Journal of Biosciences and Medicines
Vol.05 No.04(2017), Article ID:75957,20 pages
10.4236/jbm.2017.54008
Identifying Changes in Punitive Transcriptional Factor Binding Sites Created by PPARα/δ/γ SNPs Associated with Disease
Norman E. Buroker
Department of Pediatrics, University of Washington, Seattle, WA, USA

Copyright © 2017 by author and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: March 29, 2017; Accepted: April 27, 2017; Published: April 30, 2017
ABSTRACT
Single nucleotide polymorphisms (SNPs) located in the PPARα/δ/γ genes that have been previously found to be significantly associated with human disease or a condition were also found to alter the genes punitive transcriptional factor binding sites (TFBS). Two alleles (C/G) of the PPARα SNP (rs1800206) were found to generate 7 common and 8 unique punitive TFBS. One of the unique TFBS created by the minor G (0.02) allele is for the T-Box 4 (TBX4) transcription factor which is associated with heritable pulmonary arterial hypertension. Two alleles (A/G) of the PPARδ SNP (rs2016520) were found to generate 20 unique punitive TFBS while the two alleles (C/G) of the PPARδ SNP (rs9794) were found to generate 11 common and 11unique punitive TFBS. The alleles of the PPARγ SNPs (rs10865710, rs12629751, rs709158, rs1805192 and rs3856806) were found to generate 15, 12, 16, 2 and 21 common and 9, 4, 12, 4 and 7 unique punitive TFBS, respectively. These changes in TFBS are discussed with relation to alterations in gene expression that may result in disease or change in human condition.
Keywords:
PPARα/δ/γ Gene Regulation, SNPs, TFBS, Human Disease

1. Introduction
The peroxisome proliferator-activated receptors (PPARs) are ligand-activated transcriptional factors (TFs) that regulate many genes in cell differentiation and various metabolic processes including lipid and glucose homeostasis. They are nuclear hormone receptors belonging to a steroid receptor superfamily that include estrogen, thyroid hormone, vitamin D3 and glucocorticoid receptors [1] [2] [3] . The PPAR isotypes, PPARα, PPARβ/δ and PPARγ are derived from three different genes found on chromosomes 22, 6 and 3, respectively. These isotypes modulate the expression of several genes which play a central role in regulating glucose, lipid and cholesterol metabolism [4] . PPARα is expressed in the liver, skeletal muscles, heart, intestinal mucosa, and brown adipose tissue while PPARβ/δ is expressed in liver, skeletal and cardiac muscle, adipose tissue and macrophages. PPARγ occurs as three isoforms PPARγ1, PPARγ3, which are expressed in the liver, intestine and spleen, and PPARγ2, which is expressed in brown and white adipose tissue [5] . There has been much published concerning the PPARs significant involvement in the progression of human disease [1] [3] [6] [7] [8] .
There have been several PPARα/δ/γ single nucleotide polymorphisms (SNPs) significantly associated with various human diseases or conditions [9] - [17] . A SNP can alter a transcriptional factor motif sequence or a transcriptional factor binding site (TFBS) which in turn can affect the process of gene regulation [18] [19] [20] . A nucleotide change in a binding motif of a gene may increase or decrease the corresponding transcription factor’s (TF) ability to bind the DNA and thereby affect gene expression [21] resulting in a disease or change in a human condition. In this study, eight PPARα/δ/γ SNPs that have been previously associated with a disease or condition were analyzed for producing alterations in punitive TFBS (Table 1 & Table 2).
2. Materials and Methods
2.1. PPARα/δ/γ Genes and SNPs
The PPARα/δ/γ SNPs that have been shown to be significantly associated with disease or sickness [9] - [17] are listed in Table 1 as well as the SNP alleles, frequencies and mutations. SNP information was collected using dbSNP, National Center for Biotechnology Information (NCBI), U.S. National Library of Medicine. The gene symbols of conserved (black) and unique (red)punitive TFBS between the two SNP alleles are found in the Table 2. The names of the punitive TFBS can be found in the Supplement.
2.2. Identifying TFBS
The JASPAR CORE database [22] [23] and Con Site [24] were used to identify the TFBS in previous studies [25] - [36] . JASPAR is a collection of transcription factor DNA-binding preferences used for scanning genomic sequences where ConSite is a web-based tool for finding cis-regulatory elements in genomic sequences. The Vector NTI Advance 11 computer program (Invitrogen, Life Tech- nologies) was used to locate SNPs and TFBS within all genes listed in Table 2.
3. Results
The PPARα gene is mainly expressed in tissues with extensive fatty acid catabolism and its activation results in alterations in the transcription of several genes that regulate lipid and lipoprotein metabolism [6] [37] . One of its SNPs (rs18-
Table 1. PPARα/δ/γ genes whose SNPs have been shown to be associated with disease or sickness. Listed are the genes and NCBI reference sequence; their chromosome location as well as the SNP genome position and gene location; SNPs, their alleles and mutations; disease or condition associated with the SNP as well as the ethnic group. See references for information related to the disease/condition. MAF is minor allele frequency.
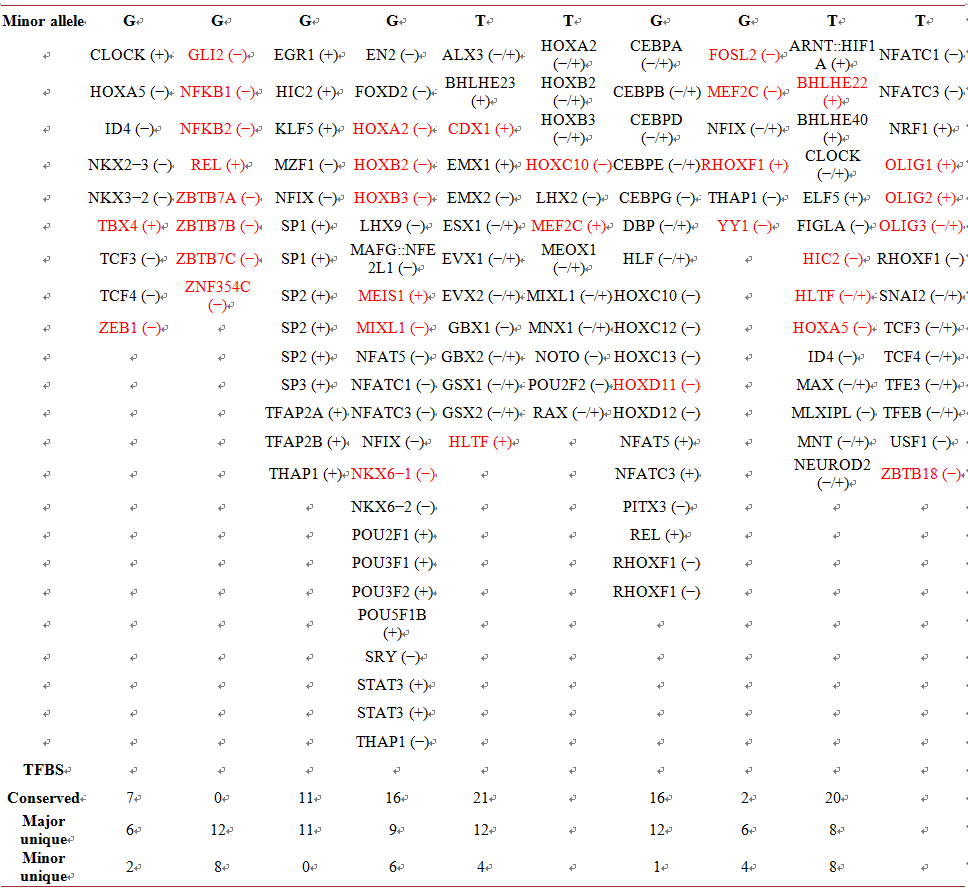
00206) has been associated with variation in lipid serum levels in Caucasian and Indian populations [38] [39] , hypertriglyceridemia risk, dyslipidemia risk and low-density lipoprotein-cholesterol risk in Han Chinese [10] [11] [16] . The two alleles (C/G) of the SNP generates seven conserved punitive TFBS between the alleles while the common C allele generates an additional six unique TFBS and the minor G allele generates two unique TFBS (Table 2). Of the minor G allele TFBS, the TBX4 transcription factor is associated with the disease heritable pulmonary arterial hypertension [40] , which may in part be responsible for the variation in lipid serum levels and disease risks found to be significantly associated with the this SNP.
The PPARδ gene is expressed in high levels in liver, kidneys, cardiac and skeletal muscle, adipose tissue, brain, colon and vasculature [41] . One of its SNPs
Table 2. Location of PPARα/δ/γ gene SNPs contained in potential TFBS. (−/+) is the DNA strand location of the TFBS. TFBS in red are only present for the given allele.
(rs2016520) has been associated with obesity risk and intracerebral hemorrhages while a second SNP (rs9794) has been found to be associated with hypertriglyceridemia and obesity in Han Chinese [9] [14] [17] . The two alleles (A/G) of the rs2016520 SNP does not generate any conserved punitive TFBS between the alleles while the common A allele generates twelve punitive unique TFBS and the minor G alleles generates eight punitive unique TFBS (Table 2). Of the common A allele TFBS, the NEUROD2 transcriptional regulator is implicated in neuronal determination and the OLIG3 TF which promotes formation and maturation of oligodendrocytes, especially within the brain could in part be associated with obesity risk and intracerebral hemorrhages (Supplement). The second SNPs (rs9794) has been associated with hypertriglyceridemia and obesity risk in Han Chinese [10] [11] [17] . The two alleles (C/G) of the rs9794 SNP generates eleven conserved punitive TFBS between the alleles while the common C allele generates eleven unique punitive TFBS and the minor G allele generates no unique TFBS (Table 2). Of the common C allele TFBS, the GCM1 and 2 TFs bind to the trophoblast specific element 2 (TSE2) of the aromatase gene enhancer and act as a master regulator of parathyroid development, respectively (Supplement). The deletion of these TFBS caused by the minor G allele for these two TFs could in part be associated with hypertriglyceridemia and obesity risk in Han Chinese [10] [17] .
The PPARγ gene is expressed at high levels in adipose tissue, lipid storage, glucose metabolism as well as the transcriptional regulation of genes involved in these metabolic processes [42] . One of its SNPs (rs10865710) has been associated with obesity risk, systemic sclerosis and low-density lipoprotein-cholesterol in Han Chinese [9] [13] [16] . The two alleles (C/G) of this SNP generates sixteen conserved punitive TFBS between the alleles while the common C allele generates nine unique punitive TFBS and the minor G allele generates six unique TFBS (Table 2). Of the common C allele unique punitive TFBS, the POU2F2 TFBS is found in immunoglobulin gene promoters and its absence from the minor G allele TFBS may in part be responsible for the association of this SNP with systemic sclerosis (Supplement). The presence of the MNX1 and RXF3 TFBS with the common C allele and the NKX6-1 TFBS with the minor G allele which are involve with pancreas development and function as well as insulin gene regulation, respectively, may in part be responsible for association of this SNP with obesity risk and low-density lipoprotein-cholesterol (Supplement).
A second PPARγ gene SNP (rs12629751) has been associated with osteoarthritis in southeast Chinese [12] . The two alleles (C/T) of this SNP generates twenty-one common punitive TFBS between the alleles while the common C allele generates twelve unique punitive TFBS and the minor T allele generates four unique TFBS (Table 2). Of the common C allele unique TFBS, the NKX3-2 TFBS occurs whose TF is a repressor that acts as a negative regulator of chondrocyte maturation which may in part be responsible for the association of this SNP with osteoarthritis (Supplement).
A third PPARγ gene SNP (rs709158) has been associated with low-density lipoprotein-cholesterol in Han Chinese [16] . The two alleles (A/G) of this SNP generates sixteen common punitive TFBS between the alleles while the common A allele generates twelve unique punitive TFBS and the minor A allele generates one unique TFBS (Table 2). Of the common A allele unique TFBS, the HOXC11 TFBS whose TF also binds to a promoter element of the lactase-phlorizin hydrolase gene which may in part be associated with low-density lipoprotein-cholesterol in Han Chinese [43] (Supplement).
A fourth PPARγ gene SNP (rs1805192) has been associated with hypertriglyceridemia, dyslipidemia and low-density lipoprotein-cholesterol in Han Chinese [10] [11] [16] . The two alleles (C/G) of this SNP generates two common punitive TFBS between the alleles while the common C allele generates six unique punitive TFBS and the minor G allele generates four unique TFBS (Table 2). Of the common C allele unique TFBS, the ERS2 TFBS whose TF is expressed in many tissues including pulmonary epithelial cells and the MEF2C TFBS generated by the minor G allele whose transcriptional activator controls cardiac morphogenesis and myogenesis, and is also involved in vascular development are in part responsible for hypertriglyceridemia, dyslipidemia, low-density lipoprotein-cho- lesterol in Han Chinese (Supplement).
A fifth PPARγ gene SNP (rs3856806) has also been associated with hypertriglyceridemia, dyslipidemia and low-density lipoprotein-cholesterol in Han Chinese [10] [11] [16] . The two alleles (C/T) of this SNP generates twenty common punitive TFBS between the alleles while the common C allele generates eight unique punitive TFBS and the minor T allele generates eight unique TFBS (Table 2). An examination of Table 2 would indicate that there are no apparent unique TFBS by either allele that would contribute to this SNPs’ association with hypertriglyceridemia, dyslipidemia and low-density lipoprotein-cholesterol in Han Chinese (Table 1). However, the TF database used in this study (Materials and Methods) represent a small portion of the nuclear TFs that interact in nucleus.
4. Discussion
The PPARα/δ/γ genes have several SNPs that have been associated with human disease or conditions [9] - [17] (Table 1). SNPs in TFBS that alter the binding ability of the respective TF and cause changes in gene expression levels are considered regulatory (r) SNPs [44] . There have been many reports on the possible outcome of such alterations by identifying punitive TFBS based on the two alleles of the SNP associated with a disease or sickness [25] - [36] . The eight SNPs among the three genes studied in this report again provide several punitive TFBS changes per SNP allele (Table 1 & Table 2).
As an example, the PPARα gene has an exon five SNP (rs1800206) that has been associated with hypertriglyceridemia risk, dyslipidemia risk and low-density lipoprotein-cholesterol risk in Han Chinese (Table 1). The minor G allele of the SNP generates a punitive TBX4 (T-Box 4) TFBS not present with the common C allele (Table 2, Supplement). The TBX4 TF has been associated with the disease heritable pulmonary arterial hypertension [40] which may in part be responsible for the disease/condition found in Han Chinese. The occurrence of the minor G allele in the Han Chinese population is 0.02. A frequency that might be expected among heritable diseases. This SNP has also been associated with changes in triglyceridemia, total cholesterol, low-density lipoprotein-cholesterol (LDL-c), high-density lipoprotein-cholesterol(HDL-c) and apolipoprotein A1 (APOA1) plasma concentrations in populations of African-Americans, Caucasians and Indians [38] [39] [45] [46] .
Another example is the PPARδ gene which has a 5’UTR SNP (rs2016520) that has been associated with obesity risk and intracerebral hemorrhages in Han Chinese (Table 1). The major A allele of the SNP generates a punitiveNEUROD2 TFBS not present with the minor G allele (Table 2, Supplement). The NEU-ROD2 TF is a regulator implicated in neuronal determination and is a critical factor essential for the repression of the genetic program for neuronal differentiation. The presence of this TFBS created by one SNP allele and not the other may in part be the reason for its association with intracerebral hemorrhages in Han Chinese [14] . PPARδ promotes inflammation and tumor growth [47] and an increase in HDL-c concentration in cardiovascular disease [48] . The SNPs major A allele generates a punitive ERG TFBS not present with the minor G allele whose TF is associated with inflammation (Table 2, Supplement) while the minor G allele generates the punitive NFKB1 & 2 and REL TFBS not found with the common A allele whose TFs are associated with inflammation and tumorigenesis (Table 2, Supplement).
A third example is the PPARγ gene which has an intron one SNP (rs1269751) that has been associated with osteoarthritis (OA) in southeast Chinese (Table 1). The major C allele of the SNP generates a punitive NKX3-2 TFBS not present with the minor T allele (Table 2, Supplement). The NKX3-2 TF is a repressor that acts as a negative regulator of chondrocyte maturation. The presence of the NKX3-2 TFBS created by one SNP allele and not the other may in part be the reason for its association with osteoarthritis in southeast Chinese. Nkx3-2 has been found to induce chondrocyte precursor cells, maintained early-stage chondrocytes and down-regulated terminal-stage chondrocytes [49] . One of the strongest environmental risk factors for knee OA is obesity [50] and is also considered to be a moderate risk factor for hip OA [51] . The PPARγ gene has a SNP (rs10865710) in its promotor that is associated with obesity risk [9] . The minor G allele of this SNP generates a punitive NKX6-1 TFBS not found with the major C allele (Table 2). The NKX6-1 TF is involved in transcriptional regulation of islet beta cells. This TF binds to the insulin promoter and is involved in the regulation of the insulin gene (Supplement). The lack of this BS with the major C allele could have a profound effect on obesity in humans. Other examples involving the presence or absence of unique SNP allele TFBS can be found in Table 2 and potential human health issues can be made.
The PPARα/δ/γ SNPs that provide changes in punitive TFBS are not only found in the promoter but in the introns, exons and the UTRs of these genes (Table 2). The nucleus of the cell is where epigenetic alterations and TFs operate to convert chromosomes into single stranded DNA for mRNA transcription while it is the cytoplasm where mRNA is processed by separating exons and introns for protein translation. Consequently, it doesn’t matter where TFs bind the DNA in the nucleus because it is only there that TFs function. There are three SNPs in the coding regions (exons) of two genes which are PPARα (rs1800206), PPARγ (rs-1805192 and rs3856806). These SNPs result in missense mutations in which p.L162V and p.P12A are non synonymous changes and p.H449H is a synonymous change in the amino acids of the proteins (Table 1). In this study, the same SNPs result in multiple punitive TFBS changes in the DNA. This raises the question: are the significant associations of these SNPs with disease or change in condition the result of nucleotide or amino acid substitutions? (Table 1). All three amino acid substitutions are considered to be clinical benign changes to human health [52] while there are reports indicating these substitution are significant associations to disease or condition [10] [11] [15] [16] . This might suggest that the nucleotide and not the amino acid substitutions are responsible for the significant associations that have been reported.
In conclusion, the alterations in punitive TFBS created by SNPs as identified in this study provides a nuclear solution as to why significant associations have been found between some SNPs and human disease or sickness. Alterations in TFBS would affect the TFs ability to regulate a gene which could result in human disease or sickness.
Cite this paper
Buroker, N.E. (2017) Identifying Changes in Punitive Transcriptional Factor Binding Sites Created by PPARα/δ/γ SNPs Associated with Disease. Journal of Biosciences and Medicines, 5, 81-100. https://doi.org/10.4236/jbm.2017.54008
References
- 1. Berger, J. and Moller, D.E (2002) The Mechanisms of Action of PPARs. Annual Review of Medicine, 53, 409-435.
https://doi.org/10.1146/annurev.med.53.082901.104018 - 2. Boitier, E., Gautier, J.C. and Roberts, R. (2003) Advances in Understanding the Regulation of Apoptosis and Mitosis by Peroxisome-Proliferator Activated Receptors in Pre-Clinical Models: Relevance for Human Health and Disease. Comparative Hepatology, 2, 3.
- 3. Harmon, G.S., Lam, M.T. and Glass, C.K. (2011) PPARs and Lipid Ligands in Inflammation and Metabolism. Chemical Reviews, 111, 6321-6340.
https://doi.org/10.1021/cr2001355 - 4. Vitale, S.G., Lagana, A.S., Nigro, A., La Rosa, V.L., Rossetti, P., Rapisarda, A.M., et al. (2016) Peroxisome Proliferator-Activated Receptor Modulation during Metabolic Diseases and Cancers: Master and Minions. PPAR Research, 2016, 651-671.
https://doi.org/10.1155/2016/6517313 - 5. Reddy, J.K. and Rao, M.S. (2006) Lipid Metabolism and Liver Inflammation. II. Fatty Liver Disease and Fatty Acid Oxidation. Gastrointestinal and Liver Physiology, 290, 852-858.
- 6. Fruchart, J.C. (2009) Peroxisome Proliferator-Activated Receptor-Alpha (PPARalpha): At the Crossroads of Obesity, Diabetes and Cardiovascular Disease. Atherosclerosis, 205, 1-8.
https://doi.org/10.1016/j.atherosclerosis.2009.03.008 - 7. Kersten, S. and Wahli, W. (2000) Peroxisome Proliferator Activated Receptor Agonists. EXS, 89, 141-151.
- 8. Desvergne, B. and Wahli, W. (1999) Peroxisome Proliferator-Activated Receptors: Nuclear Control of Metabolism. Endocrine Reviews, 20, 649-688.
https://doi.org/10.1210/er.20.5.649 - 9. Luo, W., Guo, Z., Wu, M., Hao, C., Hu, X., Zhou, Z., et al. (2013) Association of Peroxisome Proliferator-Activated Receptor Alpha/Delta/Gamma with Obesity and Gene-Gene Interaction in the Chinese Han population. Journal of Epidemiology, 23, 187-194.
- 10. Gu, S.J., Liu, M.M., Guo, Z.R., Wu, M., Chen, Q., Zhou, Z.Y., et al. (2013) Gene-Gene Interactions among Paralpha/Delta/Gamma Polymorphisms for Hypertriglyceridemia in Chinese Han population. Gene, 515, 272-176.
https://doi.org/10.1016/j.gene.2012.11.078 - 11. Gu, S.J., Guo, Z.R., Zhou, Z.Y., Hu, X.S. and Wu, M. (2014) PPAR Alpha and PPAR Gamma Polymorphisms as Risk Factors for Dyslipidemia in a Chinese Han population. Lipids in Health and Disease, 13, 23.
- 12. Zheru, D., Peiliang, F., Yuli, W., Haishan, W., Qirong, Q., Xiaohua, L., et al. (2014) Association of PPARgamma Gene Polymorphisms with Osteoarthritis in a Southeast Chinese population. Journal of Genetics, 93, 719-723.
https://doi.org/10.1007/s12041-014-0444-2 - 13. Marangoni, R.G., Korman, B.D., Allanore, Y., Dieude, P., Armstrong, L.L., Rzhetskaya, M., et al. (2015) A Candidate Gene Study Reveals Association between a Variant of the Peroxisome Proliferator-Activated Receptor Gamma (PPAR-Gamma) Gene and Systemic Sclerosis. Arthritis Research & Therapy, 17, 128.
https://doi.org/10.1186/s13075-015-0641-2 - 14. Huang, Y., Nie, S., Zhou, S., Li, K., Sun, J., Zhao, J., et al. (2015) PPARD rs2016520 Polymorphism and Circulating Lipid Levels Connect with Brain Diseases in Han Chinese and Suggest Sex-Dependent Effects. Biomedicine & Pharmacotherapy, 70, 7-11.
- 15. Lapice, E., Monticelli, A., Cocozza, S., Pinelli, M., Cocozza, S., Bruzzese, D., et al. (2015) The PPARgamma2 Pro12Ala Variant Is Protective against Progression of Nephropathy in People with Type 2 Diabetes. Journal of Translational Medicine, 13, 85.
https://doi.org/10.1186/s12967-015-0448-6 - 16. Fan, W., Shen, C., Wu, M., Zhou, Z.Y. and Guo, Z.R. (2015) Association and Interaction of PPARalpha, Delta, and Gamma Gene Polymorphisms with Low-Density Lipoprotein-Cholesterol in a Chinese Han population. Genetic Testing and Molecular Biomarkers, 19, 379-386.
- 17. Luo, W., Chen, F., Guo, Z., Wu, M., Zhou, Z. and Yao, X. (2016) A Population Association Study of PPAR Delta Gene Rs2016520 and Rs9794 Polymorphisms and Haplotypes with Body Mass Index and Waist Circumference in a Chinese Population. Annals of Human Biology, 43, 67-72.
- 18. Knight, J.C. (2003) Functional Implications of Genetic Variation in Non-Coding DNA for Disease Susceptibility and Gene Regulation. Clinical Science, 104, 493-501.
https://doi.org/10.1042/CS20020304 - 19. Wang, X., Tomso, D.J., Liu, X. and Bell, D.A. (2005) Single Nucleotide Polymorphism in Transcriptional Regulatory Regions and Expression of Environmentally Responsive Genes. Toxicology and Applied Pharmacology, 207, 84-90.
- 20. Chorley, B.N., Wang, X., Campbell, M.R., Pittman, G.S., Noureddine, M.A. and Bell, D.A. (2008) Discovery and Verification of Functional Single Nucleotide Polymorphisms in Regulatory Genomic Regions: Current and Developing Technologies. Mutation Research, 659, 147-157.
https://doi.org/10.1016/j.mrrev.2008.05.001 - 21. Buroker, N.E. (2016) Identifying Changes in Punitive Transcriptional Factor Binding Sites from Regulatory Single Nucleotide Polymoprhisms that Are Significantly Associated with Disease or Sickness. World Journal of Hematology, 5, 75-87.
- 22. Bryne, J.C., Valen, E., Tang, M.H., Marstrand, T., Winther, O., Da Piedade, I., et al. (2008) JASPAR, the Open Access Database of Transcription Factor-Binding Profiles: New Content and Tools in the 2008 Update. Nucleic Acids Research, 36, 102-106. https://doi.org/10.1093/nar/gkm955
- 23. Sandelin, A., Alkema, W., Engstrom, P., Wasserman, W.W. and Lenhard, B. (2004) JASPAR: An Open-Access Database for Eukaryotic Transcription Factor Binding Profiles. Nucleic Acids Research, 32, 91-94.
- 24. Sandelin, A., Wasserman, W.W. and Lenhard, B. (2004) ConSite: Web-Based Prediction of Regulatory Elements Using Cross-Species Comparison. Nucleic Acids Research, 32, 249-252.
https://doi.org/10.1093/nar/gkh372 - 25. Buroker, N.E. (2013) AKT3 rSNPs, Transcritional Factor Binding Sites and Human Disease. Open Journal of Blood Diseases, 3, 116-129.
https://doi.org/10.4236/ojbd.2013.34023 - 26. Buroker, N.E. (2013) ATF3 rSNPs, Transcriptional Factor Binding Sites and Human Etiology. Open Journal of Genetics, 3, 253-261.
- 27. Buroker, N.E. (2014) ADRBK1 (GRK2) rSNPs, Transcriptional Factor Binding Sites and Cardiovascular Disease in the Black Population. Journal of Cardiovascular Disease, 2.
- 28. Buroker, N.E. (2014) DIO2 rSNPs, Transcription Factor Binding Sites and Disease. British Journal of Medicine & Medical Research, 9, 1-24.
https://doi.org/10.9734/BJMMR/2015/18535 - 29. Buroker, N.E. (2016) Computational EPAS1 rSNP Analysis, Transcriptional Factor Binding Sites and High Altitude Sickness or Adaptation. Journal of Proteomics and Genomics Research, 1, 31-59.
- 30. Buroker, N.E. (2015) LIPA rSNPs (Rs1412444 and Rs2246833), Transcriptional Factor Binding Sites and Disease. British Biomedical Bulletin, 3, 281-294.
- 31. Buroker, N.E. (2016) Computational STAT4 rSNP Analysis, Transcriptional Factor Binding Sites and Disease. Bioinformatics and Diabetes, 1, 1-36.
- 32. Buroker, N.E. (2015) VEGFA rSNPs, Transcriptional Factor Binding Sites and Human Disease. The Journal of Physiological Sciences, 64, 73-76.
https://doi.org/10.1007/s12576-013-0293-4 - 33. Buroker, N.E. (2015) VEGFA SNPs (Rs34357231 & Rs35569394), Transcriptional Factor Binding Sites and Human Disease. British Journal of Medicine & Medical Research, 10, 1-11.
- 34. Buroker, N E., Ning, X.H., Li, K., Zhou, Z.N., Cen, W.J., Wu, X.F., Zhu, W.Z., Scott, C.R. and Chen, S.H. (2015) SNPs, Linkage Disequilibrium and Transcriptional Factor Binding Sites Associated with Acute Mountain Sickness among Han Chinese at the Qinghai-Tibetan Plateau. International Journal of Genomic Medicine, 3.
- 35. Buroker, N.E., Ning, X.H., Zhou, Z.N., Li, K., Cen, W.J., Wu, X.F., et al. (2013) VEGFA SNPs and Transcriptional Factor Binding Sites Associated with High Altitude Sickness in Han and Tibetan Chinese at the Qinghai-Tibetan Plateau. Journal of Physiological Sciences, 63, 183-193.
https://doi.org/10.1007/s12576-013-0257-8 - 36. Buroker, N.E. (2014) SNP (rs1570360) in Transcriptional Factor Binding Sites of the VEGFA Promoter Is Associated with Hypertensive Nephropathy and Diabetic Retinopathy. Austin Journal of Endocrinology and Diabetes, 2, 5.
- 37. Shah, A., Rader, D.J. and Millar, J.S. (2010) The Effect of PPAR-Alpha Agonism on Apolipoprotein Metabolism in Humans. Atherosclerosis, 210, 35-40.
- 38. Lacquemant, C., Lepretre, F., Pineda Torra, I., Manraj, M., Charpentier, G., Ruiz, J., et al. (2000) Mutation Screening of the PPARalpha Gene in Type 2 Diabetes Associated with Coronary Heart Disease. Diabetes & Metabolism, 26, 393-401.
- 39. Vohl, M.C., Lepage, P., Gaudet, D., Brewer, C.G., Betard, C., Perron, P., et al. (2000) Molecular Scanning of the Human PPARa Gene: Association of the L162v Mutation with Hyperapobetalipoproteinemia. The Journal of Lipid Research, 41, 945-52.
- 40. Austin, E.D., Loyd, J.E. and Phillips, J.A. (1993) Heritable Pulmonary Arterial Hypertension. In: Pagon, R.A., Adam, M.P., Ardinger, H.H., Wallace, S.E., Amemiya, A., Bean, L.J.H., et al., Eds., Gene Reviews, Seattle.
- 41. Robinson, E. and Grieve, D.J. (2009) Significance of Peroxisome Proliferator-Activated Receptors in the Cardiovascular System in Health and Disease. Pharmacology & Therapeutics, 122, 246-263.
https://doi.org/10.1016/j.pharmthera.2009.03.003 - 42. Semple, R.K., Chatterjee, V.K. and O'Rahilly, S. (2006) PPAR Gamma and Human Metabolic Disease. The Journal of Clinical Investigation, 116, 581-589.
- 43. Wagh, K., Bhatia, A., Alexe, G., Reddy, A., Ravikumar, V., Seiler, M., et al. (2012) Lactase Persistence and Lipid Pathway Selection in the Maasai. PLOS ONE, 7, e44751.
https://doi.org/10.1371/journal.pone.0044751 - 44. Bahreini, A., Levine, K., Santana-Santos, L., Benos, P.V., Wang, P., Andersen, C., et al. (2016) Non-Coding Single Nucleotide Variants Affecting Estrogen Receptor Binding and Activity. Genome Medicine, 8, 128.
- 45. Flavell, D.M., Pineda Torra, I., Jamshidi, Y., Evans, D., Diamond, J.R., Elkeles, R.S., et al. (2000) Variation in the PPARalpha Gene Is Associated with Altered Function In Vitro and Plasma Lipid Concentrations in Type II Diabetic Subjects. Diabetologia, 43, 673-680.
https://doi.org/10.1007/s001250051357 - 46. Shin, M.J., Kanaya, A.M. and Krauss, R.M. (2008) Polymorphisms in the Peroxisome Proliferator Activated Receptor Alpha Gene Are Associated with Levels of Apolipoprotein CIII and Triglyceride in African-Americans but Not Caucasians. Atherosclerosis, 198, 313-319.
- 47. Wang, D., Fu, L., Ning, W., Guo, L., Sun, X., Dey, S.K., et al. (2014) Peroxisome Proliferator-Activated Receptor Delta Promotes Colonic Inflammation and Tumor Growth. Proceedings of the National Academy of Sciences, 111, 7084-7089.
https://doi.org/10.1073/pnas.1324233111 - 48. Aberle, J., Hopfer, I., Beil, F.U. and Seedorf, U. (2006) Association of the T+294C Polymorphism in PPAR Delta with Low HDL Cholesterol and Coronary Heart Disease Risk in women. International Journal of Medical Sciences, 3, 108-111.
- 49. Choi, S.W., Jeong, D.U., Kim, J.A., Lee, B., Joeng, K.S., Long, F., et al. (2012) Indian Hedgehog Signalling Triggers Nkx3.2 Protein Degradation during Chondrocyte Maturation. Biochemical Journal, 443, 789-798.
https://doi.org/10.1042/BJ20112062 - 50. Blagojevic, M., Jinks, C., Jeffery, A. and Jordan, K.P. (2010) Risk Factors for Onset of Osteoarthritis of the Knee in Older Adults: A Systematic Review and Meta-Analysis. Osteoarthritis Cartilage, 18, 24-33.
- 51. Lievense, A.M., Bierma-Zeinstra, S.M., Verhagen, A.P., Van Baar, M.E., Verhaar, J.A. and Koes, B.W. (2002) Influence of Obesity on the Development of Osteoarthritis of the Hip: A Systematic Review. Rheumatology, 41, 1155-1162.
https://doi.org/10.1093/rheumatology/41.10.1155 - 52. Adzhubei, I.A., Schmidt, S., Peshkin, L., Ramensky, V.E., Gerasimova, A., Bork, P., et al. (2010) A Method and Server for Predicting Damaging Missense Mutations. Nat Methods, 7, 248-249.
https://doi.org/10.1038/nmeth0410-248
Supplement
Transcriptional factors (TF), protein name and their description or function.