Journal of Biomedical Science and Engineering
Vol.10 No.05(2017), Article ID:76949,11 pages
10.4236/jbise.2017.105B010
Automated Diabetic Retinopathy Detection Using Bag of Words Approach
Monzurul Islam, Anh V. Dinh, Khan A. Wahid
Department of Electrical and Computer Engineering, University of Saskatchewan, Saskatoon, Canada



Received: April 22, 2017; Accepted: June 13, 2017; Published: June 16, 2017
ABSTRACT
Imaging and computer vision systems offer the ability to study quantitatively on human physiology. On contrary, manual interpretation requires tremendous amount of work, expertise and excessive processing time. This work presents an algorithm that integrates image processing and machine learning to diagnose diabetic retinopathy from retinal fundus images. This automated method classifies diabetic retinopathy (or absence thereof) based on a dataset collected from some publicly available database such as DRIDB0, DRIDB1, MESSIDOR, STARE and HRF. Our approach utilizes bag of words model with Speeded Up Robust Features and demonstrate classification over 180 fundus images containing lesions (hard exudates, soft exudates, microaneurysms, and haemorrhages) and non-lesions with an accuracy of 94.4%, precision of 94%, recall and f1-score of 94% and AUC of 95%. Thus, the proposed approach presents a path toward precise and automated diabetic retinopathy diagnosis on a massive scale.
Keywords:
Diabetic Retinopathy, Bag of Words, Speeded Up Robust Features, Hard Exudates, Soft Exudates, Microaneurysms, Haemorrhages and SVM

1. Introduction
World Health Organization on global report on diabetes, 2016, estimates that the number of people with diabetes has risen from 108 million in 1980 to 422 million in 2014. One of the major complications associated with diabetes is diabetic retinopathy (DR) which leads to visual impairment in long term. One out of three diabetic person demonstrates signs of DR [1] and one out of ten suffers from its most severe and vision threatening forms [2]. Diabetic retinopathy results in formation of new retinal blood vessels and also leakage from retinal tissues and blood vessels. Diabetic retinopathy is characterized by group of lesions such as hard exudates, soft exudates, microaneurysms, and haemorrhages. Hard exudate, often arranged in clumps or rings, appears as yellowish-white deposits with sharp margins. The debris accumulation appearing as fluffy white lesions in the retinal Nerve Fibre Layer is called soft exudates or cotton wool spots. The red spots with irregular margin and sharp edges are called hemorrhages and microaneurysms respectively. Figure 1 illustrates some samples of normal and DR images.
Due to the increasing number of diabetic patients and the requirement of continuous monitoring on retinal status, the limited number of ophthalmologists cannot satisfy the huge demand. In order to assist in reducing the burden on retina specialists and providing the mass population with retina checking facility, the automation in the screening process is a demand of time. A computer vision assisted automatic system can help to reduce the specialist's burden and the excessive processing time. It can narrow down the search to the most severe and meticulous one and then based on computer feedback, human expert can perform screening more efficiently.
Over the years, several methods have been developed for the automatic detection of diabetic retinopathy in color fundus images. Abràmoff et al. proposed an early treatment diabetic retinopathy study protocol for automatic DR detection using 10,000 fundus images and obtained an area under curve (AUC) of 0.84 [3]. According to the guideline of National Institute for Clinical Excellence, UK, Usher et al. developed a DR screening tool with 1273 patients with a sensitivity of 95.1% and specificity of 46.3% [4].
For automated detection of microaneurysms (MA), several attempts have been made based on morphological operations and top-hat transformation by


Figure 1. Samples of retinal fundus images: (a) (b) and (c) are normal and (d) (e) and (f) contain lesions of diabetic retinopathy.
structuring element [5] [6]. An algorithm to detect the optic disk (OD), blood vessels and fovea was developed in [7] [8]. Perumalsamy et al. have developed Aravind Diabetic Retinopathy Screening 3.0 (ADRES3.0) CAD system using 210 images and achieved an accuracy of 81.3% by comparing the performance with ophthalmologists grading [9]. In recent years, many researchers proposed system for the automatic identification of features for DR to aid the automated diagnosis [10] [11] [12]. However, there is still a need for further improvement in diagnostic accuracy.
This proposed algorithm demonstrates the discrimination between normal and diabetic retinal images without prior need for segmentation of blood vessel and optic disk. The rest of the paper is organized as follows: Section 2 describes the dataset of our work and the proposed approach is explained in Section 3. The experimental results are presented in Section 4 and finally Section 5 concludes the paper including some future directions.
2. Materials
Retinal fundus images, captured clinically by intricate microscope, are not easily accessible as they contain confidential information related to the subjects (patients). However, only for clinical research and experimental purposes, some retinal fundus databases have been made publicly available. Our experimental dataset is composed of normal and diabetic retinal fundus images collected from the following publicly available databases:
・ DRIDB0 and DRIDB1: http://www.fer.unizg.hr/ipg/resources/image_database
・ MESSIDOR: http://messidor.crihan.fr
・ STARE: http://www.ces.clemson.edu/~ahoover/stare/
・ HRF: http://www5.cs.fau.de/research/data/fundus-images/
3. Methodology
The proposed method is based on the bag of words (BOW) approach to automatically discriminate between normal and diabetic retinal fundus images. In an image, every local patch or region contains descriptor which lands a point in high dimensional feature space. In this work, the Speeded Up Robust Features (SURF) interest points are detected and SURF features are extracted from the preprocessed images. Those features are allocated to some clusters by applying K-means clustering algorithm. The cluster centers represent visual words and these words altogether form the vocabulary or bag of words. Now, each individual feature in the image is quantized to the nearest word (cluster center) and thus an entire image is represented by a histogram where each bin indicates the frequency of a word within that vocabulary, i.e., bag of words. The number of bins of the resultant histogram is as the same as the number of words in the vocabulary. On the next step, every histogram is encoded to a one dimensional feature array and then fed to the classifier. The flowchart of proposed algorithm is presented in Figure 2.
Figure 2. Algorithm overview.
3.1. Preprocessing
As the experimental images under study belong to different databases, they contain significant variations in size and resolution. So at the first step of preprocessing, all images are resized to 512 × 512 resolution. From literature review, it is found that the discriminating characteristics are mostly visible in green color channel. So all further processing are performed only on the green channel of the experimental images. Resized and green channel extracted retinal images are shown in Figure 3 and Figure 4, respectively. On the next step, the images are contrast enhanced by applying adaptive histogram equalization technique. As both the diabetic and non-diabetic images contain some significant structural similarities such as optic disk and blood vessels, so it is crucial only to highlight the discriminating lesions to make them easily identifiable by our feature detector. So to achieve this task, some morphological structural element operations are performed over all images. Morphological dilation is performed by a rectangular structuring element of size 25 × 25 over the contrast enhanced image. Then by using a disk-shaped structuring element of radius 25, dilation is performed over the green channel image. Dilated images after both operations are shown in Figure 5. From the rectangular window dilated image, the contrast enhanced image is subtracted. Then this resulting image is again subtracted from the disk window dilated image. After this operation, the final image contains highlighted lesions (for DR) which can be localized efficiently by the feature detector. Final images of preprocessing are presented in Figure 6.
3.2. Speeded up Robust Features (SURF)
SURF is a scale- and rotation-invariant interest point detector and descriptor

Figure 3. Resized images: (a) normal and (b) DR.

Figure 4. Extracted green color channel images: (a) normal and (b) DR.


Figure 5. Resulting images after application of dilation by rectangular and disk structural elements respectively: (a) and (c) are normal and (b) and (d) are DR.

Figure 6. Final preprocessed images: (a) normal and (b) DR
which is computationally fast, while not sacrificing performance. SURF exploits the salient features of integer approximation of the determinant of Hessian matrix and thus it detects the blob interest points. Using the Haar wavelet response around the point of interest, the feature descriptor is constructed. Both feature detection and description is performed by utilizing some predetermined integral images. Integral images can be created by Equations (1):
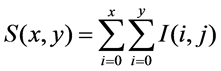 (1)
(1)
where I(i, j) is the brightness values of the pixel in original image and S(x,y) is the integral image. For a point p(x, y) in an image I, the Hessian matrix H(p,s) at point p(x, y) and scale s is denoted by Equations (2):
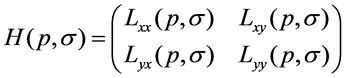 (2)
(2)
By utilizing SURF over the preprocessed images, the lesions were detected and corresponding features were extracted. Figure 7 and Figure 8 show the detected strongest SURF feature points on retinal images.
3.3. Vocabulary Generation by K-Means Clustering
K-means is an unsupervised learning algorithm that solves the well-known clustering problem. The main idea is to define k centroids into the space represented by the objects that are being clustered. Each object is assigned to the group that has the closest centroid. When no point is pending, the first step is completed and an early grouping is done. At this point, we need to re-calculate k new centroids as barycenters of the clusters resulting from the previous step. In this manner, the k centroids change their location step by step until no more changes are done.
Assuming the set of features extracted from the training set can be expressed as {x1, x2, ….,xM} where, xm Î RD, the goal is to partition this feature set into K clusters {d1, d2, ….,dk}, dk Î RD. If we consider for each feature xm, there is a corresponding set of binary indicator variables rmk Î {0, 1}. If xm is appointed to cluster K, then rmk = 1 and rmj = 0 for j ¹ k. The objective function can be defined by Equation (3).

Figure 7. Detection of Surf Feature over retinal images: (a) normal and (b) DR


Figure 8. Representative retinal images: (a) normal and (b); (c) and (d) DR. SURF features on corresponding normal (e) and DR (f); (g) and (h)
 (3)
(3)
The main idea is to find values for both rmk and dk to minimize the objective function H. By utilizing this K-means clustering algorithm, the features extracted by SURF are transformed to K = 100 visual words and then the codebook, i.e., vocabulary, is generated.
3.4. Feature Encoding
Histogram from each image is encoded to a one dimensional feature array. The main idea is that firstly we consider X be a set of D-dimensional descriptors such as X = {x1, x2, ….,xN} Î RDxN. Given a visual dictionary (the one already computed in the previous step) with K visual words, i.e., D = {d1, d2,….,dK} Î RDxK . The purpose of the encoding step is to compute a code for input x with D. Thus, each feature descriptor xn is allocated to the nearest visual word in the dictionary by satisfying arg mink ||xn − dk||2.
3.5. Classification by SVM
Support Vector Machines (SVMs) are supervised learning models which construct an optimal hyperplane to classify data into different classes. Lines drawn parallel to this separating line are the supporting hyperplanes and the distance between them is called the margin. Width of the margin is constrained by support vectors which are the data points that are closest to the separating hyperplane. Since, the optimal hyperplane is the one that separates the high probability density areas of two classes with maximum possible margin between them, the goal is to determine the direction that provides the maximum margin. It needs the solution of following optimization problem of Equation (4) for a given training set of instance label pairs (xi, yi), l = 1, 2,….,i where xi Î Rn and yi Î {1, − 1}i. subjected to
 (4)
(4)
4. Result and Discussion
The proposed algorithm is applied on a database of 180 retinal fundus images collected from some publicly available databases. Our experimental dataset consists of fundus images of 93 of normal and 87 of DR. During the experiment, the database was divided into two sets: the training set containing 126 images (65%) and the testing set containing 54 images (35%). For classification purpose, support vector machine with “linear” Kernel was utilized. For performance evaluation of the classification model, performance parameters such as accuracy, sensitivity, recall, and F1-score were calculated. At the 65% - 35% train-test split, testing accuracy of the classification is 94.44%. Moreover, to make the model more robust, 10-fold cross validation was applied over whole dataset while 94% mean accuracy was achieved. The performance measures for the proposed approach are tabulated in Table 1. In addition, this proposed work is also compared to some other recent works published in 2016 and 2017 in Table 2.
Table 1. Performance Scores of the proposed algorithm.
Table 2. Comparison on various algorithms on normal-DR classification.
For medical image processing, high classification performance is crucial as a false prediction may lead to life and death situation. In term of performance measures, our proposed method outperforms the other state-of-the-art algorithms of diabetic retinopathy detection. The method proposed by Morales et al. in [3] utilize Local Binary Pattern and perform Normal-DR classification with a True Positive Rate of 85.6% and True Negative Rate of 89.7%. Seoud et al. per
form Diabetic Retinopathy detection based on only Red lesions over Messidor database with an AUC of 89.9% by exploiting dynamic shape features [14]. By using Minimum Intensity Maximum Solidity (MinIMaS) algorithm, Saranya and Selvarani in [15] achieve a sensitivity of 89% for grouping of bright lesion and a sensitivity of 82% for classifying bright lesion. Our proposed method can detect DR containing any lesions such as hard exudates, soft exudates, microaneurysms and hemorrhages and with a better accuracy, precision, sensitivity and AUC score. It is to be noted that the obtained results are not directly comparable because the dataset used in each case is different.
Figure 9 shows the graphical representation of the confusion matrix containing test data for normal (class 0) and DR (class 1). Figure 10 illustrates that the area under the ROC curve for our classification is 95% that indicates excellent discrimination capabilities of our classifier.
5. Conclusion
In this work, bag of words model with support vector machines is utilized to develop an automated and easily accessible diagnosis system. The leading cause of vision-loss among adults, diabetic retinopathy, is screened with a little computational effort. The proposed approach will offer ophthalmologists a feasible, efficient, and timesaving way of diabetic retinopathy detection. The bag of words approach along with surf feature descriptor demonstrates excellent discriminat-
Figure 9. Plot of confusion matrix for test data.
Figure 10. ROC analysis for classification.
ing capabilities with an accuracy of 94.4%. In the future, more number of featuredetector and descriptors such as ASIFT, HOG and LBP and more experimental images to be integrated into the system. Furthermore, work on automatically estimating the severity of diabetic retinopathy is to be implemented.
Cite this paper
Islam, M., Dinh, A.V. and Wahid, K.A. (2017) Automated Diabetic Retinopathy Detection Using Bag of Words Approach. J. Biomedical Science and Engineering, 10, 86-96. https://doi.org/10.4236/jbise.2017.105B010
References
- 1. Ding, J. and Wong, T.Y. (2011) Current Epidemiology of Diabetic Retinopathy and Diabetic Macular Edema. Curr. Diabetes Rep., 12, 346-354. https://doi.org/10.1007/s11892-012-0283-6
- 2. Yau, J.W.Y., et al. (2012) Global Prevalence and Major Risk Factors of Diabetic Retinopathy. Diabetes Care, 35, 556-564. https://doi.org/10.2337/dc11-1909
- 3. Abramoff, M.D., Niemeijer, M., Suttorp-Schulten, M.S.A., Viergever, M.A., Russell, S.R. and van Ginneken, B. (2008) Evaluation of a System for Automatic Detection of Diabetic Retinopathy from Color Fundus Photographs in a Large Population of Patients with Diabetes. Diabetes Care, 31, 193-198. https://doi.org/10.2337/dc07-1312
- 4. Usher, D., Dumskyj, M., Himaga, M., Williamson, T.H., Nussey, S. and Boyce, J. (2004) Automated Detection of Diabetic Retinopathy in Digital Retinal Images: A Tool for Diabetic Retinopathy Screening. Diabetic Med., 21(1), 84-90. https://doi.org/10.1046/j.1464-5491.2003.01085.x
- 5. Frame, A.J., et al. (1998) A Comparison of Computer Based Classification Methods Applied to the Detection of Microaneurysms in Ophthalmic Fluorescein Angiograms. Comput. Biol. Med., 28, 225-238. https://doi.org/10.1016/S0010-4825(98)00011-0
- 6. Niemeijer, M., van Ginneken, B., Staal, J., Suttorp-Schulten, M.S.A., and Abràmoff, M.D. (2005) Automatic Detection of Red Lesions in Digital Color Fundus Photographs. IEEE Trans. Med. Imag., 24, 584-592. https://doi.org/10.1109/TMI.2005.843738
- 7. Sinthanayothin, C. (1999) Image Analysis for Automatic Diagnosis of Diabetic Re-tinopathy. PhD Thesis, King’s College of London.
- 8. Hsiao, H.-K., Liu, C.-C., Yu, C.-Y., Kuo, S.-W. and Yu, S.-S. (2012) A Novel Optic Disc Detection Scheme on Retinal Images. Expert Syst. Appl. 39, 10600-10606. https://doi.org/10.1016/j.eswa.2012.02.157
- 9. Perumalsamy, N., Prasad, N.M., Sathya, S. and Ramasamy, K. (2007) Software for Reading and Grading Diabetic Retinopathy: Aravind Diabetic Retinopathy Screening 3.0. Diabetes Care, 30, 2302-2306. https://doi.org/10.2337/dc07-0225
- 10. Larsen, M., Godt, J., Larsen, N., Lund-Andersen, H., Sjolie, A.K., Agardh, E., Kalm, H., Grunkin, M. and Owens, D.R. (2003) Automated Detection of Fundus Photographic Red Lesions in Diabetic Retinopathy, Invest. Ophthalmol. Vis. Sci., 44, 761-766. https://doi.org/10.1167/iovs.02-0418
- 11. Usher, D., Dumskyj, M., Himaga, M., Williamson, T.H., Nussey, S. and Boyce, J. (2004) Automated Detection of Diabetic Retinopathy in Digital Retinal Images: A Tool for Diabetic Retinopathy Screening, Diabetic Med., 21, 84-90. https://doi.org/10.1046/j.1464-5491.2003.01085.x
- 12. Zhang, X. and Chutatape, A. (2004) Detection and Classification of Bright Lesions in Color Fundus Images. ICIP’04, 2004 International Conference on Image Processing, 131, 139-142.
- 13. Morales, S., Engan, K., Naranjo, V. and Colomer, A. (2017) Retinal Disease Screening through Local Binary Patterns. IEEE Journal of Biomedical and Health Informatics, 21, 184-192. https://doi.org/10.1109/JBHI.2015.2490798
- 14. Seoud, L., Hurtut, T., Chelbi, J., Cheriet, F. and Langlois, J.M.P. (2016) Red Lesion Detection Using Dynamic Shape Features for Diabetic Retinopathy Screening. IEEE Trans-actions on Medical Imaging, 35, 1116-1126. https://doi.org/10.1109/TMI.2015.2509785
- 15. Saranya, M., and Grace Selvarani, A. (2016) Fundus Image Screening for Diabetic Retinopathy. Indian Journal of Science and Technology, 9. https://doi.org/10.17485/ijst/2016/v9i25/90183





