Journal of Biosciences and Medicines
Vol.06 No.04(2018), Article ID:84102,11 pages
10.4236/jbm.2018.64008
Antibiotics Resistance Pattern and Plasmid Profiling of Edwardsiella tarda Isolated from Heterobranchus longifilis
F. C. Ogbonne, E. R. Ukazu, F. C. Egbe
Nigerian Institute for Oceanography and Marine Research, Lagos, Nigeria
Copyright © 2018 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: March 17, 2018; Accepted: April 24, 2018; Published: April 27, 2018
ABSTRACT
A study was carried out to investigate antibiotic resistance patterns and plasmid profiling of Edwardsiella tarda isolated from farmed-cultured Heterobranchus longifilis in Lagos State, Southwest of Nigeria. A total of 44 Edwardsiella isolates were recovered from 80 fish samples collected from the 10 fish farms using selective random stratification. It was observed that Edwardsiella tarda isolates were 100% resistant to Amoxicillin, Chloranphenicol, Levofloxacin, Streptomycin and 90% resistant to Nalidixic Acid respectively. All the isolates were 100% susceptible to Spectinomycin and Ciprofloxacin, while Ofloxacin, Gentamycin, and Pefloxacin vary in their level of susceptibility with 90%, 80% and 70% sensitivity respectively. Conversely, 8 out of 10 fish farm locations studied were observed to have antibiotic-resistant strains, and 5 out of 8 drug-resistant strains were found to carry plasmid and the sizes of the plasmid ranges between 20.027 kb to 23.130 kb. The plasmid after treatment with mitomycin C and ethidium bromide were lost during the process of plasmid curing confirming that the multiple drug resistant exhibited by the isolates was plasmid mediated. There are fewer studies on antibiotic resistance in Edwardsiella tarda from aquaculture enterprises and this study provides further support to the view that there is a potential risk of transfer of resistant bacteria and their genes to human pathogen through the food chain. Although, in Nigeria there is no antibiotic product registered for aquaculture usage, yet fish farmers use them off-label for bacterial diseases prevention.
Keywords:
Edwardsiella tarda, Plasmid Profiling, Antibiotic Resistance, Heterobranchus longifilis

1. Introduction
Antibiotic resistance is a major public health concern that has emerge over the recent years as several microbes that were previously susceptible to antibiotics have evolved to become insensitive to drugs even at higher concentrations perhaps due to mutation and apparent indiscriminate use and exposure or abuse of such drugs by fish farmers that use them for diseases prevention. The emergence of antibiotic resistance in microbes such as Enterococci, Salmonella, Escherichia coli, Aeromonas and Edwardsiella tarda in fish poses a potential threat of transfer of their resistance genes to human pathogen through the food chain [1] [2] . Antibiotic resistance represents one of the greatest current threats to public health, and is predicted to overtake cancer as a cause of death by 2050 [3] . Nevertheless, there has been little or no attention paid to the potential use of antibiotics in aquaculture industries. In addition to transfer of antibiotics resistant microorganisms and their genes through consumption of contaminated fish, there is significant risk of environmental contamination consequently due to practice of using medicated feeds.
Edwardsiella tarda is a gram-negative, facultative anaerobic, short, rod-shaped and motile bacterium of the family Enterobacteriaceae, and was first characterized in 1965 [4] . Its motility is due to peritrichous flagella. They are normal gut micro flora of fish and occasionally opportunistic pathogen in human, causing gastroenteritis and diarrhea [5] . E. tarda is an important pathogen in terms of public health, since it progresses as an epizootic and zoonotic bacterium. E. tarda is an enteric bacterial pathogen that present in a world-wide distribution and a high variety of hosts [6] [7] . Moreover, Edwardsiellosis caused by E. tarda is a devastating fish disease in freshwater and marine aquaculture industries throughout the world and accounts for severe economic losses [8] . It has also been reported as the causative agent of the systemic diseases known as Emphysematous Putrefactive Disease of catfish (EPDC).
One of the significant challenges threatening sustainable aquaculture industry today is bacterial fish diseases caused by Edwardsiella tarda, Flavobacterium, Aeromonas sp., Vibrio sp. and Streptococcus iniae, hence the need for antibiotic use, although many countries no longer permit the use of antibiotic for aquaculture purposes. In 1998, Alderman and Hastings [9] highlighted that control of antibiotic use in aquaculture vary widely from country to country. In developed nations such as United State of America, Canada, Norway and other European Union members, there are a limited number of products with strong regulatory control and use of antibiotics is on the decline rate as a result of improvements in the management and development of effective vaccines [10] . However, ninety percent of aquaculture production occurs in developing countries [11] where there is little or no regulatory control of antibiotics use appears to be widespread. In Nigeria and other ECOWAS countries with small but rapidly developing aquaculture industries and with rigorous regulatory control, no antibiotics are registered for aquaculture usage. Unfortunately, restrictions on the availability of registered products can lead to pressure to use antibiotics off-label or even completely illegitimately. Antibiotics resistance has been reported in shrimps [12] , eels [13] , and aquaculture environments [14] [15] . Some reports have observed that resistance emerges within few years of treating infections with antibacterial drugs [16] and this is a factor, which limits their value in the control of bacterial fish diseases [17] . Apart from any public health concerns, recognition of the resistance issue has led to calls for intensified surveillance of antibiotic use and antibiotic resistance [18] . This study was aimed at determining the resistance of Edwardsiella tarda from Heterobranchus longifilis to antibiotics commonly used in aquaculture operations and to investigate their plasmid profile.
2. Materials and Methods
2.1. Sample Collection and Processing
80 fish samples were collected from 10 different fish farm locations using selective random stratification within Lagos State of Nigeria for a period of six months. The samples were transported to aquaculture laboratory of Nigerian Institute for Oceanography and Marine Research for diseases examination and microbiological analysis. Fish samples were aseptically dissected using a dissecting kit.
2.2. Isolation and Identification of Edwardsiella tarda
Swabs were taken aseptically from the liver, skin, intestine, gills and kidney of the dissected fish samples, and were streaked on petridish containing Tryptone soy agar (LAB M Ltd., United Kingdom) that was prepared according to manufacturers’ instructions. The plates were incubated at 37˚C for 24 hours and were observed for bacterial growth and distinct colonies were further sub-cultured on freshly prepared tryptone soy agar to obtain pure culture of the isolates. The isolates were then identified using gram staining technique, morphology, motility, biochemical reaction and sugar fermentation according to (Bergey’s Manual of determinative bacteriology, 7th Edition).
2.3. Antibiotic Susceptibility Testing
A commercially available antimicrobial susceptibility test single disc from Oxoid Ltd, Basingstoke, RG24 8PW, United Kingdom, was used. Pure culture of Edwardsiella tarda were picked from Tryptone soy broth using a sterile wire loop and transferred to tubes each containing 5 ml of sterile physiological saline. The suspension was vortexed and adjusted to match 0.5 McFarland turbidity standards. Sterile swab stick was then dipped, rotated and pressed firmly on the tube walls above the culture to remove excess inoculums from the swabs. This was then evenly swabbed on the dried surface of Mueller-Hinton agar (Oxoid Ltd., England) plates ensuring even distribution of the bacterium. 10 different antimicrobial susceptibility single discs were placed on the plates using sterile forceps and were incubated at temperature of 37˚C for 18 to 24 hours, after which the diameter of zone of inhibition were measured with white transparent meter rule in millimeter. The methods describe above and the interpretations of results were done using standard recommended by Clinical Laboratory Standard Institute [19] . The ten antibiotics used have the following concentrations: Nalidixic Acid (30 µg), Gentamycin (10 µg), Pefloxacin (5 µg), Ofloxacin (5 µg), Streptomycin (25 µg), Levofloxacin (5 µg), Chloranphenicol (30 µg), Spectinomycin (10 µg), Ciprofloxacin (10 µg), Amoxycillin (25 µg).
2.4. Plasmid Extraction
Plasmid was isolated from overnight pure culture of Edwardsiella tarda following a protocol of Fast-n-Easy Plasmid Mini-Prep Kit (Column based isolation of plasmid DNA) from Jena Bioscience, Germany. The kit contains Lysis buffer, Neutralization buffer (containing RNase A), Activation buffer, washing buffer (containing ethanol), and Elution buffer. The bacterial cells were harvested from the culture and centrifuged. The pelleted cells were resuspended in 300 µl Lysis Buffer by vortexing for 1 minute. 300 µl of Neutralization Buffer (containing RNase A) was added to the sample and then mix gently by inverting the tube 4 - 6 times, then Centrifuge at 10,000 g for 5 minutes at room temperature in a microcentrifuge until the colour of the binding mixture change to bright yellow indicating a pH of 7.5 required for optimal DNA binding. A binding Column was place into a 2 ml collection tube and 100 µl of Activation Buffer was added into the Binding Column, then Centrifuge at 10,000 g for 30 seconds in a micro-centrifuge. Thereafter, the supernatant was applied into the activated Binding column by decanting, and Centrifuge again at 10,000 g for 30 seconds. The DNA loaded Binding Colum was place into the 2 ml collection tube and 500 µl of Washing Buffer was applied to the Binding Column for washing. Thereafter, centrifuge at 10,000 g for 30 seconds, and 700 µl of Washing Buffer was added to the Binding Column, and Centrifuge again for 2 minutes to remove residual Washing Buffer. Finally, the elution was carried out by placing the Binding Column into a clean 1.5 ml microtube with addition of 50 µl Elution Buffer to the center of the column membrane, and then incubated for 1 minute at room temperature and Centrifuge at 10,000 g for 1 minute to elute DNA.
2.5. Agarose Gel Electrophoresis
Plasmid was separated on a 1.5% agarose gel and electrophoresis was carried out at 80 V for 1 hour 30 minutes. After electrophoresis, the gels were stained for 2 hours in 0.5 µg of ethidium bromide solution, destained in water, and photographed with Polaroid MP4 camera, using type 55 P/N instant film and 23A and 2B filters.
2.6. Plasmid Curing Procedure
Plasmid curing was conducted following a method described by Alicia et al. [20] . 24 hour culture were grown in a broth containing 0.5 and 0.05 µg of mitomycin C per ml. ethidium bromide concentrations used were 0.4 and 0.04 mg/ml. after treatment, bacterial cells were plated on a solid medium to observe their growth. The plates were replicated into a fresh medium containing appropriate antibiotics. Both colonies which grew on medium with antibiotics and those that fails to grow were picked randomly from the master plates to test for plasmid content and antibiotic susceptibility.
2.7. Statistical Analysis
The correlation of plasmids to antibiotic resistance patterns was calculated using Statistical Packages for Social Sciences (SPSS) software, version 20 (IBM).
3. Results
The bacteria were isolated from the kidney, gills and intestines of Heterobranchus longifilis from 10 different fish farms location in Ikorodu, Epe and Ajah area of Lagos State, Nigeria. Table 1 and Plate 1 shows morphological, biochemical characteristics and gram staining photomicrograph of the isolates, which revealed that the isolates were Edwardsiella tarda. A total of 44 Edwardsiella isolates were recovered from 80 fish samples collected from the 10 fish farms studied. Results shows that Edwardsiella tarda isolates were 100% resistant to Amoxicillin, Chloranphenicol, Levofloxacin, Streptomycin and 90% resistant to Nalidixic Acid respectively (Figure 1). All the isolates were 100% susceptible to Spectinomycin and Ciprofloxacin. Conversely, other antibiotics used in this study shows variable level of sensitivity and they include: Ofloxacin (90%), Gentamycin (80%) and Pefloxacin (70%) respectively (Figure 1). Figure 2 shows the plasmid profiling of the isolates that were resistant to the tested antibiotics.
Table 1. Morphological and biochemical characteristics of the isolated Edwardsiella tarda.
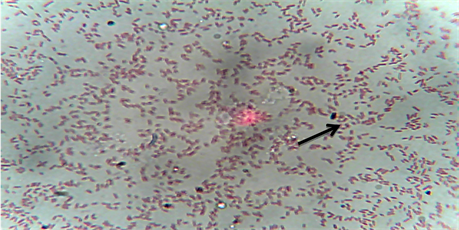
Plate 1. Gram stain photomicrograph of the isolates. Edwardsiella tarda is indicated with an arrow.
Figure 1. The percentage of antibiotics sensitivity and resistant patterns of Edwardsiella tarda isolates.
However, 5 out of 8 drug-resistant strains were found to carry plasmid and the sizes of the plasmid ranges between 20.027 kb to 23.130 kb. Figure 3 shows the plasmid curing of drug-resistant isolates to determine changes in plasmid content associated with antibiotic resistance pattern after the plasmid responsible for the isolates drug-resistant has been knocked off.
The relationship between the antibiotics resistance patterns and all the Edwardsiella strains isolated in this study are shown in Table 2.
4. Discussion
Edwardsiellosis is one of the most significant bacterial diseases in fishes and it causes massive mortalities in the various populations and age groups of fish and consequently high economic losses [21] . Therefore, antibiotic resistance by E. tarda is a call for concern. Resistance patterns observed in this study could not be attributed to frequent antibiotic use as none were actually registered for aquaculture usage in Nigeria. However, the results have some similarity to those seen in countries where antibiotics were allowed to be used for aquaculture
Figure 2. Plasmid profile of Edwardsiella tarda isolated from Heterobranchus longifilis. Lane M = DNA ladder (Contain the standard reference molecular weight). L3 = E. tarda isolated from fish farm location 3 showing no plasmid. L4 = E. tarda isolated from fish farm location 4 having plasmid size of 23.130 kb. L5 = E. tarda isolated from fish farm location 5 having plasmid size of 23.130 kb. L6 = E. tarda isolated from fish farm location 6 showing no plasmid. L7 = E. tarda isolated from fish farm location 7 showing no plasmid. L8 = E. tarda isolated from fish farm location 8 having plasmid size of 23.130 kb. L9 = E. tarda isolated from fish farm location 9 having plasmid size of 23.130 kb. L10 = E. tarda isolated from fish farm location 10 having plasmid size of 2.027 kb.
Figure 3. Loss of plasmids by Edwardsiella tarda after curing. Lane M = 564 bp DNA ladder. L4, L5, L8, L9 and L10 indicate lanes with loss of plasmids after curing. Horizontal lines indicate plasmid loss areas.
Table 2. Antibiotics resistance patterns to all Edwardsiella strains isolated in this study.
operations. Resistance to levofloxacin, streptomycin, amoxicillin, Chloranphenicol and Nalidixic acid was prevalent in this study. Resistance to amoxicillin and Chloranphenicol is not shocking as it has been reported in many studies in different countries [22] [23] [24] . In contrast, no resistance to Spectinomycin and ciprofloxacin was observed. However, our results showed resistance to more than one class of antibiotic and this is in agreement with the work of Lee et al. [25] whose study reported multiple drug resistance by E. tarda in freshwater fish. Furthermore, a total of 10 fish farms location were surveyed for antibiotics resistant strain of Edwardsiella tarda, 8 out of 10 fish farm locations studied were observed to have antibiotic-resistant strains, and 5 out of 8 drug-resistant strains have plasmid. The plasmid after treatment with mitomycin C and ethidium bromide were lost during the process of plasmid curing while trying to determine whether the drug resistant is plasmid mediated or not and this is in line with the work of Alicia et al. [20] who characterize plasmids in bacterial fish pathogens. Also, only one out of the eight isolates tested were found to carry 2 different sizes of plasmids (20.027 kb and 23.130 kb) while others have one sized plasmid of high molecular weight (23.130 kb). The number of plasmids isolated from the Edwardsiella strains used in this study did not show any significant correlation to antibiotic resistance patterns P (>0.05).
The presence of high molecular weight plasmids in fish farms and environments has been reported [26] and transferable R-plasmids conferring resistance to various antimicrobials including chloramphenicol, streptomycin and Nalidixic acid were reported many years ago [27] . Most of these plasmids are capable of being transferred to other organisms as well as transferring their resistance genes [28] . The use of antibiotics in aquaculture has been for the purpose of managing and treating fish disease. These synthetic drugs effectively inhibit bacterial cell wall synthesis, protein synthesis, and DNA function [29] . However, the indiscriminate use of the antibiotics in fish farming has caused the development of antibiotics resistant in fish pathogen [30] . Studies from other researchers has shown that the reservoirs of antibiotic resistance can interact between different ecological systems and the possibility of transfer of resistant bacteria or their genes from animals to humans may occur through the food chain [31] [32] . Furushita and other researchers [26] , have noted that tetracycline resistance genes from fish farm isolates were similar to those detected in clinical isolates. Nevertheless, the use of antibiotics in aquaculture operations should be totally discouraged and adequate fish health management system and quality control should be adopted.
Cite this paper
Ogbonne, F.C., Ukazu, E.R. and Egbe, F.C. (2018) Antibiotics Resistance Pattern and Plasmid Profiling of Edwardsiella tarda Isolated from Heterobranchus longifilis. Journal of Biosciences and Medicines, 6, 95-105. https://doi.org/10.4236/jbm.2018.64008
References
- 1. Angulo, F.J., Nargund, V.N. and Chiller, T.C. (2004) Evidence of an Association between Use of Antimicrobial Agents in Food Animals and Antimicrobial Resistance among Bacteria Isolated from Humans and the Human Health Consequences of Such Resistance. Journal of Veterinary Medicine B, 51, 374-379. https://doi.org/10.1111/j.1439-0450.2004.00789.x
- 2. Stobberingh, E.E. and Van den Bogaard, A.E. (2000) Spread of Antibiotic Resistance from Food Animals to Man. Acta Veterinaria Scandinavica, 93, 47-52.
- 3. WHO (World Health Organisation) (2002) Use of Antimicrobials outside Human Medicine and Resultant Antimicrobial Resistance in Humans. Fact Sheet Number 268, 2.
- 4. Health Canada (2008) Edwardsiella tarda—Material Safety Data Sheets (MSDS). Public Health Agency of Canada. http://www.phac-aspc.gc.ca/msds-ftss/msds57e-eng.php
- 5. Verjan, N., Ikuo, H. and Takashi, A. (2005) Genetic Loci of Major Antigenic Protein Genes of Edwardsiella tarda. Journal of Applied Environmental Microbiology, 71, 5654-5658. https://doi.org/10.1128/AEM.71.9.5654-5658.2005
- 6. Austin, B. and Austin, D.A. (2007) Bacterial Fish Pathogens: Diseases of Farmed and Wild Fish. Praxis Publishing Ltd., Chichester.
- 7. Plumb, J.A. (1993) Edwardsiella Septicemia. In: Inglis, V., Roberts, R.J. and Bromage, N.R., Eds., Bacterial Diseases of Fish, Blackwell Scientific Publications, Oxford, 61-79.
- 8. Mohanty, B.R. and Sahoo, P.K. (2007) Edwardsiellosis in Fish: A Brief Review. Journal of Bioscience, 32, 1331-1344. https://doi.org/10.1007/s12038-007-0143-8
- 9. Alderman, D.J. and Hastings, T.S. (1998) Antibiotic Use in Aquaculture: Development of Antibiotic Resistance—Potential for Consumer Health Risks. International Journal of Food Science and Technology, 33, 139-155. https://doi.org/10.1046/j.1365-2621.1998.3320139.x
- 10. Lillehaug, A., Lunestad, B.T. and Grave, K. (2003) Epidemiology of Bacterial Diseases in Norwegian Aquaculture—A Description Based on Antibiotic Prescription Data for the Ten-Year Period 1991 to 2000. Journal of Diseases of Aquatic Organisms, 53, 115-125. https://doi.org/10.3354/dao053115
- 11. Bondad-Reantaso, M.G., Subasinghe, R.P., Arthur, J.R., Ogawa, K., Chinabut, S., Adlard, R., Tan, Z. and Shariff, M. (2005) Disease and Health Management in Asian Aquaculture. Journal of Veterinary Parasitology, 132, 249-272. https://doi.org/10.1016/j.vetpar.2005.07.005
- 12. Tjahjadi, M.R., Angka, S.L. and Suwanto, A. (1994) Isolation and Evaluation of Marine Bacteria for Bio-control of luminous Bacterial Disease in Tiger Shrimp Larvae (Penaeus monodon, Fab.). Asian-Pacific Journal of Molecular Biology and Biotechnology, 2, 347-352.
- 13. Alcaide, A., Blasco, M.D. and Esteve, C. (2005) Occurrence of Drug-Resistant Bacteria in Two European Eel Farms. Journal of Applied Environmental Microbiology, 71, 3348-3350. https://doi.org/10.1128/AEM.71.6.3348-3350.2005
- 14. Chelossi, E., Vezzulli, L., Milano, A., Branzoni, M., Fabiano, M., Ricarrdi, G. and Banat, I.M. (2003) Antibiotic Resistance of Benthic Bacteria in Fish Farm and Control Sediments of Western Mediterranean. Journal of Aquaculture, 219, 83-97. https://doi.org/10.1016/S0044-8486(03)00016-4
- 15. Kim, S.R., Nonaka, L. and Suzuki, S. (2004) Occurrence of Tetracycline Resistance Genes tet (M) and tet (S) in Bacteria from Marine Aquaculture Sites. FEMS Microbiology Letter, 237, 147-156. https://doi.org/10.1111/j.1574-6968.2004.tb09690.x
- 16. Sorum, H. (1999) Antibiotic Resistance in Aquaculture. Acta Veterinaria Scandinavica. Supplementum, 92, 29-36.
- 17. Smith, P., Hiney, M. and Samuelson, O. (1994) Bacterial Resistance to Antimicrobial Agents Used in Fish Farming: A Critical Evaluation of Method and Meaning. Annual Review Fish Disease, 4, 273-313. https://doi.org/10.1016/0959-8030(94)90032-9
- 18. Aoki, T. (1992) Present and Future Problems Concerning the Development of Resistance in Aquaculture. In: Michel, C. and Alderman, D., Eds., Chemotherapy in Aquaculture: From Theory to Reality, Office International des Epizooties, Paris, 254-262.
- 19. Clinical Laboratory Standards Institute (2016) Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute, Wayne.
- 20. Alicia, E.T., Juan, L.B., Rita, R.C. and Frank, M.H. (1983) Characterization of Plasmids in Bacterial Fish Pathogens. Journal of Infection and Immunity, 39, 184-192.
- 21. Jun, Z. and Yin, L. (2006) Functional Studies of a Type III and a Novel Secretion System in E. tarda. PhD Thesis, National University of Singapore, Singapore.
- 22. Hatha, M., Vivekanandam, A.A., Joice, G.J. and Christol, G.J. (2005) Antibiotic Resistance Pattern of Motile Aeromonads from Farm Raised Freshwater Fish. International Journal of Food Microbiology, 98, 131-134. https://doi.org/10.1016/j.ijfoodmicro.2004.05.017
- 23. Ho, S.P., Hsu, T.Y., Chen, M.H. and Wang, W.S. (2000) Antibacterial Effects of Chloramphenicol, Thiamphenicol and Florfenicol against Aquatic Animal Bacteria. Journal of Veterinary Medical Science, 62, 479-485. https://doi.org/10.1292/jvms.62.479
- 24. Mirand, C.D. and Zemelman, R. (2002) Antimicrobial Multi-Resistance in Bacteria Isolated from Freshwater Chilean Salmon Farms. Science of the Total Environment, 293, 207-218. https://doi.org/10.1016/S0048-9697(02)00022-0
- 25. Lee, S.W., Najiah, M., Chuah, T.S., Noor azhar, M.S., Wendy, W., Nadira, M. and Mohd, E.A.W. (2011) Antibiogram and Plasmid Profiling from Edwardsiella tarda Isolated from Freshwater Fish in East Coast Malaysia. Journal of Sustainability Science and Management, 6, 19-27.
- 26. Furushita, M., Shiba, T., Maeda, T., Yahata, M., Kaneoka, A., Takahashi, Y., Torii, K. and Hasegawa, T. (2003) Similarity of Tetracycline Resistance Genes Isolated from Fish Farm Bacteria to Those from Clinical Isolates. Journal of Applied Environmental Microbiology, 69, 5336-5342. https://doi.org/10.1128/AEM.69.9.5336-5342.2003
- 27. Aoki, T. and Takahashi, A. (1987) Class D Tetracycline Resistance Determinants of R-Plasmids from Fish Pathogens Aeromonas hydrophila, Edwardsiella tarda and Pasteurella piscisida. Journal of Antimicrobial Agents and Chemotherapy, 31, 1278-1280. https://doi.org/10.1128/AAC.31.8.1278
- 28. Aoki, T. (1988) Drug Resistance Plasmids from Fish-Pathogens. Journal of Microbiological Science, 5, 219-223.
- 29. Harold, N.C. (1992) The Crisis in Antibiotics Resistance. Science, 257, 1064-1073.
- 30. Sarter, S., Nguyen, H.N.K., Hung, L.T., Lazard, J. and Montet, D. (2007) Antibiotic Resistance in Gram Negative Bacteria Isolated from Farmed Catfish. Journal of Food Control, 18, 1391-1396. https://doi.org/10.1016/j.foodcont.2006.10.003
- 31. Teuber, M. (2001) Veterinary Use and Antibiotic Resistance. Current Opinion in Microbiology, 4, 493-499. https://doi.org/10.1016/S1369-5274(00)00241-1
- 32. Van den Bogaard, A.E. and Stobberingh, E.E. (2000) Epidemiology of Resistance to Antibiotics Links between Animals and Humans. International Journal of Anti-microbial Agents, 14, 327-335. https://doi.org/10.1016/S0924-8579(00)00145-X





