American Journal of Molecular Biology
Vol.2 No.1(2012), Article ID:16548,13 pages DOI:10.4236/ajmb.2012.21008
Comparative gene expression analysis in stems of Dolichos purpureus and Arabidopsis thaliana
![]()
1Life Sciences College, South-Central University for Nationalities, Wuhan, China
2CAS Key Laboratory of Genome Science and Information, Beijing Institute of Genomics, Chinese Academy of Science, Beijing, China
Email: #haiyuan988@yahoo.com
Received 29 August 2011; revised 14 October 2011; accepted 23 October 2011
Keywords: Stem Epidermis; Signal Transduction; Environment; Development
ABSTRACT
The outermost layer epidermis of a plant stem plays an important role in protection and environmentsensing. The mechanisms of sensing and response to the environment through the stem epidermis remain unclear. Here we report enriched expression of genes involved in stress resistance and signal transduction functions in the stem epidermis of both D. purpureus and A. thaliana by cDNA cloning and QPCR in D. purpureus and by analysis using dataset from a genome-wide comparison with cDNAs differentially expressed between the epidermis and inner parts of top and base stem in A. thaliana. Among 188 cDNAs from the stem epidermis of D. purpureus, 13% and 17% were related to signal transduction and defense respectively. Most of them were up-regulated more in the stem epidermis than the inner stem, as well as in A. thaliana. Also, the distribution of the numbers and specificities of up-regulated genes related to signal transduction and regulatory networks in the epidermis and inner stem revealed the possibility of positional differences in regulation. The results revealed the importance of the epidermis in signal transduction and plant defence.
1. INTRODUCTION
The epidermis is a protective layer covering the entire plant, usually comprising one to a few layers of cells: the outmost cuticle layer and the epidermis layer composed of basic cells, trichomes and guard cells. The epidermis begins to differentiate as early as the 8-cell stage, and differentiation of epidermal cell types appears later in embryo development [1-3]. In growing seedlings and mature plants, the epidermal cells of a fully developed plant not only continue to divide and differentiate into specific cell types at specific sites within the epidermis [4], but also secrete cutin and wax to cover the surface, forming a protective layer [5,6]. Also, within the epidermal cells, secondary substances resistant to pathogens and physical stress are produced [7]. The epidermis is also the site at which mechanical and other environmental stimuli are sensed [8-10]. The expression profile of genes in the epidermis changes with developmental stage [11-13] and cell type [7,12-13].
The structure and chemical components of the epidermis layer of the stem are adapted to resist various stresses [7,14]. As the barrier and the site of direct contact between plant and environment, the epidermis is also the most important site for environmental signal perception. How signals are perceived and transduced in the epidermis and conveyed to adjacent tissues has not drawn attention. It remains unknown whether there are receptors sensing stress factors and resistance mechanisms specifically located in the epidermis.
D. purpureus L. which has a purple stem epidermis, grows from spring to early winter. In early winter, the plants become senescent with the cold. Up to that stage, they have experienced pathogens, cold, and various other environmental stresses, so they may have developed some stress responses.
In this study, we sequenced 188 cDNAs from a purple epidermis cDNA library of D. purpureus stems collected during early winter and used bioinformatics methods to analyze gene function. In addition,, we grouped, compared and analyzed genes related to signal transduction pathways expressed in the epidermis and the inner stem of both top and base stem using the Arabidopsis stem epidermis gene expression data [15], depicting the spatial and developmental distribution profiles of signal transduction pathway components, thus obtaining indications of a profile of signal perception and transduction in the stem.
2. MATERIAL AND METHODS
2.1. Stem Epidermis cDNA Library
Total RNA was isolated from the purple epidermis of lentil stem (ca. 1000 µg, collected in November, 2006 in Hangzhou) using the classic guanidine isothiocyanate method [16] followed by treatment with Dnase (1 unit RQ1 Dnase; Promega). Poly(A+) RNA was purified using a biotinylated oligo(dT) primer (Promega), followed by reverse transcription and first strand synthesis. The terminal of the cDNA was blunted and added by an EcoR I adapter (5’-EcoR I-GGCACGAGG-3’). After EcoR I end phosphorylation and digestion with Xho I (5’-Xho I-CTCGAG-3’), all cDNA inserts longer than 500 bp were cloned into a pBluescript® II XR plasmid vector and electroporated into E. coli strain DH10B after recollecting the cDNA fragments longer than 500 bp from agarose gel.
2.2. Sequencing and Sequence Annotation
Plasmid DNA was extracted and sequenced using ABI3730XL sequencers. The sequencing primer used was 5’-ATTAACCCTCATCTAAAGGGA-3’. Base-calling was performed with phred. Vector sequences were trimmed using Cross-match. Low-quality bases (quality score <20) were trimmed from both ends of the sequences using Qualtrim and Simpletrim. ESTs longer than 200 bp after both vector and quality trimming were considered “highquality” ESTs. The repeat sequences in these ESTs were then masked using the RepeatMasker program. The masked sequences were further screened for bacterial chromosomal DNA, RNA, insect viral DNA, rRNA, and mitochondrial DNA using BLASTN [17]. Further screens for possible contaminants were conducted by BLASTN searches of the Non-Redundant Nucleotide Sequences (nt), EST_human, EST_mouse, and EST_others databases. Raw EST data and contigs were compared using BLASTX against the UniProtKB/Swiss-Prot database and BLASTN against EST/N database. The BLAST search was filtered with an e-value set at ≤0.001.
2.3. Quantitative RT PCR
RNA was extracted from triplicate samples of the inner stem and epidermis (each 50 mg, by pealing stem surface skin from stem, the surface skin is the epidemis part, the rest are inner stem) of red stems of D. purpureus L. collected in November, 2009 in Wuhan, using TRIZOL reagent (BioTEKE). The mRNAs were reverse transcribed with a reverse transcription kit (BioTEKE). Set I quantitative PCR was conducted by StarGene Co. using a Fluorescent Quantitative Detection System, Model: FQD- 48A(A4) (Bioer). The reaction mixture contained cDNAspecific primers (Supplementary file 1), dNTP, Taq polymerase with buffer (Takara) and SYBRGreen I 1x. The reaction conditions were: 95˚C 3 min, 72˚C 10 s, 95˚C 30 s, Tm (Supplementary file 1) 30 s , 72˚C 30 s, 35 cycles; then 72˚C, 10 min. Set II quantitative PCR was conducted in the CAS Key Laboratory of Genome Science and Information, Beijing Institute of Genomics, using TECHNE TC-512. The reaction mixture contained Primer-F (10 p) and Primer-R (10 p) 0.65 ml each, cDNA 1 ml , SYBR Green/Mix 9 ml, Taq polymerase 0.2 ml, dd H2O to 20 ml. The reaction procedure is described in (Supplementary file 1). Each data is an average of three repeats.
2.4. Analysis of Data from A. thaliana
We divided the data from supplementary table 2 in Suh et al. [15] into two sets: top and base stem. Genes involved in different signal transduction pathways were selected and compared on the basis of the functions provided. The up-regulation standard was set as the mean ratio of expression level in two tissues >0.5 to cover all possible genes expressed. We defined up-regulation as a signal ratio > 0.5 and down-regulation as a signal ratio < 0.5. Thus, we grouped the data at the whole transcriptome level into four classes: ratio of epidermis to inner stems (epi/inner) > 0.5; <0.5 in both top and base epidermis; and either >0.5 or <0.5 in the top or base epidermis. We also analyzed the genes with up-regulation ratio of epidermis/stem larger than 2.
3. RESULTS AND DISCUSSION
As the defense barrier, the epidermis layer has developed specific surface structures and intracellular chemicals [18, 19,15]. These processes are controlled and directed by internal and environmental signals. The gene expression profile in the stem epidermis reflects the activities of this layer and thus the function of this tissue. In this research, we analyzed the functional pattern of genes expressed in stem epiderm of D. purpureus L., further confirmed by QPCR on some representative genes, and compared with the similar genes from A. thaliana. Finally, from expression distribution pattern of genes related to signal transduction in the top and base stem, we try to deduct the response pathway of plant to environment.
3.1. Functional Pattern of Genes Expressed in Stem Epidermis of D. purpureus
As many as 32% of the 188 sequenced cDNAs in the stem epidermis of D. purpureus L. were related to signal transduction and regulation (Figure 1), including groups of signal transduction, DNA and RNA processing, regulation which involved in the signal transduction process,
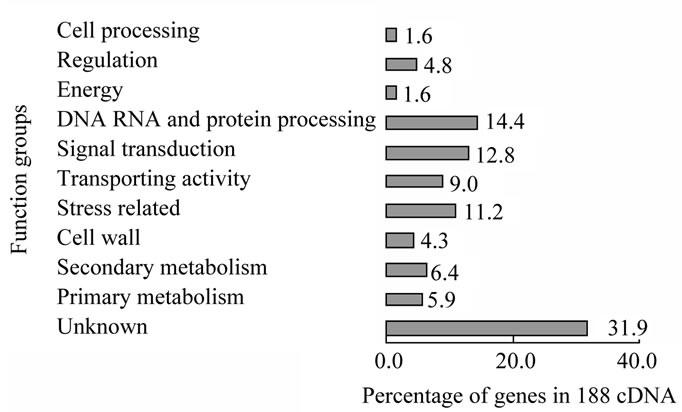
Figure 1. Gene expression profile of 188 cDNAs from purple stem of D. purpureus L. Note: some genes are included in different groups, such as cell wall gene CW14s which is water stress related gene, so are included in stress related gene group. Some genes not included in signal transduction group which also play function in final biology effect in pathway of regulation are also considered as in the signal transduction related genes, such as transcription factors (in regulation group), MAPKs (in stress related group), some protein kinase or proteases (in DNA, RNA, protein group).
indicating the importance of signal transduction in this tissue. This result is consistent with that in the stem epidermis of A. thaliana [15].
In the stem epidermis of D. purpureus, among known cDNA expressed, the top three classes of expressed cDNAs are stress related, DNA/ RNA/protein processing related and signal transduction related cDNAs. The ratio of them are 17% (6% secondary metabolism plus 11% stress directly related), 14%, and 13% respectively. Transporting related genes are the fourth abundant transcripts in stem epidermis of D. purpureus (Figure 1). This profile shows that signal transduction and stress resistance are two major functions in stem epidermis. Genes in 13% of the cDNAs expressed in the stem epidermis of D. purpureus related to signal transduction were: receptors and membrane proteins, G proteins, MAPK pathway kinases, protein kinases, calcium-signal related, and hormone-signal related (Table 1). 17% of the 188 genes expressed in the stem epidermis are directly related to the stress response, among which 6% in secondary pathways and 11% encoding stress resistance proteins (Figure 1, Table 2).
Expressed genes that function in the regulation of transcription or RNA level constituted 4.8% of the 188 cDNAs of D. purpureus. They include HDZIPIII family genes, MYB103, WD family genes, Knox gene, C2H2 zinc finger family genes, and various genes encoding RNA binding proteins. MYB and WD family proteins are known to form complexes and play key roles in the differentiation of the epidermis [20].
Genes related to various stress resistance include genes encoding the Avr9 elicitor response protein (GR463609), which is similar to At1g77810 (87%), LRR Cf2/Cf5 disease resistance protein (GR463639), which is 50% similar to AT5G53890, ERD4 responding to dehydration (GR463575), with 60% similarity to A. thaliana ERD4 At1g30360, MLP-like proteins (GR463493, GR- 463520, GR463625, GR463645), which are mostly similar to MLP43 in A. thaliana (At1g70890), etc. More genes related to stress response such as AMMECR1 (GR- 463522 GR463629), Steroid sulfotransferase (STF, GR- 463507 GR463615), purple acid phosphatase (PAP, GR- 463521 GR463627), Chitinase class I (GR463543 GR- 463595), CW14 (GR463593), Xyloglucan endotransferae (GR463567), Isoflavone reductase (IFR, GR463578), and Chalcone reductase (CHR, GR463527, GR463635) are expressed in stem epidermis of D. purpureus.
Cf genes are elicited in response to avirulence (Avr) peptides secreted by the pathogen Cladosporium fulvum. They encode transmembrane glycoproteins carrying extracytoplasmic Leu-rich repeats (LRRs). Avr9 is one of the Avr peptides; it elicits a set of genes through Cf9 in the hypersensitivity response (HR). These genes are pivotal in the initial defense response characterized by Avr9/Cf9-elicited effects including changes in ion flux, activation of MAPK and a CDPK, and production of active oxygen [21].
ERD4 was induced by various stresses [22,23], and is involved in the brassinosteroid [24], ABA and ethylene [25] hormone regulation pathways.
In humans, AMMECR1 is possibly related to Alport syndrome, mental retardation, midface hypoplasia and eliptocytosis, which involve a contiguous gene deletion syndrome. AMMECR1 is believed to function in RNA base modification [26]. Its sequence is highly similar to ESTs from MOO Vigna unguiculata, Phaseolus vulgaris and Glycine max in plants, which may therefore have a similar catalytic function. GO annotation of AMMECR1 from A. thaliana (AT2G38710) shows that it responds to salt stress.
Steroid sulfotransferase (STF) catalyzes the sulfonation of steroids. It is reported to be up-regulated in response to cold, NaCl, pathogens, and cell death-triggering reagents [27-30].
The main function of vacular purple acid phosphatase (PAP) is reported to be the release of inorganic phosphates as nutrients from phosphate esters [31,32], cell wall PAP play roles in cell elongation [33,34], some specific PAPs have also been implicated in response to stresses such as drought, heat (At3g17790) [35] and NaCl (GmPAP3) [36,37].
The cell wall is a structural defense layer. Genes for cell wall components were expressed in the epidermis of both D. purpureus and A. thaliana (Table 2). Chitinases are enzymes synthesized by plants that directly target
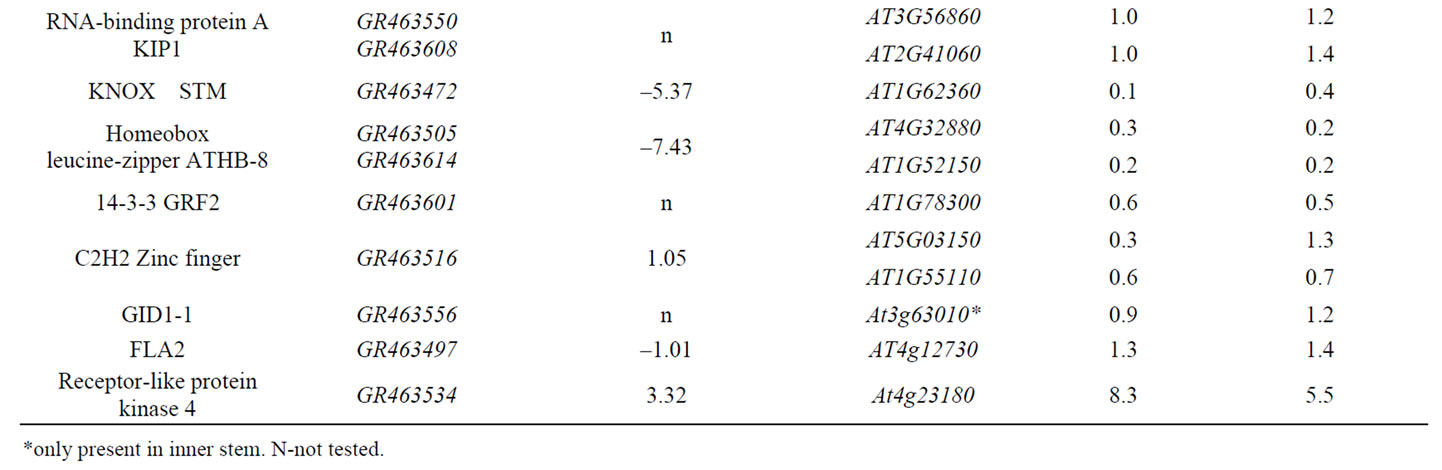
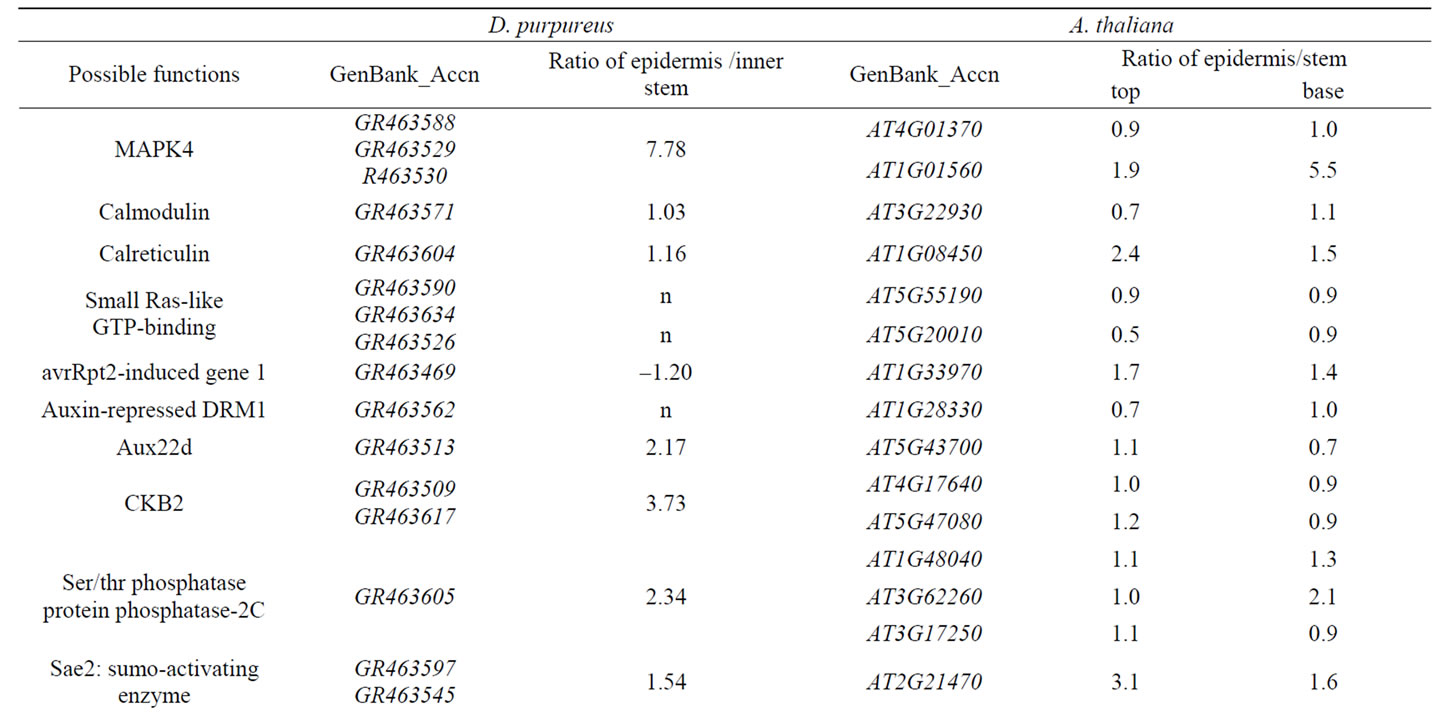
Table 1. Genes expressed in stem epidermis putatively related to signal transduction in D. purpureus and A. thaliana.

Table 2. Genes expressed in epidermis of D. purpureus L. var. and A. thaliana stem putatively related to stress response.
and digest chitin in the cell walls of pathogens and function through both constitutive and inducible pathways as enzyme defenses [38-42]. The structure, function and distribution of chitinases have co-evolved with plantpathogen interactions, facilitating adaptation to the environment [43]. Sinapyl alcohol dehydrogenase (SAD), catalyzing the reduction of sinapyl aldehyde to sinapyl alcohol, which is a component in lignification [44], together with xyloglucan endotransglycosylase, functions in cell elongation and growth.
It has been reported that UV, environmental stresses and developmental factors stimulate the activation of promoters of chalcone synthase [45,46]. Chalcone synthase is the first step in the flavonoid-specific branch pathway. This enzyme was up-regulated only in the epidermis, indicating that the signals for synthesis of CHS are from the environment, the epidermis being the major site for signal perception and early responses. This is also true for ERD4.
Flavonoids function in various plant development processes such as pigmentation, pollen germination, auxin transport, UV protection, interactions with microbes in the roots of legumes [47-50], and in response to wounding and pathogen invasion [51]. Chalcone reductase (CHR) is a key enzyme in flavonoid and isoflavonone synthesis. Isoflavonone reductase (IFR) is a key enzyme in pathways for the synthesis of phytoalexins and other stress response products. CHR works in harmony with CHS, controlling the direction of the flavonoid pathway [52]. CHR and IFR were expressed in the epidermis of D. purpureus. IFR is an NAD(P)H-dependent oxidoreductase, functioning in the synthesis of isoflavonones, which mostly act in plant defense. IFR expression is related to plant resistance to pathogens, UV and other stresses [53-56].
3.2. Comparison Expression Analysis of Genes Related to Signal Transduction and Stress in Stem Epidermis and Inner Stem D. purpureus and A. thaliana
The expression level of genes related to signal transduction and stress from D. purpureus were examined with QPCR method in both stem epidermis and inner stem. Altogether, they are compared with the expression data from A. thaliana (Table 1 and Table 2). The results show that most of these genes are up-regulated in stem epidermis.
Receptors and membrane proteins located on stem epidermis cell surfaces are important in responses to environmental signals. Comparison of the differential expression ratio of 13 genes related to signal transduction expressed in stem epidermis to inner stem from D. purpureus to the similar genes expressed in stem epidermis to inner stem from A. thaliana, 11 genes have similar differential expression tendency (Table 1). With expression ratio in stem epidermis to inner stem larger than 0.5, nine of them are up-regulated in stem epidermis of D. purpureus, takes up 69% of the 13 genes. Among 26 genes related to signal transduction expressed in stem of A. thaliana listed in Table 1, 22 genes in top stem and 23 genes in base stem are up-regulated in stem epidermis at the ratio of 0.5, take up 85% and 88% of 26 genes respectively, only two genes down regulated in stem epidermis of both D. purpureus and A. thaliana. The two down-regulated genes are STM (GR463472, At1g62360) and a Homeobox gene (GR463505, At4g32880), which are both transcription factors function in transcription regulation. STM is a gene essencial for maintenance of meristem structural organization, which is inside of the epidermis. The function of At4g32880 is unknown. Most of genes up-regulated in stem epidermis in table 1 are intermediate components such as kinases, phosphatases, protein interacting proteins, RNA-binding proteins, and down-regulated genes are transcription factors. In table 2, All listed genes are up-regulated in stem epidermis than inner stem, most of them are functional enzymes, receptor or other proteins in cytosol. These results indicate that in epidermis responding to environment earlier or more activity than inner stem.
Most of genes related to stress from D. purpureus are up-regulated in epidermis than the inner part, and all their orthologs in A. thaliana are expressed in both stem epidermis and markedly up-regulated in the epidermis than the inner part (Table 2). One Avr9-responsive protein with a glycosyl group-transferring domain encoded by gene GR463609 was down-regulated in the stem epidermis of D. purpureus relative to the inner stem, but its homolog AT1G77810 was up-regulated in the stem epidermis of A. thaliana (Table 2). Whether this is a consequence of differences in developmental stage or of some other cause requires further study.
3.3. Expression Analysis of Distribution of Genes Related to Signal Transduction, Stress Response and Development in A. thaliana
Up-regulation profile of genes related to signal transduction, stress response, and biotic or abiotic stimulation in stem epidermis and stem from A. thaliana were shown in Figure 2. It has significant difference between numbers of genes up-regulated in stem epidermis and inner stem. Much more genes related to signal transduction and stress were up-regulated in both top and base stem epidermis than in inner stem.
Distribution of genes related to signal transduction and stress in stem were analyzed by comparing their numbers with expression ratios of signals of epidermis/stem among
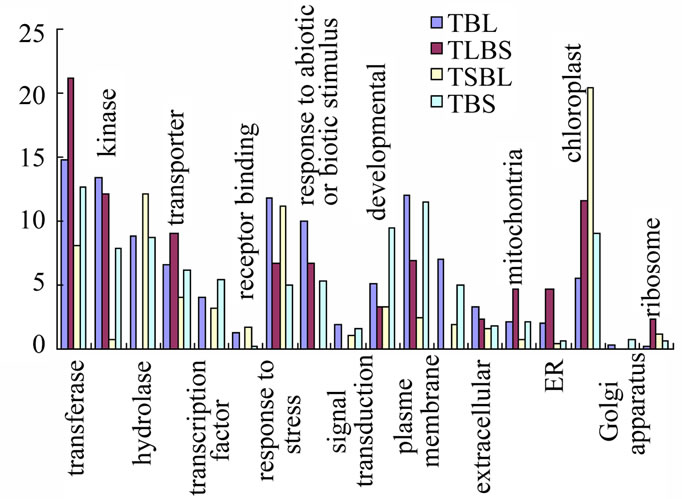
Figure 2. Functional distributions of genes in Arabidopsis stem Note. Y-axial is the percentage of genes up-regulated with ratio of epidermis/stem in each group.
four groups: 1) in the epidermis to that in inner stems (epidermis/inner) of both top and base stem larger than 0.5 and 2, desiginated as TBl, 2) in top epidermis larger than 0.5 and 2, in base epidermis less than 0.5 labeled as TlBs,and 3) in top epidermis less than 0.5, in base epidermis larger than 0.5 and 2, labeled as TsBl and 4) in both top and base epidermis less than 0.5; labeled as TBs in base epidermis, (Tables 3-5). The expression ratios of signals of epidermis/stem is large than 0.5, indicate the expression preferred in epidermis, otherwise preferred in inner stem. Thus the four groups represent genes of four types of gene expression preferences: 1) genes prefer expressed in both top and base stem epidermis, 2) genes prefer expressed in both top and base inner stem, 3) genes prefer expressed in top epidermis and base stem and 4) genes prefer expressioned in top stem and base epidermis.
Table 3 and Figure 3 listed genes involved in hormone and light signal transduction pathway. The signigicantly more genes prefer expressed in stem epidermis than in inner stem.
In the database, among 13589 genes present in macroarray hybridization, the numbers of up-regulated genes with the ratio of epidermis/inner stem larger than 2 in a, b, c, d four groups of genes are 921, 37, 96 and 363 respectively, takes up 6.78%, 0.27%, 0.71% and 2.67% in 13589 genes respectively. In each group, the function of genes are categoried and compared between four groups as in Figure 2.
In Figure 2, functional groups were picked up from three different categories by GO annotation using four groups of genes with expression ratio larger than 2 respectively and combined together for comparison and analysis.
Among four groups, TBL group has highest expression ratio of genes in stress responses (11.8%), biotic and
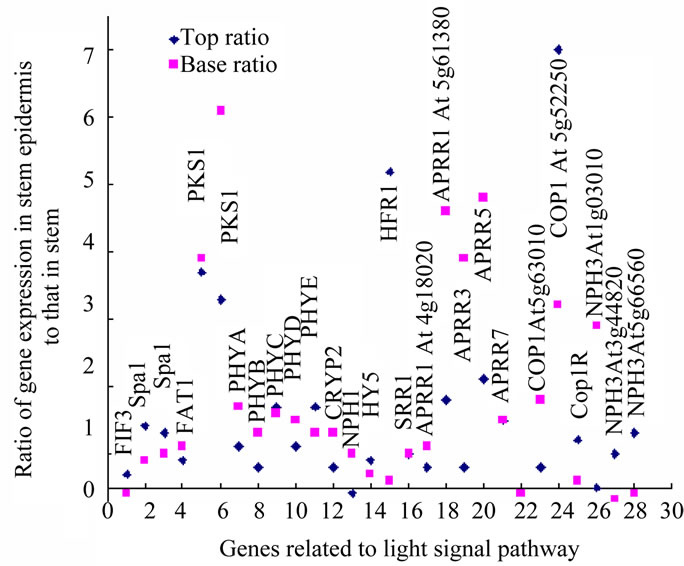
Figure 3. Expression pattern of genes in light signal pathway in Atabidopsis stem.
abiotic responses (9.9%), plasma membrane (12%), kinases (13.3%), signal transduction (1.9%), extracellular (3.4%), cell wall (7%). The less enrichment of TBL group of genes are in transporters (6.5%), hydrolases (8.8%), transferases (14.8%), receptor binding (1.3%), transcription factors (4.1%), developmental (5.1%). TLBs group has highest expression ratio of genes in transferases (21.2%), transporters (9.1%), mitochontria (4.7%), ER (4.7%), ribosomes (2.3%), receptor binding (1.6%). The less enrichment of TLBs group of genes are in kinases (12.1%), response to abiotic or biotic stimulus (6.7%), extracellular (2.3%), chloroplast (11.6%). TSBL group has highest expression ratio of genes in hydrolases (12.1%) and chloroplast (20.4%). The less enrichment of TsBL group of genes are in response to stress (11.2%). TBs group has highest expression ratio of genes in transcription factors (5.3%), Golgi apparatus (0.8%). The less enrichment of TBs group of genes are in plasma membrane (11.5%), hydrolase (8.7%). The types of genes enriched in epidermis of both top and base stem are more likely function in defense and signal transduction.
Among auxin related genes, the only one auxin binding gene, four auxin effllux carrieries are up-regulated in both top and base stem epidermis, one auxin import transporter PIN3 is specifically expressed in both top and base stem epidermis, and two auxin import transporters PIN7 prefer up-regulated in top stem, base stem and base epidermis respectively (Table 3). PIN3 was reported lateral located at epidermis and cortex of apical hooks and mediate the tropism in Arabidopsis [57], but PIN7 seems has stronger signal at epidermis than in cortex at apical hooks of Arabidopsis. It is possible that the distribution of PIN3 and PIN7 are under regulation of complex factors. Among 17 genes which expression up-regulation ratio of epidermis/stem is larger than 2, 6 are IAA6, 6 are
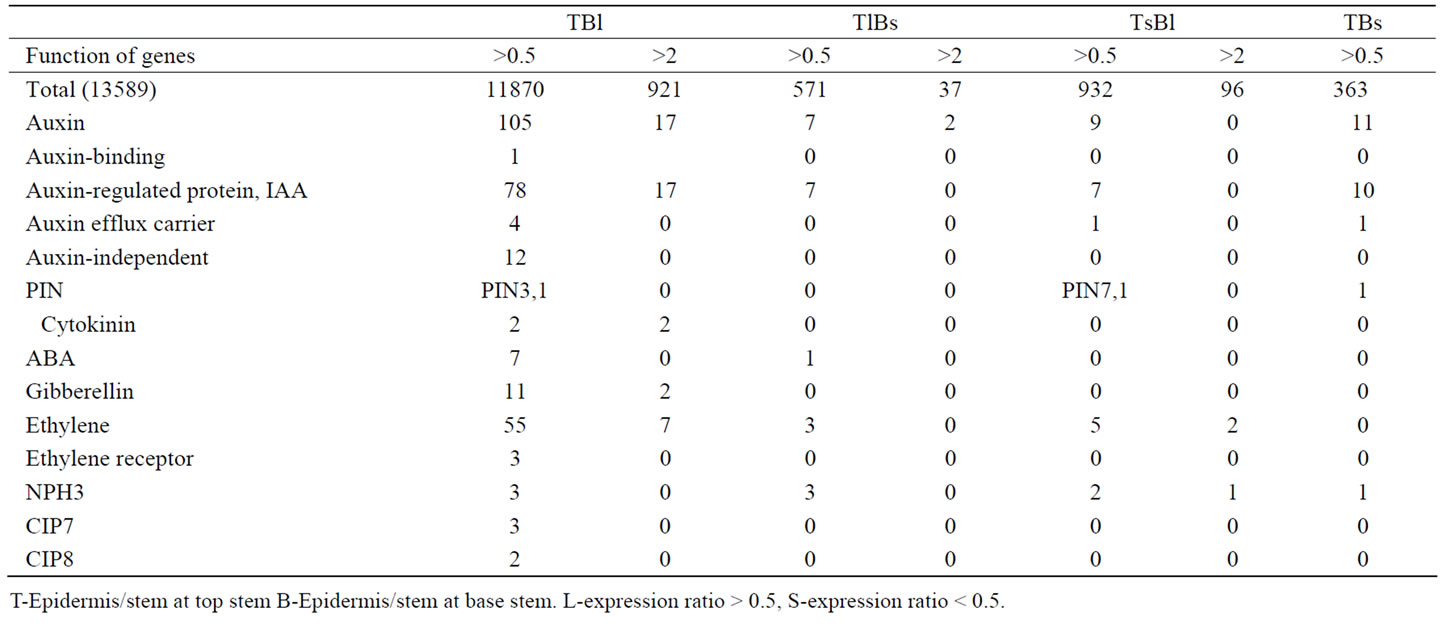
Table 3. Distribution of genes expressed in stem related to hormones and light from A. thaliana with up-regulation ratio > 0.5 and >2 [15].

Table 4. Distribution of some genes expressed in stem in signal transduction pathway from A. thaliana with upregulation ratio > 0.5 and >2 [15].
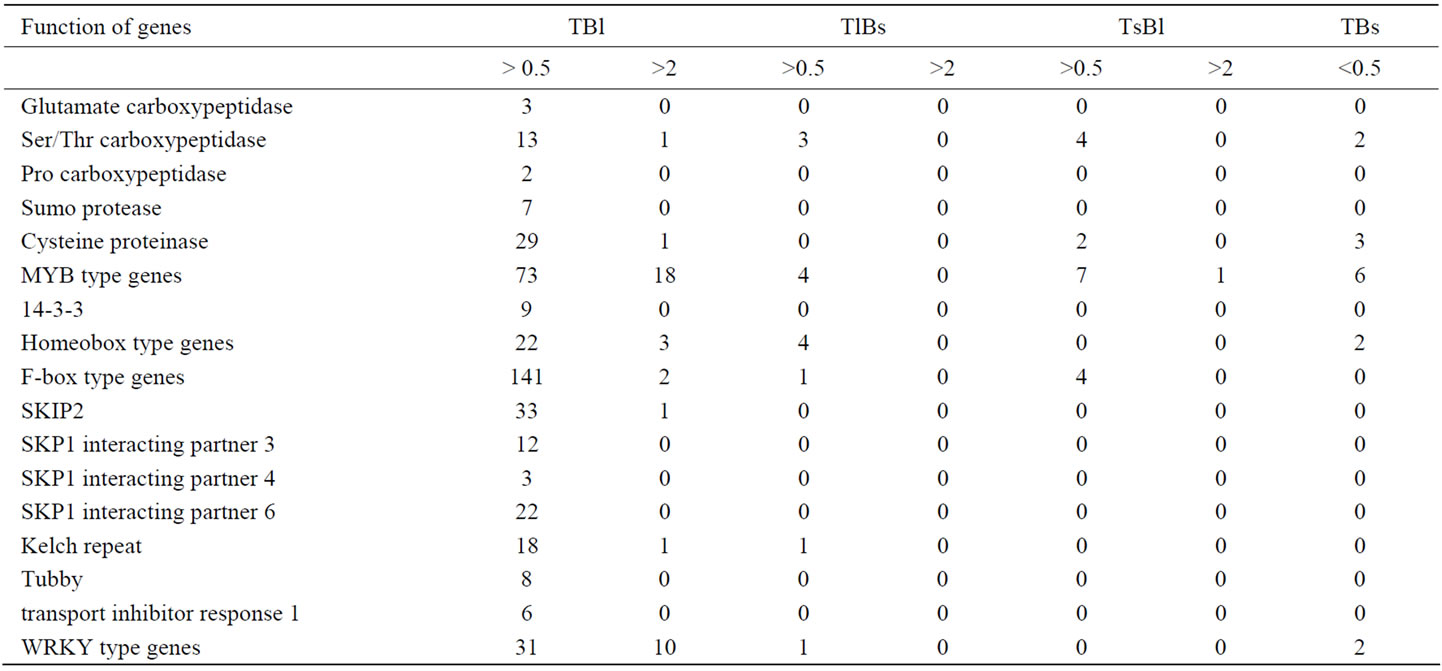
Table 5. Distribution of other components in signal transduction and regulation network in stem of A. thaliana with up-regulation ratio > 0.5 and >2 [15].
SAURs, 2 ARG7, 1 ARG, 1 GH3, 1 AIR12.
Two cytokinin related genes are specifically up-regulated only in epidermis of both top and stem, no other cytokinin related genes up-regulated in other parts. One is At5g05440 with low similarity to cytokinin-specific binding protein (Vigna radiata GI:4190976). The cytokinin-related gene At3g55950 is specifically up-regulated in the epidermis of both top and base stem, similar to the cytokinin-regulated kinase 1 gene (Nicotiana tabacum) AAG25966. Cell growth mainly due to expansion can be controlled by a signal (s) initiated from the epidermis, and the epidermis could be a barrier to inner tissue growth; growth activity is coordinated between the shoot epidermis and the inner part [58]. These genes are possibly related to growth regulation.
Three abscisic acid responsive elements-binding factor (ABF) At4g34000, At3g19290, At1g49720, two abscisic acid-activated protein kinases At3g50500, At4g33950, and two abscisic acid-induced proteins At4g24960, At1g74520 are up-regulated in only epidermis of both top and base stem. One RD20 protein encoding gene At2g33380, a putative transmembrane channel protein gene, induced by abscisic acid during dehydration, is up-regulated in top stem epidermis and base inner stem. This position specificity indicates the regulation of stem growth by environmental factors.
Twenty gibberellin (GA) related genes are expressed in stem. 11(55%) genes are up-regulated in epidermis of both top and base stem, 2 are up-regulated in only top stem epidermis, not present in base stem epidermis. These genes include gibberellin response modulators At1g- 14920 (GAI/RGA2), At1g66350 (RGAL), At2g01570 (RGA1), gibberellin signal transduction protein (SPINDLY) At3g11540, GAST1-related protein At1g74670, At- 1g22690, gibberellin 20-oxidase At3g60290, At4g25420, and GA2-oxidase At2g34555. Two genes specifically expressed in top epidermis are gibberellin 2oxidase At1g- 30040 and gibberellin response modulator putative member of the VHIID domain transcription factor family RGAL At5g17490. GA2-oxidase function in inactivation of Gibberellin.
All three ethylene receptors present in stem are upregulated in both top and base stem epidermis. In 63 genes related to ethylene response present in stem, 55 genes (87%) are up-regulated in both top and base stem epidermis, only 3 (4.7%) and 5 (7.9%) genes are in groups TLBS and TSBL respectively. No genes related to ethylene response only up-regulated in both top and base inner stem (Table 3).
Similarly, in the light signal pathway, the initial steps of light signal transduction in the phytochrome pathway seem mainly to be up-regulated in the stem epidermis. All the five types of phytochromes, the A, B, C, D and E and cryptochrome 2 were absolutely up-regulated in the both top and base epidermis, not in the inner stem (Figure 3). Correspondingly, the immediate substrate of PhyA, PKS1, Phytochrome A signal transduction 1 (PAT1), has the same regulation tendency as PhyA; Spa1, APRR1, 3, 5, 7, SRR1, HFR1, COP1, 9, 10, CIP7, 8, COP1R and HY5, the other components in light signal pathway are also up-regulated only in epidermis of both top and base stem. PIF3, which migrates into the nucleus to regulate DNA transcription, is up-regulated in the top stem epidermis and inner parts of base stem (Figure 3) .
Blue light receptors CRY1 and 2 interact with COP1, releasing HY5, a transcription factor that directly targets the light-responsive promoters from negative regulation by COP1 [59]. In A. thaliana stem, two genes At5g63010 and At5g52250 encoded COP1 have opposite expression pattern in top and base stem. At5g63010 encoded gene has reverse pattern as HY5, possibly function in the same signal pathway (Figure 3). The function of far-red light receptors PhyA, PhyC, PhyD and PhyE, are associated with signal transduction by other phytochromes [60,61]. Photoreceptors may function as groups, cooperatively. PhyA is known to be up-regulated in the dark and downregulated under light; PhyB shows the reverse pattern. The level of phytochromes is reported to be 23 times lower in the light than in the dark, the A:B:C:D:E ratios being 85:10:2:1.5:1.5 in the dark and 5:40:15:15:25 under light [62]. During A. thaliana stem growth, Phy A acts in the initiation stage after light stimulation, and this is followed by PhyB function; they act sequentially and in coordination [63]. In the genome-wise expression data of A. thaliana stem, PhyA, B, D, Cryp2, NPH1 have similar expression distribulation trend between top and base stem, and PhyC, E have similar expression distribulation trend between top and base stem, the same as PIF3, SPA1, PAT1, HY5, COP1R, At5g52250 encoded COP1 (Figure 3).
NPHs are factors in phototropin signal pathways that are involved in blue light-induced movement. The distribution of NPH3 in four groups are 3, 1, 3, 2, they are encoded by different genes; for NPH1 is 0, 0, 0, 1. Similarly, the distribution of up-regulated genes reflected the stages or timings of other components in signal transduction pathways. PKS1 (phytochrome kinase substrate 1) is located only in the cytosol, binding directly to Phy A and NPH1 (phototropin1, phot1) [64]. It is up-regulated only in the epidermis of both top and base stem, the position closest to the light source. NPH1 is reportedly expressed in both epidermis and inner stem, but the expression level is very weak in the epidermis and strong in inner stem cells [65]. Correspondingly, NPH1 is up-regulated in the inner stem at the top stem, the site with active growth and differentiation activities, but at base stem, up-regulated in stem epidermis (Figure 3). NPH1 is located on the inner side of the plasma membrane in the dark, phosphorylated and dissociated from the membrane, then translocated into the cytoplasm under blue light. The NPH1 level in the cells was down-regulated by 24 hours light exposure [65]. NPH3 interacts with NPH1 and is involved in auxin-regulated organogenesis [66-68]. NPH3 encoding genes At1g03010, At1g30440 was upregulated in epidermis at base stem and inner parts of the top stem, similar to NPH1 (Figure 3), in agreement with the optical observations of phototropin1 location in A. thaliana [65]. NPH3 encoding At3g44820 and At5g- 66560 was up-regulated in epidermis of top stem and inner part of base stem, has opposite direction as NPH1, togerher with other NPH3s which constantly preferred at epidermis or inner parts ot both top and base stem, are components of complex regulation network.
The light energy absorbance levels are PhyA< PhyB < Cry2 < NPH1; correspondingly, according to expression ratios of signals of epidermis/stem, the the photoreceptors distribution show the up-regulation tendency as sequences as the top epidermis, top inner, base epidermis, and base inner of the stem (Figure 3). Here, two elements could be distinguished: lateral position (or distance from the environment) and developmental stage (top and base). Low energy light receptors are located on the outside of the stem, and higher energy light receptors on the inside. It is possible that light of a higher energy level can penetrate deeper into the plant tissue and that the distribution of up-regulated genes indicates this coordination and adaptation. The distribution of up-regulated receptors may indicate the adaptation of receptors to light penetration.
The distributions of up-regulated light receptors and the components of their signal transduction pathways reveal that the direction of signal transduction in the stem is from the outer layer to the inner part.
Receptors, membrane receptors, calcium related, CDPK, MAPK are important components in many signal transduction pathway, the significantly more numbers of these genes are up-regulated in stem epidermis of both top and base stem (Table 4).
The number of total receptors expressed in stem is 243, the number of total calcium related genes expressed in stem is 123, the number of MAPK genes expressed in stem is 27. 28 (11.5%) receptor genes, 17 (6.9%) of LRR type receptors, 13 (11%) of calcium related genes and 3(11%) of MAPK genes are up-regulated in epidermis of both top and base stem at ratio of epidermis/stem larger than 2. Much fewer genes are in TLBs and TSBL types of up-regulation pattern for all these class of genes (Table 4). Non-membrane receptors include small molecule receptors such as glutamate, sugar, steroid, S-locus protein, light, retrovirus, protein organelle targeting, probein binding, signal transduction etc., involved in various precess of environmental signal, hormone signal transduction and development process. Somatic embryogenesis receptors CLV1 (At1g60800), CLV2 (At1g- 65380), light repressible receptor (At1g48270), and other major receptors all up-regulated in epidermis of both top and base stem.
Table 5 show the maginificent number of genes upregulated in epidermis of both top and base stem. They are all function in various developmental and defense process. Among total 37 of homeobox genes, 203 of Fbox genes, 136 of MYB, 42 of WRKY genes expressed in stem, 22 (59.4%), 141 (69%), 73 (54%), and 31 (74%) were up-regulated in epidermis of both top and base stem, but the genes are up-regulated in epidermis of only top or base stem are much fewer. This suggests that the epidermis layer is not only a very important location for plant sensing environmental stimulation, but also for sensing signals from inner tissues, integreting the effector from various site, directing and initiate morphogenesis of various new organs of the plant. It is a key place for both defense and developmental decision in plant. The much fewer genes up-regulated in epidermis of only top or base stem indicates that the common properties or function of genes required in epidermis function, and those fewer genes up-regulated only in epidermis of top or base stem probably have specific function in top stem or base stem.
The top stem is where the inflorescence rises and flowers and other organs are initiated. Genes that are more up-regulated in the top epidermis of the stem may therefore be related to these initiation and organogenesis activities. Efremova et al. [69] showed that class B homeotic genes in Antirrhinum and A. thaliana control the organ identity by expression initiated from the L1 meristematic layer and, later, to the epidermis and subepidermis of the differentiating organs. Among genes up-regulated in epidermis of both top and base stem with upregulation ratio of epidermis/stem larger than 2, there are At1g05230, At1g79840 (GLABRA2, GL2), At4g21750 (ATML1), At1g01380 (CPC), At2g46410 (CPC) [2,15,70, 71], which have been reported specifically located in epidermis and function in meristem organization or initiation of new organs and patterning of epidermis cells. This expression pattern is similar to the distribution pattern of top epidermis, inner top stem, base epidermis, inner base stem noted for some of the genes.
In addition, the sex determination protein tassel seed 2-like genes (At3g26770, At4g03140) and septum sitedetermining MinD gene (At5g24020) are expressed in both top and base stem. This indicates that signaling and early events in flower development may also be located at certain sites in the stem, and this may be related to the regulation of stem growth and organogenesis by a conjunction of light signals and other environmental signals and hormone signals, hinting at positional decisions for organ formation and flower development on the stem.
The stem epidermis is a barrier between the plant and its environment. It is important not only in protection and defense, but also as the site for sensing and initiating responses to various signals from environment, including those for development, resistance, and others. The important functions of the epidermis in plant protection and development are far from clear and need more detailed study.
4. CONCLUSIONS
The differential gene expression profiles in epidermis and inner part of stem from both D. purpureus L. and A. thaliana reaveal that defense and signal transduction is important function in stem epidermis. 13% and 17% of genes from 188 sequenced cDNAs expressed in stem epidermis of D. purpureus L. are belong to signal transduction and defense pathways respectively. The results from D. purpureus L. were confirmed by QPCR assay. Macroarray data analysis with A. thaliana suggest that receptors and components in light and hormone signal pathway are preferred expressed in stem epidermis, reveals that the stem epidermis is not only the barrier between the plant and its environment, but also the sensor for environmental stimuli and signals. In D. purpureus L. and A. thaliana stems, the gene expression profiles and the distributions of up-regulated genes displayed the predominance of signal transduction processes in stem epidermis function.
5. ACKNOWLEDGEMENTS
This research was supported with a research fund from South-Central University for Nationalities, No. YZZ06004, NSFC: 40606035 and the Knowledge Innovation Program of the Chinese Academy of Sciences. Partial RTQPCR was performed by the service of Stargene Sci-Tech development Co. Ltd. and Partial was done in CAS Key Laboratory of Genome Science and Information, Beijing Institute of Genomics, Chinese Academy of Science.
REFERENCES
- Costa, S. and Dolan, L. (2003) Epidermal patterning genes are active during embryogenesis in Arabidopsis. Development, 130, 2893-2901. doi:10.1242/dev.00493
- Tominaga, R., Iwata, M., Okada, K. and Wada, T. (2007) Functional analysis of the epidermal-specific MYB genes CAPRICE and WEREWOLF in Arabidopsis. Plant Cell, 19, 2264-2277. doi:10.1105/tpc.106.045732
- Wang, S.C., Hubbard, L., Chang, Y., Guo, J.J., Schiefelbein, J. and Chen, J.G. (2008) Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biology, 8, 81.
- Chlyah, H. and Van, M.T. (1975) Distribution pattern of cell division centers on the epidermis of stem segments of torenia fournieri during de novo bud formation. Plant Physiology, 56, 28-33. doi:10.1104/pp.56.1.28
- Kunst, L. and Samuels, A.L. (2003) Biosynthesis and secretion of plant cuticular wax. Progress in Lipid Research, 42, 51-80. doi:10.1016/S0163-7827(02)00045-0
- Pollard, M., Beisson, F., Li, Y.H. and Ohlrogge, J.B. (2008) Building lipid barriers: Biosynthesis of cutin and suberin. Trends in Plant Science, 13, 236-246. doi:10.1016/j.tplants.2008.03.003
- Murata, J., Roepke, J., Gordon, H. and De Luca, V. (2008) The leaf epidermome of Catharanthus roseus reveals its biochemical specialization. Plant Cell, 20, 524-542. doi:10.1105/tpc.107.056630
- Haley, A., Russell, A.J., Wood, N., Allan, A.C., Knight, M., Campbell, A.K. and Trewavas, A.J. (1995) Effects of mechanical signaling on plant-cell cytosolic calcium. Proceeding of the National Academy of Science of the United States of America, 92, 4124-4128.
- Shabala, S. and Newman, I.I. (1999) Light-induced changes in hydrogen, calcium, potassium, and chloride ion fluxes and concentrations from the mesophyll and epidermal tissues of bean leaves. Understanding the ionic basis of light-induced bioelectrogenesis. Plant Physiology, 119, 1115-1124. doi:10.1104/pp.119.3.1115
- Hardham, A.R., Takemoto, D. and White, R.G. (2008) Rapid and dynamic subcellular reorganization following mechanical stimulation of Arabidopsis epidermal cells mimics responses to fungal and oomycete attack. Bmc Plant Biology, 8, 63.
- Clark, A.M., Verbeke, J.A. and Bohnert, H.J. (1992) Epidermis-specific gene expression in Pachyphytum. Plant Cell, 4, 1189-1198.
- Kryvych, S., Nikiforova, V., Herzog, M., Perazza, D. and Fisahn, J. (2008) Gene expression profiling of the different stages of Arabidopsis thaliana trichome development on the single cell level. Plant Physiology and Biochemistry, 46, 160-173. doi:10.1016/j.plaphy.2007.11.001
- Negri, A.S., Prinsi, B., Rossoni, M., Failla, O., Scienza, A., Cocucci, M. and Espen, L. (2008) Proteome changes in the skin of the grape cultivar Barbera among different stages of ripening. BMC Genomics, 9, 378.
- Panikashvili, D., Savaldi-Goldstein, S., Mandel, T., Yifhar, T., Franke, R.B., Hofer, R., Schreiber, L., Chory, J.and Aharoni, A. (2007) The Arabidopsis DESPERADO/ AtWBC11 transporter is required for cutin and wax secretion. Plant Physiology, 145, 1345-1360. doi:10.1104/pp.107.105676
- Suh, M.C., Samuels, A.L., Jetter, R., Kunst, L., Pollard, M., Ohlrogge, J. and Beisson, F. (2005) Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiology, 139, 1649-1665. doi:10.1104/pp.105.070805
- Sambrook, J. F.E. and Maniatis, T. (1989) Molecular cloning: A laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor.
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J.H., Zhang, Z., Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research, 25, 3389-3402. doi:10.1093/nar/25.17.3389
- Clark, A.M. and Bohnert, H.J. (1993) Epidermis-specific transcripts. Nucleotide sequence of a full-length cDNA of EPI12, encoding a putative lipid transfer protein. Plant Physiology, 103, 677-678.
- Thomas, R., Fang, X., Ranathunge, K., Anderson, T.R., Peterson, C.A. and Bernards, M.A. (2007) Soybean root suberin: Anatomical distribution, chemical composition, and relationship to partial resistance to Phytophthora sojae. Plant Physiology, 144, 299-311. doi:10.1104/pp.106.091090
- Ramsay, N.A. and Glover, B.J. (2005) MYB-bHLHWD40 protein complex and the evolution of cellular diversity. Trends Plant Science, 10, 63-70. doi:10.1016/j.tplants.2004.12.011
- Rowland, O., Ludwig, A.A., Merrick, C.J., Baillieul, F., Tracy, F.E., Durrant, W.E., Fritz-Laylin, L., Nekrasov, V., Sjolander, K., Yoshioka, H. and Jones, J.D.G. (2005) Functional analysis of Avr9/Cf-9 rapidly elicited genes identifies a protein kinase, ACIK1, that is essential for full Cf-9-dependent disease resistance in tomato. Plant Cell, 17, 295-310. doi:10.1105/tpc.104.026013
- Taji, T., Seki, M., Yamaguchi-Shinozaki, K., Kamada, H., Giraudat, J. and Shinozaki, K. (1999) Mapping of 25 drought-inducible genes, RD and ERD, in Arabidopsis thaliana. Plant Cell Physiology, 40, 119-123
- Liu, J., Elmore, J.M., Fuglsang, A.T., Palmgren, M.G., Staskawicz, B.J. and Coaker, G. (2009) RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biology, 7, e1000139. doi:10.1371/journal.pbio.1000139
- Lisso, J., Steinhauser, D., Altmann, T., Kopka, J. and Mussig, C. (2005) Identification of brassinosteroidrelated genes by means of transcript co-response analyses. Nucleic Acids Research, 33, 2685-2696. doi:10.1093/nar/gki566
- Huang, D.Q., Wu, W.R., Abrams, S.R. and Cutler, A.J. (2008) The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. Journal of Experimental Botany, 59, 2991-3007. doi:10.1093/jxb/ern155
- Balaji, S. and Aravind, L. (2007) The RAGNYA fold: A novel fold with multiple topological variants found in functionally diverse nucleic acid, nucleotide and peptidebinding proteins. Nucleic Acids Research, 35, 5658-5671. doi:10.1093/nar/gkm558
- Kreps, J.A., Wu, Y.J., Chang, H.S., Zhu, T., Wang, X., and Harper, J.F. (2002) Transcriptome changes for arabidopsis in response to salt, osmotic, and cold stress. Plant Physiology, 130, 2129-2141. doi:10.1104/pp.008532
- Lee, B.H., Kapoor, A., Zhu, J.H. and Zhu, J.K. (2006) STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. Plant Cell, 18, 1736-1749. doi:10.1105/tpc.106.042184
- Guan, Y. and Nothnagel, E.A. (2004) Binding of arabinogalactan proteins by Yariv phenylglycoside triggers wound-like responses in Arabidopsis cell cultures. Plant Physiology, 135, 1346-1366. doi:10.1104/pp.104.039370
- Hiraoka, Y., Ueda, H. and Sugimoto, Y. (2009) Molecular responses of Lotus japonicus to parasitism by the compatible species Orobanche aegyptiaca and the incompatible species Striga hermonthica. Journal of Experimental Botany, 60, 641-650. doi:10.1093/jxb/ern316
- Veljanovski, V., Vanderbeld, B., Knowles, V.L., Snedden, WA. and Plaxton, W.C. (2006) Biochemical and molecular characterization of AtPAP26, a vacuolar purple acid phosphatase up-regulated in phosphate-deprived Arabidopsis suspension cells and seedlings. Plant Physiology, 142, 1282-1293. doi:10.1104/pp.106.087171
- Li, D. and Wang, D. (2003) Responses of putative purple acid phosphatase genes in Arabidopsis thaliana (AtPAPs) to phosphorus starvation. Life Science Research , 7, 65-69
- Kaida, R., Hayashi, T. and Kaneko, T.S. (2008) Purple acid phosphatase in the walls of tobacco cells. Phytochemistry, 69, 2546-2551. doi:10.1016/j.phytochem.2008.07.008
- Irshad, M., Canut, H., Borderies, G., Pont-Lezica, R. and Jamet, E. (2008) A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana: Confirmed actors and newcomers. BMC Plant Biology, 8, 94.
- Rizhsky, L., Liang, H.J., Shuman, J., Shulaev, V., Davletova, S. and Mittler, R. (2004) When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiology, 134, 1683- 1696. doi:10.1104/pp.103.033431
- Bhalerao, R., Keskitalo, J., Sterky, F., Erlandsson, R., Bjorkbacka, H., Birve, S.J., Karlsson, J., Gardestrom, P., Gustafsson, P., Lundeberg, J. and Jansson, S. (2003) Gene expression in autumn leaves. Plant Physiology, 131, 430-442. doi:10.1104/pp.012732
- Liao, H., Wong, F.L., Phang, T.H., Cheung, M.Y., Li, W.Y., Shao, G., Yan, X. and Lam, H.M. (2003) GmPAP3, a novel purple acid phosphatase-like gene in soybean induced by NaCl stress but not phosphorus deficiency. Gene, 318, 103-111. doi:10.1016/S0378-1119(03)00764-9
- Ponstein, A.S., Bres-Vloemans, S.A., Sela-Buurlage, M.B., van den Elzen, P.J., Melchers, L.S. and Cornelissen, B.J. (1994) A novel pathogenand wound-inducible tobacco (Nicotiana tabacum) protein with antifungal activity. Plant Physiology, 104, 109-118. doi:10.1104/pp.104.1.109
- Wu, C.T., Leubner-Metzger, G., Meins, F., Jr. and Bradford, K.J. (2001) Class I beta-1,3-glucanase and chitinase are expressed in the micropylar endosperm of tomato seeds prior to radicle emergence. Plant Physiology, 126, 1299-1313. doi:10.1104/pp.126.3.1299
- Wu, C.T. and Bradford, K.J. (2003) Class I chitinase and beta-1,3-glucanase are differentially regulated by wounding, methyl jasmonate, ethylene, and gibberellin in tomato seeds and leaves. Plant Physiology, 133, 263-273. doi:10.1104/pp.103.024687
- Hietala, A.M., Kvaalen, H., Schmidt, A., Johnk, N., Solheim, H. and Fossdal, C.G. (2004) Temporal and spatial profiles of chitinase expression by norway spruce in response to bark colonization by Heterobasidion annosum. Applied and Environmental Microbiology, 70, 3948-3953. doi:10.1128/AEM.70.7.3948-3953.2004
- Taira, T., Ohdomari, A., Nakama, N., Shimoji, M. and Ishihara, M. (2005) Characterization and antifungal activity of gazyumaru (Ficus microcarpa) latex chitinases: Both the chitin-binding and the antifungal activities of class I chitinase are reinforced with increasing ionic strength. Bioscience Biotechnology and Biochemistry, 69, 811-818. doi:10.1271/bbb.69.811
- Bishop, J.G., Dean, A.M. and Mitchell-Olds, T. (2000) Rapid evolution in plant chitinases: Molecular targets of selection in plant-pathogen coevolution. Proceeding of the National Academy of Science of the United States of America, 97, 5322-5327.
- Li, L.G., Cheng, X.F., Leshkevich, J., Umezawa, T., Harding, S.A. and Chiang, V.L. (2001) The last step of syringyl monolignol biosynthesis in angiosperms is regulated by a novel gene encoding sinapyl alcohol dehydrogenase. Plant Cell, 13, 1567-1585.
- Schmid, J., Doerner, P.W., Clouse, S.D., Dixon, R. A. and Lamb, C.J. (1990) Developmental and environmental regulation of a bean chalcone synthase promoter in transgenic tobacco. Plant Cell, 2, 619-631.
- Fuglevand, G., Jackson, J.A. and Jenkins, G.I. (1996) UV-B, UV-A, and blue light signal transduction pathways interact synergistically to regulate chalcone synthase gene expression in Arabidopsis. Plant Cell, 8, 2347-2357.
- Jacobs, M. and Rubery, P.H. (1988) Naturally-Occurring Auxin Transport Regulators. Science, 241, 346-349. doi:10.1126/science.241.4863.346
- Koes, R.E., Quattrocchio, F. and Mol, J.N.M. (1994) The Flavonoid Biosynthetic-Pathway in Plants—Function and Evolution. Bioessays, 16, 123-132. doi:10.1002/bies.950160209
- Shirley, B.W. (1996) Flavonoid biosynthesis: “New” functions for an “old” pathway. Trends in Plant Science, 1, 377-382. doi:10.1016/1360-1385(96)10040-6
- Subramanian, S., Stacey, G. and Yu, O. (2006) Endogenous isoflavones are essential for the establishment of symbiosis between soybean and Bradyrhizobium japonicum. Plant Journal, 48, 261-273. doi:10.1111/j.1365-313X.2006.02874.x
- Sallaud, C., Elturk, J., Breda, C., Buffard, D., Dekozak, I., Esnault, R. and Kondorosi, A. (1995) Differential Expression of Cdna Coding for Chalcone Reductase, a Key Enzyme of the 5-Deoxyflavonoid Pathway, under Various Stress Conditions in Medicago-Sativa. Plant Science, 109, 179-190. doi:10.1016/0168-9452(95)04179-X
- Joung, J.Y., Kasthuri, G.M., Park, J.Y., Kang, W.J., Kim, H.S., Yoon, B.S., Joung, H. and Jeon, J.H. (2003) An overexpression of chalcone reductase of Pueraria montana var. lobata alters biosynthesis of anthocyanin and 5’-deoxyflavonoids in transgenic tobacco. Biochemical and Biophysical Research Communications, 303, 326- 331. doi:10.1016/S0006-291X(03)00344-9
- Petrucco, S., Bolchi, A., Foroni, C., Percudani, R., Rossi, G.L. and Ottonello, S. (1996) A maize gene encoding an NADPH binding enzyme highly homologous to isoflavone reductases is activated in response to sulfur starvation. Plant Cell, 8, 69-80.
- Baldridge, G.D., O’Neill, N.R. and Samac, D.A. (1998) Alfalfa (Medicago sativa L.) resistance to the root-lesion nematode, Pratylenchus penetrans: Defense-response gene mRNA and isoflavonoid phytoalexin levels in roots. Plant Molecular Biology, 38, 999-1010. doi:10.1023/A:1006182908528
- Lers, A., Burd, S., Lomaniec, E., Droby, S. and Chalutz, E. (1998) The expression of a grapefruit gene encoding an isoflavone reductase-like protein is induced in response to UV irradiation. Plant Molecular Biology, 36, 847-856. doi:10.1023/A:1005996515602
- Kim, S.T., Cho, K.S., Kim, S.G., Kang, S.Y. and Kang, K.Y. (2003) A rice isoflavone reductase-like gene, OsIRL, is induced by rice blast fungal elicitor. Molecular Cells, 16, 224-231.
- Friml, J., Wisniewska, J., Benkova, E., Mendgen, K. and Palme, K. (2002) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature, 415, 806-809. doi:10.1038/415806a
- 58. Savaldi-Goldstein, S. and Chory, J. (2008) Growth coordination and the shoot epidermis. Current Opinion in Plant Biology, 11, 42-48.
- Chattopadhyay, S., Ang, L.H., Puente, P., Deng, X.W. and Wei, N. (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell, 10, 673-683
- Franklin, K.A., Davis, S.J., Stoddart, W.M., Vierstra, R.D. and Whitelam, G.C. (2003) Mutant analyses define multiple roles for phytochrome C in Arabidopsis photomorphogenesis. Plant Cell, 15, 1981-1989. doi:10.1105/tpc.015164
- Halliday, K.J. and Whitelam, G.C. (2003) Changes in photoperiod or temperature alter the functional relationships between phytochromes and reveal roles for phyD and phyE. Plant Physiology, 131, 1913-1920. doi:10.1104/pp.102.018135
- Sharrock, R.A. and Clack, T. (2002) Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiology, 130, 442-456. doi:10.1104/pp.005389
- Parks, B.M. and Spalding, E.P. (1999) Sequential and coordinated action of phytochromes A and B during Arabidopsis stem growth revealed by kinetic analysis. Proceeding of the National Academy of Science of the United States of America, 96, 14142-14146
- Lariguet, P., Schepens, I., Hodgson, D., Pedmale, U.V., Trevisan, M., Kami, C., de Carbonnel, M., Alonso, J.M., Ecker, J.R., Liscum, E. and Fankhauser, C. (2006) PHYTOCHROME KINASE SUBSTRATE 1 is a phototropin 1 binding protein required for phototropism. Proceeding of the National Academy of Science of the United States of America, 103, 10134-10139. doi:10.1073/pnas.0603799103
- Sakamoto, K. and Briggs, W.R. (2002) Cellular and subcellular localization of phototropin 1. Plant Cell, 14, 1723-1735. doi:10.1105/tpc.003293
- Motchoulski, A. and Liscum, E. (1999) Arabidopsis NPH3: A NPH1 photoreceptor-interacting protein essential for phototropism. Science, 286, 961-964. doi:10.1126/science.286.5441.961
- Haga, K., Takano, M, Neumann, R. and Iino, M. (2005) The rice coleoptile phototropism gene encoding an ortholog of Arabidopsis NPH3 is required for phototropism of coleoptiles and lateral translocation of auxin. Plant Cell, 17, 103-115. doi:10.1105/tpc.104.028357
- Cheng, Y., Qin, G., Dai, X. and Zhao, Y. (2007) NPY1, a BTB-NPH3-like protein, plays a critical role in auxinregulated organogenesis in Arabidopsis. Proceeding of the National Academy of Science of the United States of America, 104, 18825-18829. doi:10.1073/pnas.0708506104
- Efremova, N., Perbal, M.C., Yephremov, A., Hofmann, W.A., Saedler, H. and Schwarz-Sommer, Z. (2001) Epidermal control of floral organ identity by class B homeotic genes in Antirrhinum and Arabidopsis. Development, 128, 2661-2671.
- Hung, C.Y., Lin, Y., Zhang, M., Pollock, S., Marks, M.D. and Schiefelbein, J. (1998) A common position-dependent mechanism controls cell-type patterning and GLABRA2 regulation in the root and hypocotyl epidermis of Arabidopsis. Plant Physiology, 117, 73-84. doi:10.1104/pp.117.1.73
- Gordon, S.P., Heisler, M.G., Reddy, G.V., Ohno, C., Das, P. and Meyerowitz, E.M. (2007) Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development, 134, 3539-3548. doi:10.1242/dev.010298
NOTES
*These authors contributed equally to this work.
#Corresponding author.

