottlieb, “Efficacy and Safety of Infliximab Monotherapy for Plaque-Type Psoriasis: A Randomised Trial,” Lancet, Vol. 357, No. 9271, 2001, pp. 1842-1847. doi:10.1016/S0140-6736(00)04954-0
- A. B. Gottlieb, “Etanercept for the Treatment of Psoriasis and Psoriatic Arthritis,” Dermatology and Therapy, Vol. 17, 2004, pp. 401-408. doi:10.1016/S0140-6736(00)02530-7
- A. B. Gottlieb, F. Chamian, S. Masud, I. Cardinale, M. V. Abello, M. A. Lowes, F. Chen, M. Magliocco and J. G. Krueger, “TNF Inhibition Rapidly Down-Regulates Multiple Proinflammatory Pathways in Psoriasis Plaques,” The Journal of Immunology, Vol. 175, No. 4, 2005, pp. 2721-2729.
- R. Bissonnette, V. Ho and R. G. Langley, “Safety of Conventional Systemic Agents and Biologic Agents in the Treatment of Psoriasis,” Journal of Cutaneous Medicine and Surgery, Vol. 13, Suppl. 2, 2009, pp. 67-76.
- S. Amladi, C. Nayak, R. Torsekar, V. Vishalakshi, U. Khopkar, V. Saraf, S. Kulkarni, A. Shindikar, A. Patil, S. Patil, H. Parikh, A. Suthar, V. Chauhan, M. Padigaru, D. Chakrabarti and S. Sharma, “Sphaeranthus indicus (East Indian Globe Thistle)—A Promising Natural Remedy for Psoriasis,” Program of the 68th Annual Meeting of the American Academy of Dermatology, Miami Beach, 5-9 March 2010.
- L. C. Fonseca, S. S. Dadarkar, A. S. Lobo, A. C. Suthar, V. S. Chauhan, S. Chandrababu, S. D. Sharma, N. M. Dagia and M. Padigaru, “7-Hydroxyfrullanolide, a Sesquiterpene Lactone, Inhibits Pro-Inflammatory Cytokine Production from Immune Cells and Is Orally Efficacious in Animal Models of Inflammation,” European Journal of Pharmacology, Vol. 644, No. 1-3, 2010, pp. 220-229. doi:10.1016/j.ejphar.2010.06.052
- D. C. Rio, M. Ares Jr., G. J. Hannon and T. W. Nilsen, “Purification of RNA Using TRIzol (TRI Reagent),” Cold Spring Harbor Protocols, Vol. 10, No. 6, 2010. doi:10.1101/pdb.prot5439
- A. Khanna, K. Mahalingam, D. Chakrabarti and G. Periyasamy, “Ets-1 Expression and Gemcitabine Chemoresistance in Pancreatic Cancer Cells,” Cellular & Molecular Biology Letters, Vol. 16, No. 1, 2011, pp. 101-113. doi:10.2478/s11658-010-0043-z
- S. Rozen and H. Skaletsky, “Primer3 on the WWW for General Users and for Biologist Programmers,” Methods in Molecular Biology, Vol. 132, No. 3, 2000, pp. 365- 386.
- K. J. Livak and T. D. Schmittgen, “Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method,” Methods, Vol. 25, No. 4, 2001, pp. 402-408. doi:10.1006/meth.2001.1262
- E. Staub, J. Groene, M. Heinze, D. Mennerich, S. Roepcke, I. Klaman, B. Hinzmann, E. Castanos-Velez, C. Pilarsky, B. Mann, T. Brummendorf, B. Weber, H. J. Buhr and A. Rosenthal, “Genome-Wide Expression Patterns of Invasion Front, Inner Tumor Mass and Surrounding Normal Epithelium of Colorectal Tumors,” Molecular Cancer, Vol. 6, No. 1, 2007, p. 79. doi:10.1186/1476-4598-6-79
- H. Mi and P. Thomas, “PANTHER Pathway: An Ontology-Based Pathway Database Coupled with Data Analysis Tools,” Methods in Molecular Biology, Vol. 563, Part 2, 2009, pp. 123-140. doi:10.1007/978-1-60761-175-2_7
- A. Subramanian, P. Tamayo, V. K. Mootha, S. Mukherjee, B. L. Ebert, M. A. Gillette, A. Paulovich, S. L. Pomeroy, T. R. Golub, E. S. Lander, J. P. Mesirov, “Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 102, No. 43, 2005, pp. 15545-15550. doi:10.1073/pnas.0506580102
- P. M. Campeau, M. Rafei, M. N. Boivin, Y. Sun, G. A. Grabowski and J. Galipeau, “Characterization of Gaucher Disease Bone Marrow Mesenchymal Stromal Cells Reveals an Altered Inflammatory Secretome,” Blood, Vol. 114, No. 15, 2009, pp. 3181-3190. doi:10.1182/blood-2009-02-205708
- F. Liang, J. Liang, W. Q. Wang, J. P. Sun, E. Udho and Z. Y. Zhang, “PRL3 Promotes Cell Invasion and Proliferation by Down-Regulation of Csk Leading to Src Activation,” The Journal of Biological Chemistry, Vol. 282, No. 8, 2007, pp. 5413-5419. doi:10.1074/jbc.M608940200
- L. C. Fonseca, S. S. Dadarkar, A. S. Lobo, P. B. Mishra, A. D. Thakkar, S. Chandrababu and M. Padigaru, “NFKappaB-Mediated Anti-Inflammatory Activity of the Sesquiterpene Lactone 7-Hydroxyfrullanolide,” European Journal of Pharmacology, Vol. 657, No. 1-3, 2011, pp. 41-50. doi:10.1016/j.ejphar.2011.01.050
- T. Banno, A. Gazel and M. Blumenberg, “Effects of Tumor Necrosis Factor-Alpha (TNF Alpha) in Epidermal Keratinocytes Revealed Using Global Transcriptional Profiling,” The Journal of Biological Chemistry, Vol. 279, No. 31, 2004, pp. 32633-32642. doi:10.1074/jbc.M400642200
- J. Raingeaud and J. Pierre, “Interleukin-4 Downregulates TNFα-Induced IL-8 Production in Keratinocytes,” FEBS Letters, Vol. 579, No. 18, 2005, pp. 3953-3959. doi:10.1016/j.febslet.2005.06.019
- B. J. Nickoloff, R. S. Mitra, J. Varani, V. M. Dixit and P. J. Polverini, “Aberrant Production of Interleukin-8 and Thrombospondin-1 by Psoriatic Keratinocytes Mediates Angiogenesis,” American Journal of Pathology, Vol. 144, No. 4, 1994, pp. 820-828.
- H. Mizutani, Y. Ohmoto, T. Mizutani, M. Murata and M. Shimizu, “Role of Increased Production of Monocytes TNF-α, IL-1β and IL-6 in Psoriasis: Relation to Focal Infection, Disease Activity and Responses to Treatments,” Journal of Dermatological Science, Vol. 14, No. 2, 1997, pp. 145-153. doi:10.1016/S0923-1811(96)00562-2
- K. Boniface, C. Diveu, F. Morel, N. Pedretti, J. Froger, E. Ravon, M. Garcia, E. Venereau, L. Preisser, E. Guignouard, G. Guillet, G. Dagregorio, J. Pene, J. P. Moles, H. Yssel, S. Chevalier, F. X. Bernard, H. Gascan and J. C. Lecron, “Oncostatin M Secreted by Skin Infiltrating T Lymphocytes Is a Potent Keratinocyte Activator Involved in Skin Inflammation,” The Journal of Immunology, Vol. 178, No. 7, 2007, pp. 4615-4622.
- T. Nukui, R. Ehama, M. Sakaguchi, H. Sonegawa, C. Katagiri, T. Hibino and N. H. Huh, “S100A8/A9, a Key Mediator for Positive Feedback Growth Stimulation of Normal Human Keratinocytes,” Journal of Cellular Biochemistry, Vol. 104, No. 2, 2008, pp. 453-464. doi:10.1002/jcb.21639
- B. Adzu, S. Amos, S. D. Kapu and K. S. Gamaniel, “AntiInflammatory and Anti-Nociceptive Effects of Sphaeranthus senegalensis,” Journal of Ethnopharmacology, Vol. 84, No. 2-3, 2003, pp. 169-173. doi:10.1016/S0378-8741(02)00295-7
- U. R. Atta, M. S. Shekhani, S. Perveen, U. R. Habib, A. Yasmin, A. Z. Haque and D. Shaikh, “7-Hydroxyfrullanolide, an Antimicrobial Sesquiterpene Lactone from Sphaeranthus indicus Linn,” European Journal of Pharmacology, Vol. 644, No. 1-3, 1989, pp. 220-229. doi:10.1016/j.ejphar.2010.06.052
- J. S. Sohoni, S. R. Rojatkar, M. M. Kulkarni, N. N. Dhaneshwar, S. S. Tavale, T. N. Gururow and B. A. Nagasampagi, “A New Eudesmanolide and 2-Hydroxycostic Acid from Sphaeranthus indicus Linn: X-Ray Molecular Structure of 4-α 5-α Epoxy-7-α-Hydroxyeudesmanolide,” Journal of the Chemical Society, Perkin Transactions 1, Vol. 2, 1988, pp. 157-160
- S. C. Gupta, C. Sundaram, S. Reuter and B. B. Aggarwal, “Inhibiting NF-KappaB Activation by Small Molecules as a Therapeutic Strategy,” Biochimica et Biophysica Acta, Vol. 1799, No. 10-12, pp. 775-787. doi:10.1016/j.bbagrm.2010.05.004
- P. F. Lizzul, A. Aphale, R. Malaviya, Y. Sun, S. Masud, V. Dombrovskiy and A. B. Gottlieb, “Differential Expression of Phosphorylated NF-KappaB/RelA in Normal and Psoriatic Epidermis and Downregulation of NF-KappaB in Response to Treatment with Etanercept,” Journal of Investigative Dermatology, Vol. 124, No. 6, 2005, pp. 1275-1283. doi:10.1111/j.0022-202X.2005.23735.x
- Y. J. Na, Y. J. Jeon, J. H. Suh, J. S. Kang, K. H. Yang and H. M. Kim, “Suppression of IL-8 Gene Expression by Radicicol Is Mediated through the Inhibition of ERK1/2 and p38 Signaling and Negative Regulation of NF-KappaB and AP-1,” International Immunopharmacology, Vol. 1, No. 9-10, 2001, pp. 1877-1887. doi:10.1016/S1567-5769(01)00113-8
- M. MacFarlane, “TRAIL-Induced Signalling and Apoptosis,” Toxicology Letters, Vol. 139, No. 2-3, 2003, pp. 89-97. doi:10.1016/S0378-4274(02)00422-8
- G. E. Diehl, H. H. Yue, K. Hsieh, A. A. Kuang, M. Ho, L. A. Morici, L. L. Lenz, D. Cado, L. W. Riley and A. Winoto, “TRAIL-R as a Negative Regulator of Innate Immune Cell Responses,” Immunity, Vol. 21, No. 6, 2004, pp. 877-889. doi:10.1016/j.immuni.2004.11.008
- S. Peternel, L. Prpic-Massari, T. Manestar-Blazic, I. Brajac and M. Kastelan, “Increased Expression of TRAIL and Its Death Receptors DR4 and DR5 in Plaque Psoriasis,” Archives of Dermatological Research, Vol. 303, No. 6, 2011, pp. 389-397.
- L. C. Zaba, J. Fuentes-Duculan, N. J. Eungdamrong, L. M, Johnson-Huang, K. E, Nograles, T. R, White, K. C, Pierson, T. Lentini, M. Suarez-Farinas, M. A. Lowes and J. G. Krueger, “Identification of TNF-Related Apoptosis-Inducing Ligand and Other Molecules that Distinguish Inflammatory from Resident Dendritic Cells in Patients with Psoriasis,” Journal of Allergy and Clinical Immunology, Vol. 125, No. 6, 2010, pp. 1261-1268. doi:10.1016/j.jaci.2010.03.018
- D. J. Dauer, B. Ferraro, L. Song, B. Yu, L. Mora, R. Buettner, S. Enkemann, R. Jove and E. B. Haura, “Stat3 Regulates Genes Common to Both Wound Healing and Cancer,” Oncogene, Vol. 24, No. 21, 2005, pp. 3397-3408. doi:10.1038/sj.onc.1208469
- S. Sano, K. S. Chan and J. DiGiovanni, “Impact of Stat3 Activation upon Skin Biology: A Dichotomy of Its Role between Homeostasis and Diseases,” Journal of Dermatological Science, Vol. 50, No. 1, 2008, pp. 1-14. doi:10.1016/j.jdermsci.2007.05.016
- M. Kira, S. Sano, S. Takagi, K. Yoshikawa, J. Takeda and S. Itami, “STAT3 Deficiency in Keratinocytes Leads to Compromised Cell Migration through Hyperphosphorylation of p130(cas),” The Journal of Biological Chemistry, Vol. 277, No. 15, 2002. pp. 12931-12936. doi:10.1074/jbc.M110795200
- H. Toyoda, T. Komurasaki, D. Uchida and S. Morimoto, “Distribution of mRNA for Human Epiregulin, a Differentially Expressed Member of the Epidermal Growth Factor Family,” Biochemical Journal, Vol. 326, Part 1, 1997, pp. 69-75.
- E. Sasaki, R. Pai, F. Halter, T. Komurasaki, T. Arakawa, K. Kobayashi, T. Kuroki and A. S. Tarnawski, “Induction of Cyclooxygenase-2 in a Rat Gastric Epithelial Cell Line by Epiregulin and Basic Fibroblast Growth Factor,” Journal of Clinical Gastroenterology, Vol. 27, Suppl. 1, 1998, pp. S21-S27.
- D. S. Taylor, X. Cheng, J. E. Pawlowski, A. R. Wallace, P. Ferrer and C. J. Molloy, “Epiregulin Is a Potent Vascular Smooth Muscle Cell-Derived Mitogen Induced by Angiotensin II, Endothelin-1, and Thrombin,” Proceedings of the National Academy of Sciences, Vol. 96, No. 4, 1999, pp. 1633-1638. doi:10.1073/pnas.96.4.1633
- T. T. Huang, S. L. Feinberg, S. Suryanarayanan and S. Miyamoto, “The Zinc Finger Domain of NEMO Is Selectively Required for NF-Kappa B Activation by UV Radiation and Topoisomerase Inhibitors,” Molecular and Cellular Biology, Vol. 22, No. 16, 2002, pp. 5813-5825. doi:10.1128/MCB.22.16.5813-5825.2002
- J. T. Elder, “In This Issue,” Journal of Investigative Dermatology, Vol. 123, No. 2, 2004, pp. 6-7. doi:10.1111/j.0022-202X.2004.23240.x
- H. Onuma, C. Mastui and M. Morohashi, “Quantitative Analysis of the Proliferation of Epidermal Cells Using a Human Skin Organ Culture System and the Effect of DbcAMP Using Markers of Proliferation (BrdU, Ki-67, PCNA),” Archives of Dermatological Research, Vol. 293, No. 3, 2001, pp. 133-138. doi:10.1007/s004030000195
- M. Heenen, S. Thiriar, J. C. Noel and P. Galand, “Ki-67 Immunostaining of Normal Human Epidermis: Comparison with 3H-Thymidine Labelling and PCNA Immunostaining,” Dermatology, Vol. 197, No. 2, 1998, pp. 123-126. doi:10.1159/000017982
- M. Kamsteeg, P. A. Jansen, I. M. van Vlijmen-Willems, P. E. van Erp, D. Rodijk-Olthuis, P. G. van der Valk, T. Feuth, P. L. Zeeuwen and J. Schalkwijk, “Molecular Diagnostics of Psoriasis, Atopic Dermatitis, Allergic Contact Dermatitis and Irritant Contact Dermatitis,” British Journal of Dermatology, Vol. 162, No. 3, 2010, pp. 568- 578. doi:10.1111/j.1365-2133.2009.09547.x
- L. Rittie, S. Kansra, S. W. Stoll, Y. Li, J. E. Gudjonsson, Y. Shao, L. E. Michael, G. J. Fisher, T. M. Johnson and J. T. Elder, “Differential ErbB1 Signaling in Squamous Cell Versus Basal Cell Carcinoma of the Skin,” The American Journal of Pathology, Vol. 170, No. 6, 2007, pp. 2089- 2099. doi:10.2353/ajpath.2007.060537
Supplementary Tables

Supplementary Table 1. Primer sequences used for RTq-PCR.




Supplementary Table 2. Gene expression data and biological annotations.
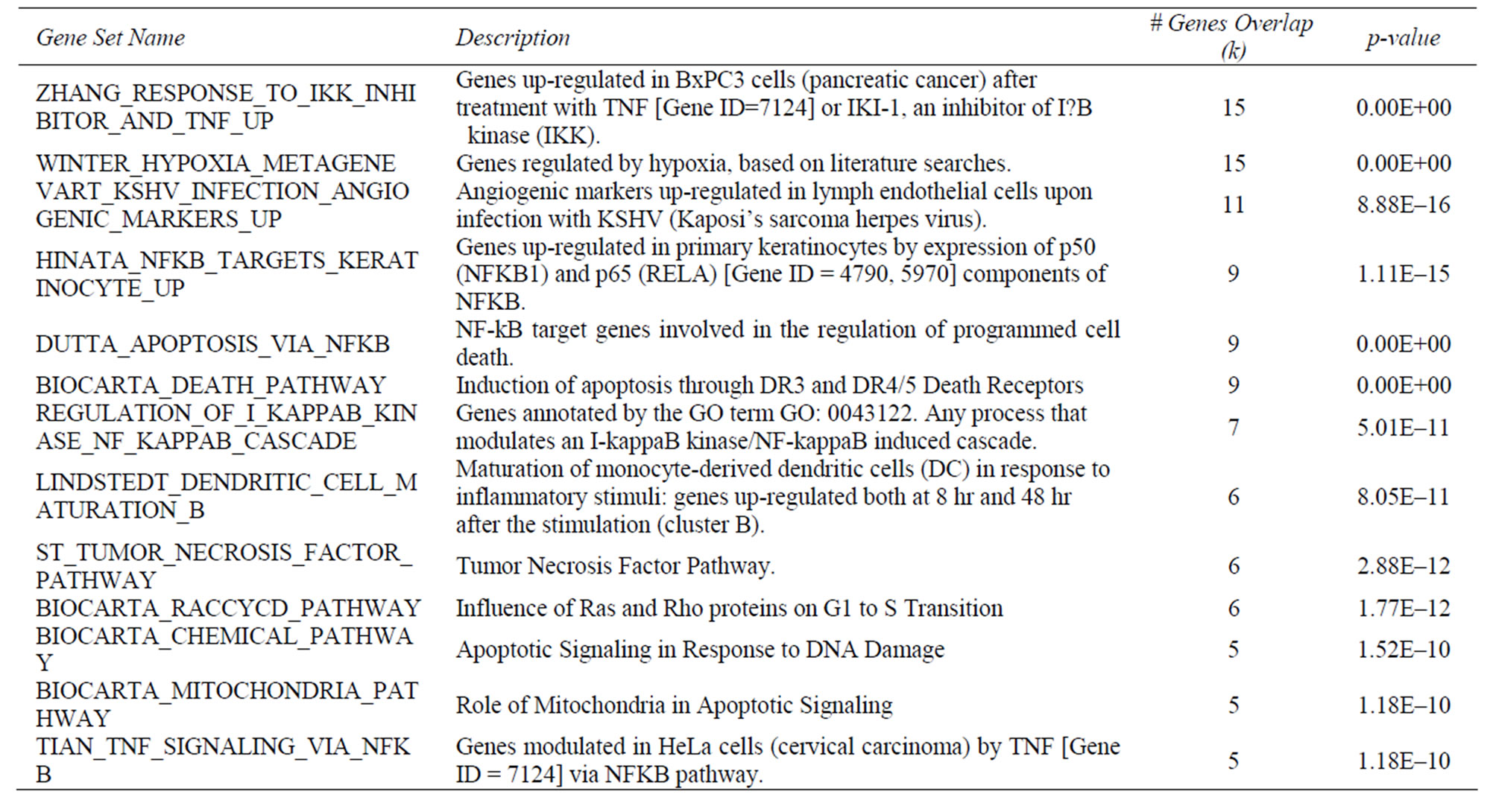
Supplementary Table 3. Selected GSEA results for gene set overlap for down-modulated transcripts.
NOTES
*Corresponding authors.

