Natural Science
Vol.4 No.1(2012), Article ID:17029,9 pages DOI:10.4236/ns.2012.41007
Expression of casein kinase genes in glioma cell line U87: Effect of hypoxia and glucose or glutamine deprivation
![]()
1Department of Molecular Biology, Palladin Institute of Biochemistry, National Academy of Science of Ukraine, Kyiv, Ukraine;
2Bogomolets National Medical University, Kyiv, Ukraine; *Corresponding Author: ominchenko@yahoo.com
Received 7 October 2011; revised 18 November 2011; accepted 25 November 2011
Keywords: mRNA Expression; Casein Kinase 1A; 1D; 1G2; 1G3; 1E; 2A1; 2A2; 2B and NUCKS1; Glioma Cells; Endoplasmic Reticulum-Nuclei-1 (ERN1; IRE-1α); Hypoxia; Glucose and Glutamine Deprivation
ABSTRACT
The endoplasmic reticulum-nuclei-1 (ERN1) sensing and signaling enzyme mediates a set of complex intracellular signaling events known as the unfolded protein response. We have studied the effect of hypoxia and ischemic conditions (glucose or glutamine deprivation) on the expression of several casein kinase-1 and -2 genes in glioma U87 cells and its subline with suppressed function of ERN1. It was shown that blockade of ERN1, the key endoplasmic reticulum stress sensor, leads to an increase in the expression levels of casein kinase-1G2, -1E, -2B and NUCKS1 mRNA, but suppresses casein kinase-1A1, -1D and -2A1. Moreover, the expression levels of casein kinase-1A1, -1D and 1G3 as well as casein kinase-2A1 and -2A2 mRNAs are significantly increased under glutamine deprivation conditions both in control and ERN1-deficient glioma cells. At the same time, casein kinase-1E, -2B and NUCKS1 mRNA expression levels are also increased under this condition, but only in cells with suppressed function of ERN1. The expression level of NUCKS1 mRNA, however, is decreased both in control glioma cells and in genetically modified cells, but casein kinase-1G2—only in control U87 cells. Cell exposure to glucose deprivation conditions enhances the expression levels of casein kinase- 1D, 1G3, -1E and -2A1 in both types of glioma cells used, but casein kinase-2B expression levels increase only in cells with suppressed function of ERN1. Hypoxia induces or suppresses the expression of most of the studied genes mainly in ERN1-knockdown cells only. Results of this study show that hypoxia as well as glutamine and glucose deprivation conditions change the expression level most of casein kinase genes and that these effects are dependent on ERN1 signaling enzyme function.
1. INTRODUCTION
The endoplasmic reticulum is a key organelle in the cellular response to ischemia, hypoxia, and some chemicals which activate a complex set of signaling pathways named the unfolded protein response. This adaptive response is activated upon the accumulation of misfolded proteins in the endoplasmic reticulum and is mediated by three endoplasmic reticulum-resident sensors named PERK (PRK-like ER kinase), ERN1 (Endoplasmic Reticulum—Nuclei-1) also known as IRE-1α (Inositol Requiring Enzyme-1α) and ATF6 (Activating Transcription Factor 6), however, ERN1 is the dominant sensor [1-5]. Activation of the unfolded protein response tends to limit the de novo entry of proteins in to the endoplasmic reticulum and facilitate both the endoplasmic reticulum protein folding and degradation to adapt cells for survival or, alternatively, to enter cell death programs through endoplasmic reticulum-associated machineries [1,2,4]. As such, it participates in the early cellular response to the accumulation of misfolded proteins in the lumen of the endoplasmic reticulum, occurring under both physiological and pathological conditions.
Two distinct catalytic domains of the bifunctional signaling enzyme ERN1 were identified: a serine/threonine kinase and an endoribonuclease which contribute to this enzyme signalling. The ERN1-associated kinase activity autophosphorylates and dimerizes this enzyme, leading to the activation of its endoribonuclease domain, initiation of the pre-XBP1 (X-box binding protein 1) mRNA splicing, and degradation of a specific subset of mRNA [6-8]. Mature XBP1 mRNA splice variant (XBP1s) encodes a transcription factor that has different C-terminal amino acid sequence and stimulates the expression of hundreds of unfolded protein response-specific genes [1,9-11] . In ERN1 gene of different human cancers single mutations were detected and encoded by this gene protein was proposed as a major contributor to tumor progression among protein kinases [12].
Moreover, the growing tumor requires the endoplasmic reticulum stress as well as hypoxia and ischemia for own neovascularization and growth and the complete blockade of ERN1 signal transduction pathway has antitumor effects [13-16] . The endoplasmic reticulum stress response-signalling pathway is linked to the neovascularization process, tumor growth and differentiation as well as cell death processes [15,17,18]. Thus, the blockade of the main unfolded protein response sensor ERN1 is important in studying the role of ERN1 signalling pathways in tumor progression, especially in malignant gliomas; it is important in developing a new understanding concerning molecular mechanisms of malignant tumors progression in relation to ischemia/hypoxia and it can help define the best targets for the design of specific inhibitors that could act as potent antitumor drugs.
It is known that the circadian clock system controls the cell proliferation process and carcinogenesis, thus circadian proteins PER1, PER2, CLOCK, BMAL1 and CRY1 function as tumor suppressors [19-21]. Deregulated expression of the PER1, PER2 and PER3 genes as well as inactivation or knockout PER1, PER2, CLOCK and BMAL1 leads to malignization, especially in the breast, as well as to the appearance of different disorders such as obesity [22-25]. In the regulation of circadian clock system casein kinases play an important role, especially casein kinase-1ε (CSNK1E) and -1δ (CSNK1D) [26-28]. The phosphorylation state of a protein is controlled dynamically by both protein kinases and also phosphatases. Phosphorylation of circadian clock proteins is an essential posttranscriptional mechanism in the regulation of circadian clocks, and several protein kinases and phosphatases have been shown to regulate key clock components in eukaryotic systems [27]. Moreover, casein kinase-1δ is the principal regulator of the clock periods compared to casein kinase-1ε, because selective inhibition of casein kinase-1ε minimally alters circadian clock period [28,29]. Inhibition of casein kinase I epsilon/delta produces phase shifts in the circadian rhythms [30].
Casein kinase-1ε is a serine/threonine protein kinase that has been suggested to participate in the regulation of various signaling pathways. Three alternatively spliced forms of CSNK1E were found in rat tissues, which are expressed with different tissue-specific expression patterns [31]. Moreover, the kinase activity of the CSNK1E-3 protein differed from that of CSNK1E-1. This data suggests that the two variants of CSNK1E may play different functional roles. There is data that casein kinase-1ε and -1δ are required for efficient removal of Rec8, the meiosis-specific alpha-kleisin subunit of cohesion, during meiosis [32]. Moreover, CSNK1E is a crucial regulator of the Wnt signaling cascades, and breast cancer-specific mutations in this gene inhibit Wnt/beta-catenin and activate the Wnt/ Rac1/JNK and NFAT pathways to decrease cell adhesion and promote cell migration, thus contributing to breast cancer development via effects on cell adhesion and migration [33,34]. It was also shown that three isoforms of casein kinase-1 (alpha, gamma-1 and delta) participate in phosphorylation of the N-terminal region of TP53 [35]. There is data that casein kinase-1 gamma 2, an inhibitor of TGF-beta signaling, regulates ligand-dependent ubiquitynation of Smad3 [36].
Casein kinase-2 is a constitutively active, ubiquitously expressed serine/threonine kinase consisting of two catalytic α/α' and two regulatory β subunits and is thought to have a regulatory function in cell proliferation, cell differentiation and apoptosis. CSNK2 regulates metabolic pathways, signal transduction, transcription, translation, and replication via phosphorylation of a number of key intracellular signaling proteins implicated in tumor suppression (TP53 and PTEN) and tumorigenesis (myc, jun, NF-kappaB). CSNK2 is also thought to influence Wnt signaling via beta-catenin phosphorylation and the PI3-K signaling pathway via the phosphorylation of Akt. Expression of CSNK2 is highly elevated in tumor cells where it protects cells from apoptosis [37]. Accordingly inhibition of CSNK2 is known to induce programmed cell death, making it a promising target for cancer therapy. Inhibitor of CSNK2 4,5,6,7-tetrabromobenzotriazole induces apoptosis in LNCaP cells by down-regulation of BCL-2 and subsequent cytochrome c release without activation of caspase 9 [37]. Instead, an activation of the endoplasmic reticulum stress response in LNCaP cells after treatment with the CSNK2 inhibitor was found. Thus, protein kinase CSNK2 inhibition results in activetion of the receptor mediated apoptosis pathway via the endoplasmic reticulum stress response and up-regulation of the death receptor DR5 [37].
A better understanding of the impact of circadian gene networks regulation on cell cycle control as well as nutrient balance at the molecular, cellular and system levels promises to shed light on the emerging association between casein kinases, circadian timing, endoplasmic reticulum stress response and cancer.
The main goal of this work is to study the role of ERN1- signaling pathways in tumor progression by investigating the expression of different variants of casein kinase-1 and casein kinase-2 in U87 glioma cells and its ERN1-deficient subline under normal, hypoxic and ischemic (glucose or glutamine deprivation) conditions.
2. MATERIALS AND METHODS
2.1. Cell Lines and Culture Conditions
The glioma cell line U87 was obtained from ATCC (USA) and grown in high glucose (4.5 g/l) Dulbecco’s modified Eagle’s minimum essential medium (DMEM; Gibco, Invitrogen, USA) supplemented with glutamine (2 mM), 10% fetal bovine serum (Equitech-Bio, Inc., USA), penicillin (100 units/ml; Gibco) and streptomycin (0.1 mg/ml; Gibco) at 37˚C in a 5% CO2 incubator. In this work we used two sublines of this glioma cell line. One subline was obtained by selection of stable transfected clones with overexpression of vector, which was used for creation of dnERN1. This untreated subline of glioma cells (control glioma cells) was used as control 1 in the study of the effect of hypoxia and glutamine or glucose deprivations on the expression level of different casein kinase genes. Second subline was obtained by selection of stable transfected clones with overexpression of ERN1 dominant/negative constructs (dnERN1) and has suppressed both protein kinase and endoribonuclease activities of this signaling enzyme [13]. The expression level of casein kinase genes in these cells was compared with cells, transfected by vector (control 1), but this subline was also used as control 2 for investigation the effect of hypoxia and glutamine or glucose deprivations on the expression level of casein kinase genes under blockade ERN1 function.
Hypoxic conditions were created in special incubator with 3% oxygen and 5% carbon dioxide levels; culture plates were exposed to these conditions for 16 hr. For glucose or glutamine deprivation the growing medium in culture plates was replaced with a medium without glucose or without glutamine and thus exposed for 16 hr.
2.2. RNA Isolation
The Total RNA was extracted from both variants of glioma U87 cells using Trizol reagent according to manufacturer protocol (Invitrogen, USA) as described previously [38]. RNA pellets was washed with 75% ethanol, dissolved in nuclease-free water and used for creation of complementary DNA (cDNA).
2.3. Reverse Transcription and Quantitative PCR Analysis
The expression of casein kinase-1A1, -1D, -1G2, -1G3, -1E, -2A1, -2A2, -2B and NUCKS1 was measured in glioma cell line U87 and its subline with ERN1-deficiency by quantitative polymerase chain reaction of cDNA using, “Stratagene Mx 3000P cycler” (USA) and SYBR Green Mix (AB gene, Great Britain). QuaniTect Reverse Transcription Kit (QIAGEN, Germany) was used for cDNA synthesis as described previously [38]. Polymerase chain reaction was performed in triplicate.
For amplification of casein kinase-1α1 (casein kinase- 1A1; CSNK1A1) cDNA we used forward (5’-ggatcttct gggacctagcc-3’ and reverse (5’-tgttgccttgtcctgttgtc-3’) primers which create 246 bp fragment. The nucleotide sequences of these primers correspond to sequences 750 - 769 and 995 - 976 of human CSNK1A1 cDNA (GenBank accession number NM_001892).
The amplification of casein kinase-1δ (casein kinase- 1D; CSNK1D) cDNA was performed using forward primer (5’-cttggagtctctgggctacg-3’) and reverse primer (5’-tggaacagattccggaaaag-3’), which create 249 bp fragment. These oligonucleotides correspond to sequences 922 - 941 and 1170 - 1151 of human CSNK1D cDNA (GenBank accession number NM_001893).
The amplification of casein kinase-1γ2 (casein kinase- 1G2; CSNK1G2) cDNA for real time RCR analysis was performed using two oligonucleotides primers: forward— 5’-caaattggagccgatcaagt-3’ and reverse—5’-ggtgtgcacatactccatgc-3’, which create 253 bp fragment. The nucleotide sequences of these primers correspond to sequences 745 - 764 and 997 - 978 of human CSNK1G2 cDNA (GenBank accession number NM_001319).
Two other primers were used for real time RCR analysis of casein kinase-1γ3 (casein kinase-1G3; CSNK1G3) cDNA expression: forward—5’-actgggtcttcatcgtctgg-3’ and reverse—5’-taccacaagggccgaaatag-3’, which create 244 bp fragment. The nucleotide sequences of these primers correspond to sequences 804 - 823 and 1047 - 1028 of human CSNK1G3 cDNA (GenBank accession number NM_004384).
For amplification of casein kinase-1ε (casein kinase- 1E; CSNK1E) cDNA we used forward (5’-tgagtatgaggctgcacagg-3’ and reverse (5’-aaactccatcgtcggttttg-3’) primers, which create 264 bp fragment. The nucleotide sequences of these primers correspond to sequences 1238 - 1257 and 1501 - 1482 of human CSNK1E cDNA (GenBank accession number NM_001894).
For real time PCR analysis of casein kinase-2α1 (casein kinase-2A1; CSNK2A1) cDNA expression we used next primers: forward—5’-acgagtcacatgtggtggaa-3’ and reverse—5’-ggttcgtgacacagggtctt-3’, which create 248 bp fragment. The nucleotide sequences of these primers correspond to sequences 353 - 372 and 600 - 581 of human CSNK2A1 cDNA (GenBank accession number NM_001895).
Two other primers were used for real time RCR analysis of casein kinase-2α2 (casein kinase-2α’ or -2A2; CSNK2A2) cDNA expression: forward—5’-catgcacagggatgtgaaac-3’ and reverse—5’-tgcgaacaagctggtcatag-3’, which create 278 bp fragment. The nucleotide sequences of these primers correspond to sequences 601 - 620 and 878 - 859 of human CSNK2A2 cDNA (GenBank accession number NM_001896).
The amplification of casein kinase-2β (casein kinase- 2B; CSNK2B) cDNA for real time RCR analysis was performed using next primers: forward—5’-tcctggatttcctggttctg-3’ and reverse—5’-cactctggttggggttgtct-3’, which create 187 bp fragment. The nucleotide sequences of these primers correspond to sequences 362 - 381 and 548 - 529 of human CSNK2B cDNA (GenBank accession number NM_001320).
Two primers were used for real time RCR analysis of nuclear ubiquitous casein kinase NUCKS1 (cyclin-dependent kinase substrate 1) cDNA expression: forward— 5’-gggcagtgaggaagaacaag-3’ and reverse—5’-ttgatgcctttgaagctgtg-3’, which create 263 bp fragment. The nucleotide sequences of these primers correspond to sequences 604 - 623 and 866 - 847 of human NUCKS1 cDNA (GenBank accession number NM_022731).
The amplification of beta-actin cDNA was performed using primers: forward—5’-CGTACCACTGGCATCGTGAT-3’ and reverse—5’-GTGTTGGCGTACAGGTCTTT-3’. The expression of beta-actin mRNA was used as control of analyzed RNA quantity. The primers were received from “Sigma” (USA).
Analysis of quantitative PCR was performed using special computer program “Differential expression calculator” and statistic analysis—in Excel program. The amplified DNA fragments were separated on a 2% agarose gel and visualized by 5× Sight DNA Stain (EUROMEDEA).
3. RESULTS
In this study, we have used the human glioma cell line U87 and a genetically modified variant of these cells (deficient in the signaling enzyme ERN1) to investigate the expression of different genes that encode different isoforms of casein kinase-1 and casein kinase-2 as well as the involvement of endoplasmic reticulum stress signaling system in the effect of hypoxia and glutamine or glucose deprivation on the expression of these genes. The level of suppression of the enzymatic activity of ERN1 was estimated by analysis of the expression of transcription factor XBP1 and its splice variant (XBP1s) in U87 glioma cells that over express a dominant-negative construct of ERN1 as compared to control glioma cells transfected with a vector. As shown in Figure 1, inductor of endoplasmic reticulum stress, tunicamycin (0.01 mg/ml), strongly induces the alternative splicing of XBP1 only in control glioma cells, while having no effect on this process in transfected by dnERN1 subline glioma cells.
We have found that casein kinase-1α1, -1δ, -1γ2, -1γ3 and -1ε mRNAs are expressed in the human glioma cell line U87 and the level of their expression is dependent on signaling enzyme ERN1 function as well as from glutamine or glucose deprivation. As shown in Figure 2, the level of casein kinase-1α mRNA expression is decreased by 28% in glioma cells, deficient in signaling enzyme ERN1, as compared to control cells. Exposure of cells for 16 hours to glutamine deprivation condition leads to an increase of CSNK1A1 mRNA expression level in control glioma cells (+20%) and ERN1 knockdown cells (+86%), but glucose deprivation condition induces this mRNA expression in ERN1 knockdown cells only (+17% and +35%, respectively, as compared to control 2).
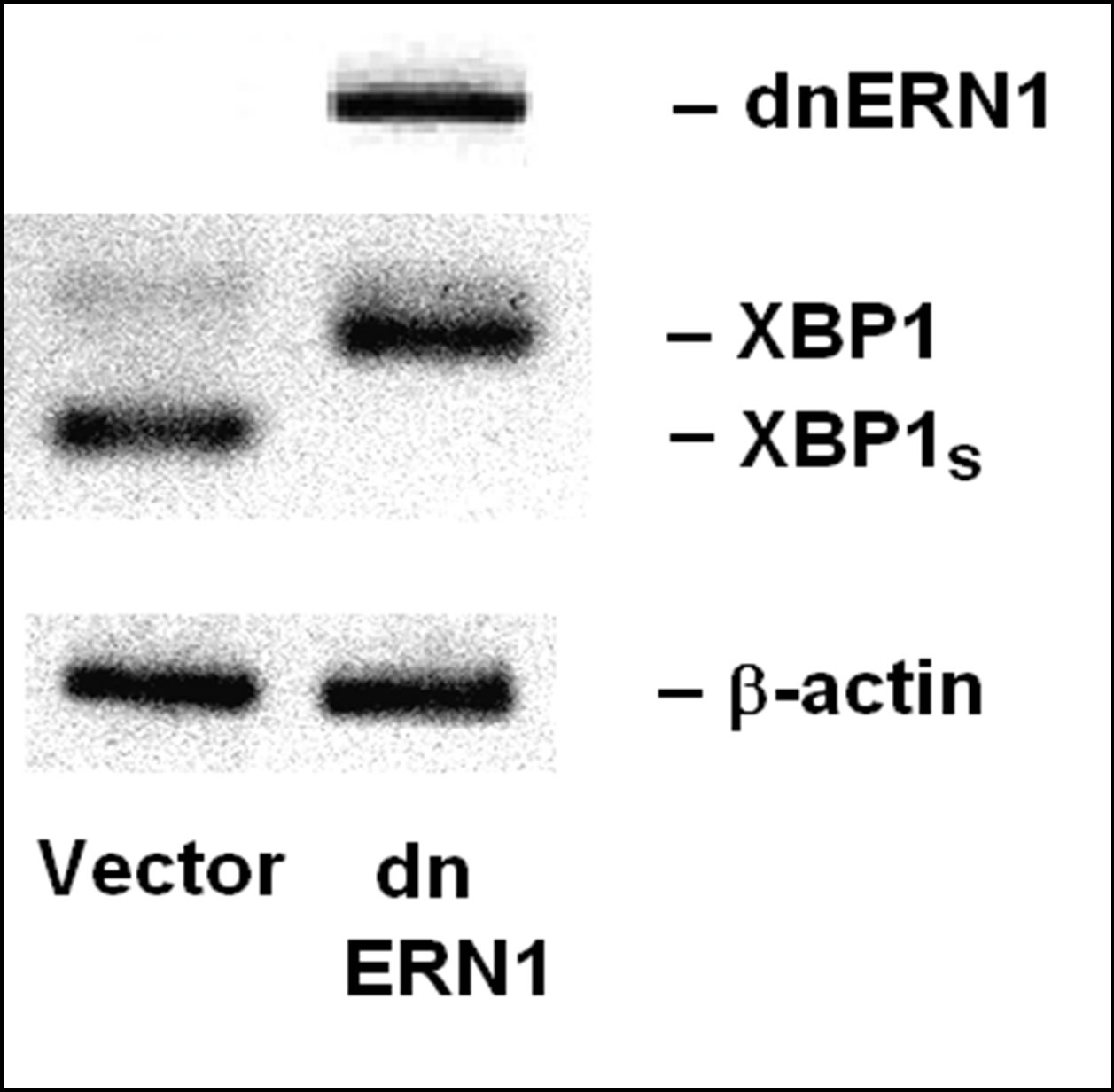
Figure 1. Expression of transcription factor XBP1, its alternative splice variant (XBP1s) and dominant-negative endoplasmic reticulum-nuclei-1 (dnERN1) mRNA in glioma cell line U87 stable transfected with vector (Vector) and its subline with blockade of signaling enzyme endoplasmic reticulum-nuclei-1 stable transfected with dnERN1 by polymerase chain reaction.
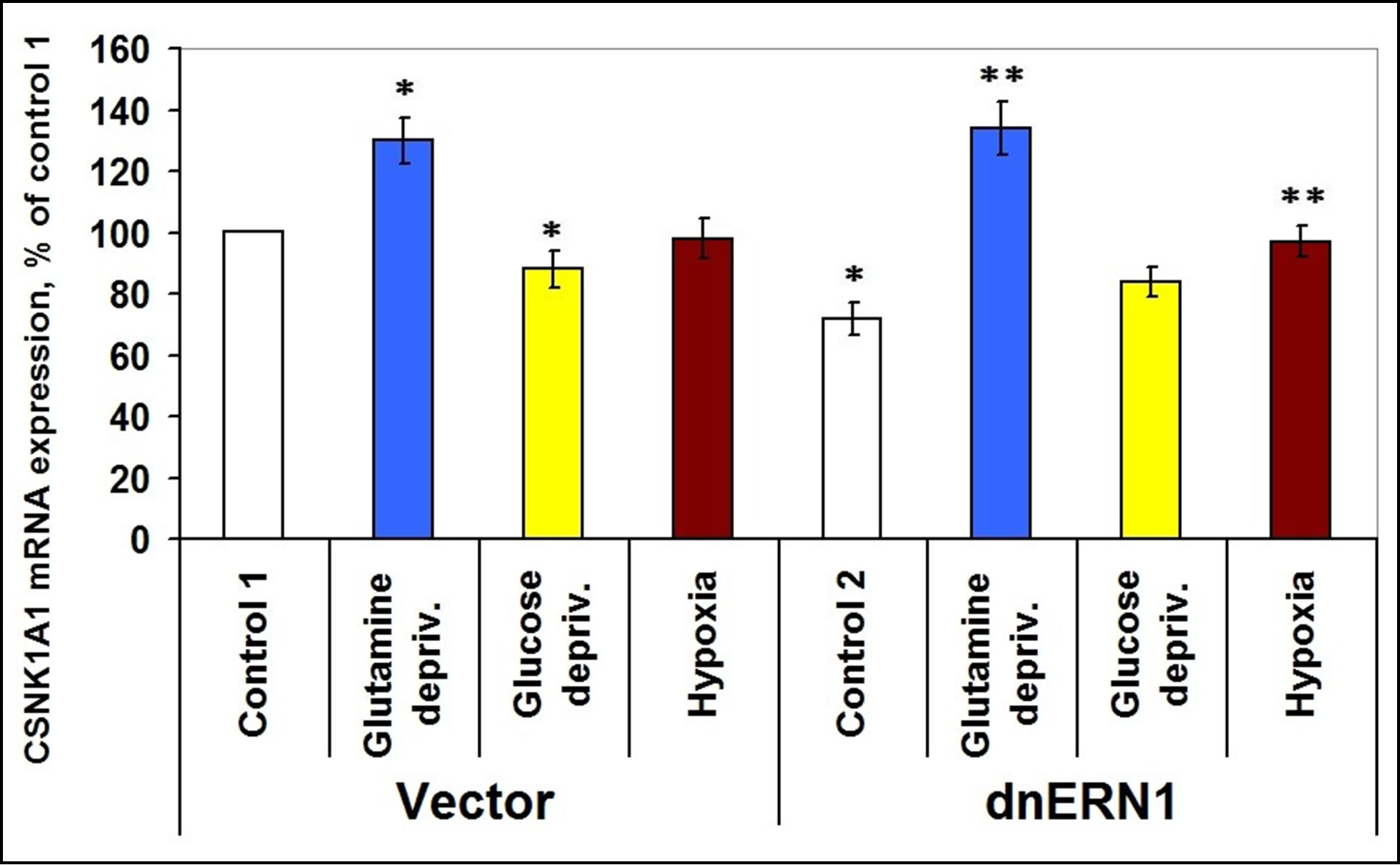
Figure 2. Effect of hypoxia and glutamine or glucose deprivation on the expression of casein kinase-1A1 (CSNK1A1) mRNA in glioma cell line U87 (control 1) and its subline with blockade of signaling enzyme endoplasmic reticulum-nuclei-1 (dnERN1) activity (control 2) measured by quantitative polymerase chain reaction. Values of CSNK1A1 mRNA expression were normalized to beta-actin mRNA expression and represent as percent for control 1 (100%); n = 4; *P < 0.05 as compared to control 1; **P < 0.05 as compared to control 2.
No significant changes in CSNK1A1 mRNA expression level were found in control glioma cells after exposure to hypoxia.
As shown in Figure 3, expression levels of casein kinase-1δ mRNA in glioma cells, deficient in signaling enzyme ERN1 are decreased by 33% as compared to control 1. Exposure of cells to hypoxia for 16 hrs leads to a decrease of this mRNA expression level, but only in glioma cells with suppressed function of ERN1 (–39%), as compared to control 2. However, under glutamine or glucose deprivation conditions expression level of casein kinase-1δ mRNA increases in control glioma cells (+38% and +23%, respectively, as compared to control 1), but much stronger in ERN1 knockdown glioma cells (+162% and +60%, respectively, as compared to control 2).
At the same time, the expression level of casein kinase-1γ3 mRNA does not change significantly, but the expression of casein kinase-1γ2 and of casein kinase-1ε mRNA is increased in ERN1 knockdown glioma cells (+38% and +23%, respectively, as compared to control 1; Figures 4-6). No significant changes were found in both types of glioma cells tested under hypoxic and ischemic conditions; only glucose deprivation conditions in cells, deficient in signaling enzyme ERN1 showed a decrease (−20%, as compared to control 2; Figure 4).
Exposure of cells for 16 hours in glutamine or glucose deprivation condition leads to an increase of CSNK1G3 mRNA expression levels in both tested glioma cell types, but no significant changes were found in CSNK1A1 mRNA expression levels in control glioma cells and in ERN1 knockdown glioma cells under hypoxia (Figure 5).
It was shown that expression level of casein kinase-1ε mRNA is increased in glutamine or glucose deprivation condition, but only in ERN1 knockdown glioma cells
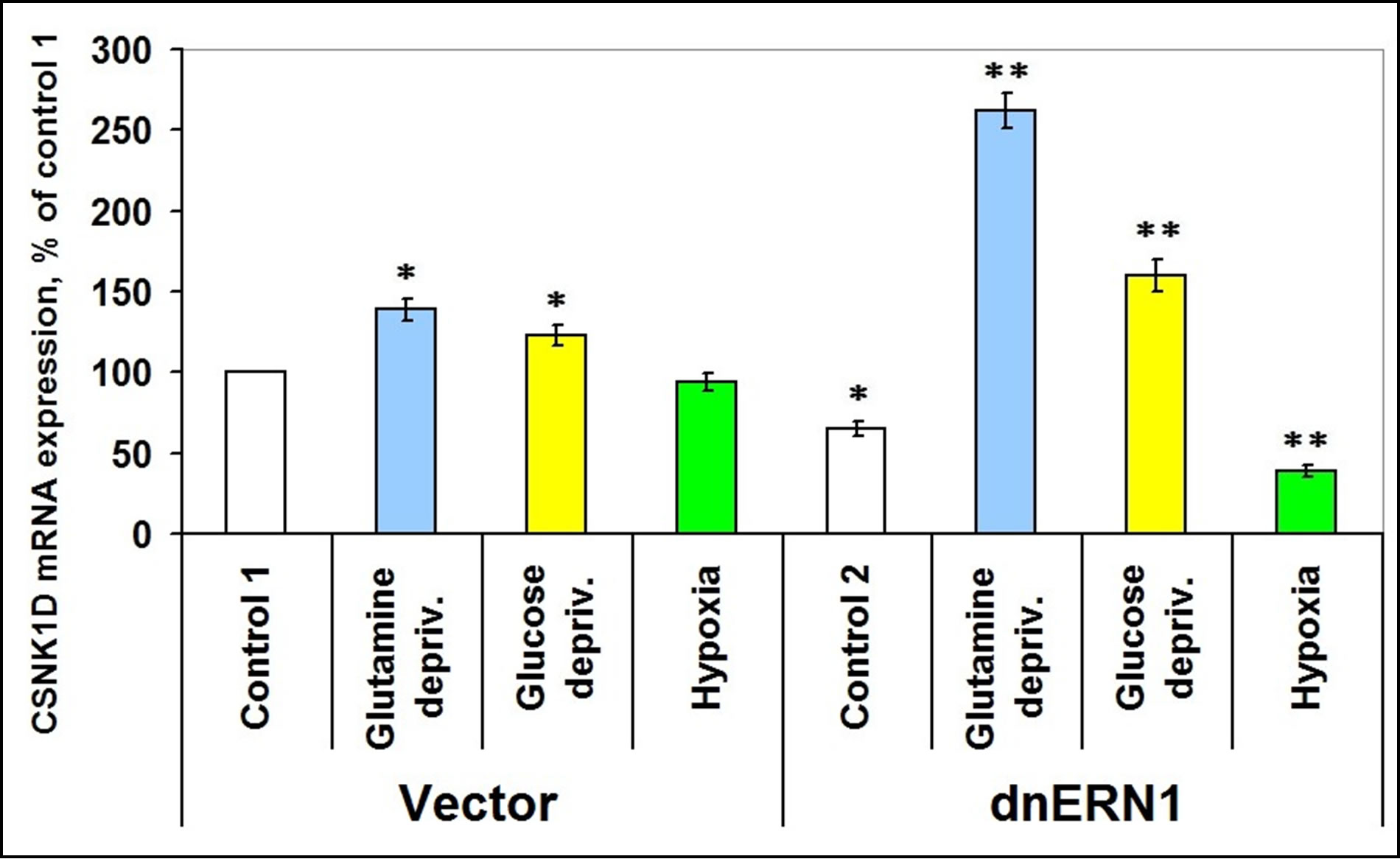
Figure 3. Effect of hypoxia and glutamine or glucose deprivation on the expression of casein kinase-1D (CSNK1D) mRNA in glioma cell line U87 (control 1) and its subline with blockade of signaling enzyme endoplasmic reticulum-nuclei-1 (dnERN1) activity (control 2) measured by quantitative polymerase chain reaction. Values of CSNK1D mRNA expression were normalized to beta-actin mRNA expression and represent as percent for control 1 (100%); n = 4; *P < 0.05 as compared to control 1; **P < 0.05 as compared to control 2.

Figure 4. Effect of hypoxia and glutamine or glucose deprivation on the expression of casein kinase-1G2 (CSNK1G2) mRNA in glioma cell line U87 (control 1) and its subline with blockade of signaling enzyme endoplasmic reticulum-nuclei-1 (dnERN1) activity (control 2) measured by quantitative polymerase chain reaction. Values of CSNK1G2 mRNA expression were normalized to beta-actin mRNA expression and represent as percent for control 1 (100%); n = 4; *P < 0.05 as compared to control 1; **P < 0.05 as compared to control 2.
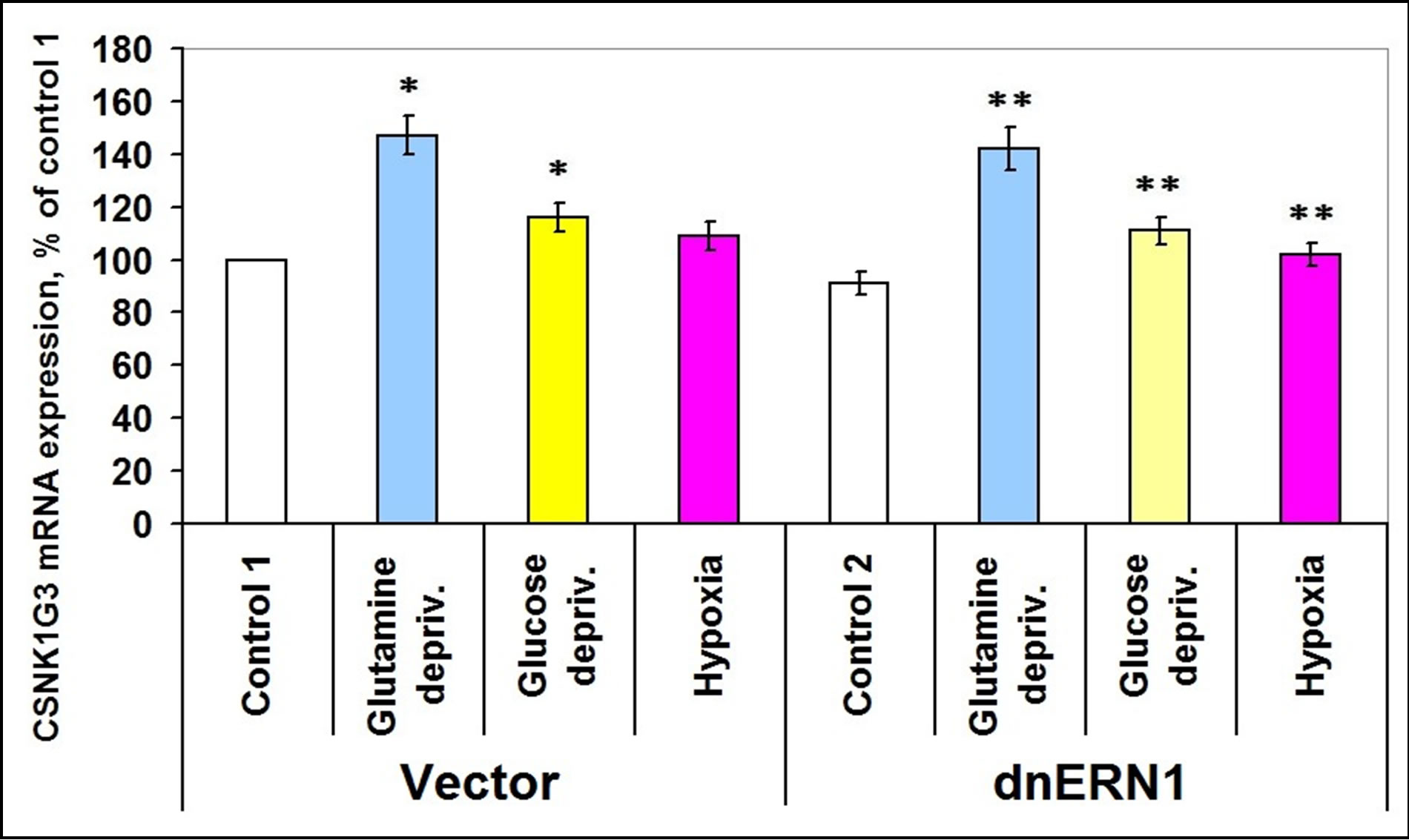
Figure 5. Effect of hypoxia and glutamine or glucose deprivation on the expression of casein kinase-1G3 (CSNK1G3) mRNA in glioma cell line U87 (control 1) and its subline with blockade of signaling enzyme endoplasmic reticulum-nuclei-1 (dnERN1) activity (control 2) measured by quantitative polymerase chain reaction. Values of CSNK1G3 mRNA expression were normalized to beta-actin mRNA expression and represent as percent for control 1 (100%); n = 4; *P < 0.05 as compared to control 1; **P < 0.05 as compared to control 2.
(+23% and +26%, respectively, as compared to control 2; Figure 6). Hypoxia, however, induces expression of this mRNA in both types of tested glioma cells (Figure 6).
Investigation of the expression levels of different subunits of casein kinase-2 (-2α1, -2α2 and -2β showed different patterns of its expression in ERN1 knockdown glioma cells: casein kinase-2α1 is decreased (–18%), casein kinase-2β is increased (+38%) and does not change significantly (Figures 7-9). Glutamine and glucose deprivation conditions lead to an increase of casein kinase-2α1 mRNA expression in both tested cell types, but the effect in ERN1 knockdown glioma cells was much stronger (+33% and +14%, respectively, in control cells, as compared to control 1, and +61% and +77%, respectively, in ERN1 knockdown glioma cells, as compared to control 2; Figure 7). At the same time, hypoxia does not change significantly the expression of CSNK2A1 in both tested cell types, as shown in Figure 7.
Glutamine deprivation condition also induces the expression of CSNK2A2 both in control glioma cells (+51%) and in ERN1 knockdown cells (+42%), but hypoxia induces this mRNA expression in control glioma cells only (Figure 8). Moreover, in glioma cells with suppressed activity of signaling enzyme ERN1 expression of CSNK2A2 significantly decreased (almost 2 times). However, expression of casein kinase-2β is increased in glutamine and glucose deprivation conditions in ERN1 knockdown glioma cells only (+11% and +16%, respectively, as compared to control 2) and does not change significantly in control glioma cells (Figure 9). As shown in Figure 10, the expression level of mRNA of nuclear ubiquitous casein kinase NUCKS1 is increased (+26%) in glioma cells with suppressed activity of signaling enzyme ERN1 as compared to control glioma cells. Investigation of the effect of glutamine deprivation condition on the expression level of NUCKS1 mRNA showed that both in control and ERN1- deficient glioma cells there was a significant decrease of the expression of this mRNA; in control cells, however, the change was slightly less profound.
The level of NUCKS1 mRNA expression is also decreased under glucose deprivation condition, but only in control glioma cells (–18%). At the same time, effect of hypoxia on the expression level of NUCKS1 mRNA was different in control glioma cells and in ERN1 knockdown cells: there was a decrease (–18%) in control glioma cells as compared to control 1 and an increase (+25%) in glioma cells with suppressed activity of signaling enzyme ERN1 (Figure 10).
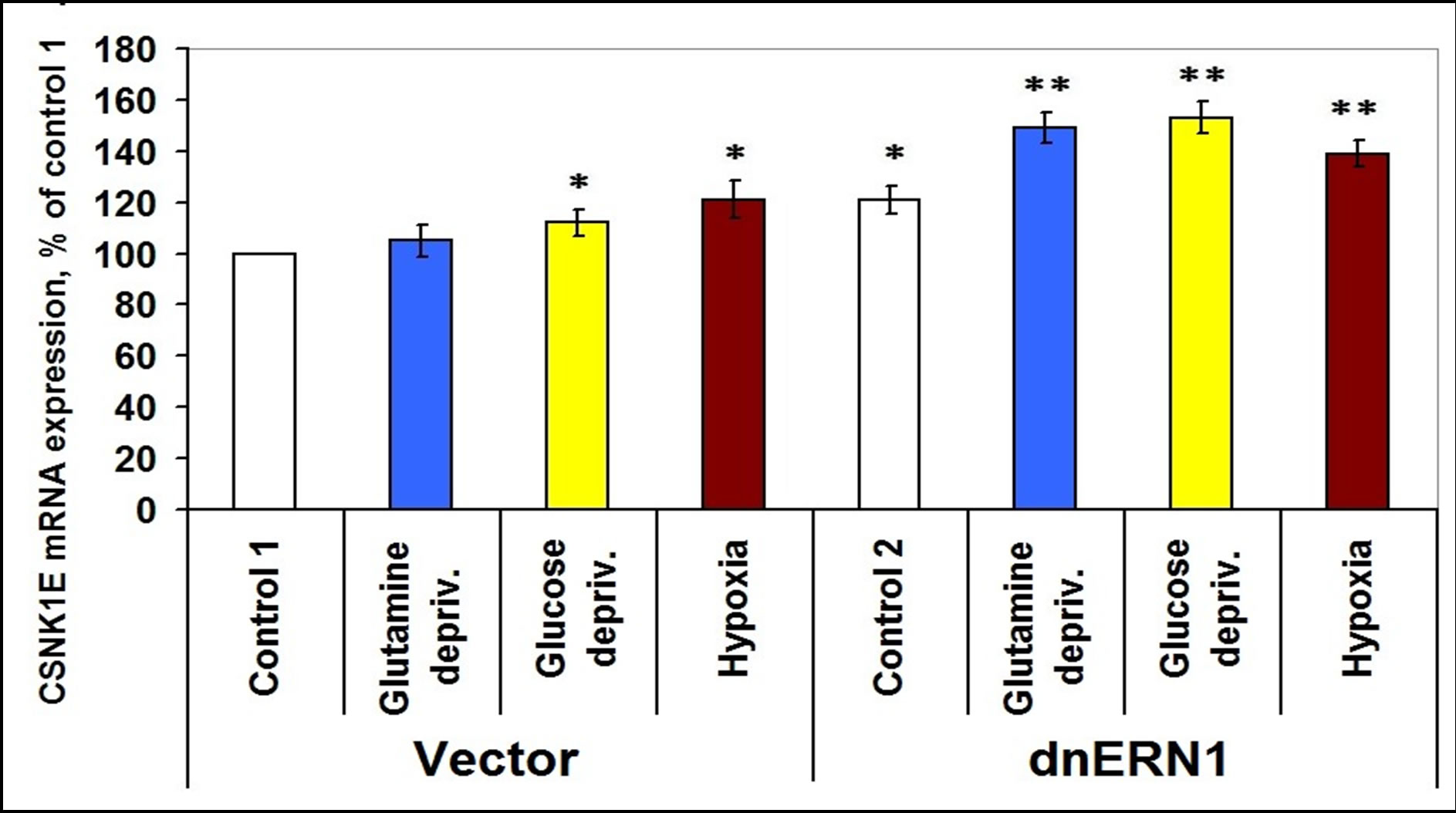
Figure 6. Effect of hypoxia and glutamine or glucose deprivation on the expression of casein kinase-1E (CSNK1E) mRNA in glioma cell line U87 (control 1) and its subline with blockade of signaling enzyme endoplasmic reticulum-nuclei-1 (dnERN1) activity (control 2) measured by quantitative polymerase chain reaction. Values of CSNK1E mRNA expression were normalized to beta-actin mRNA expression and represent as percent for control 1 (100%); n = 5; *P < 0.05 as compared to control 1; **P < 0.05 as compared to control 2.
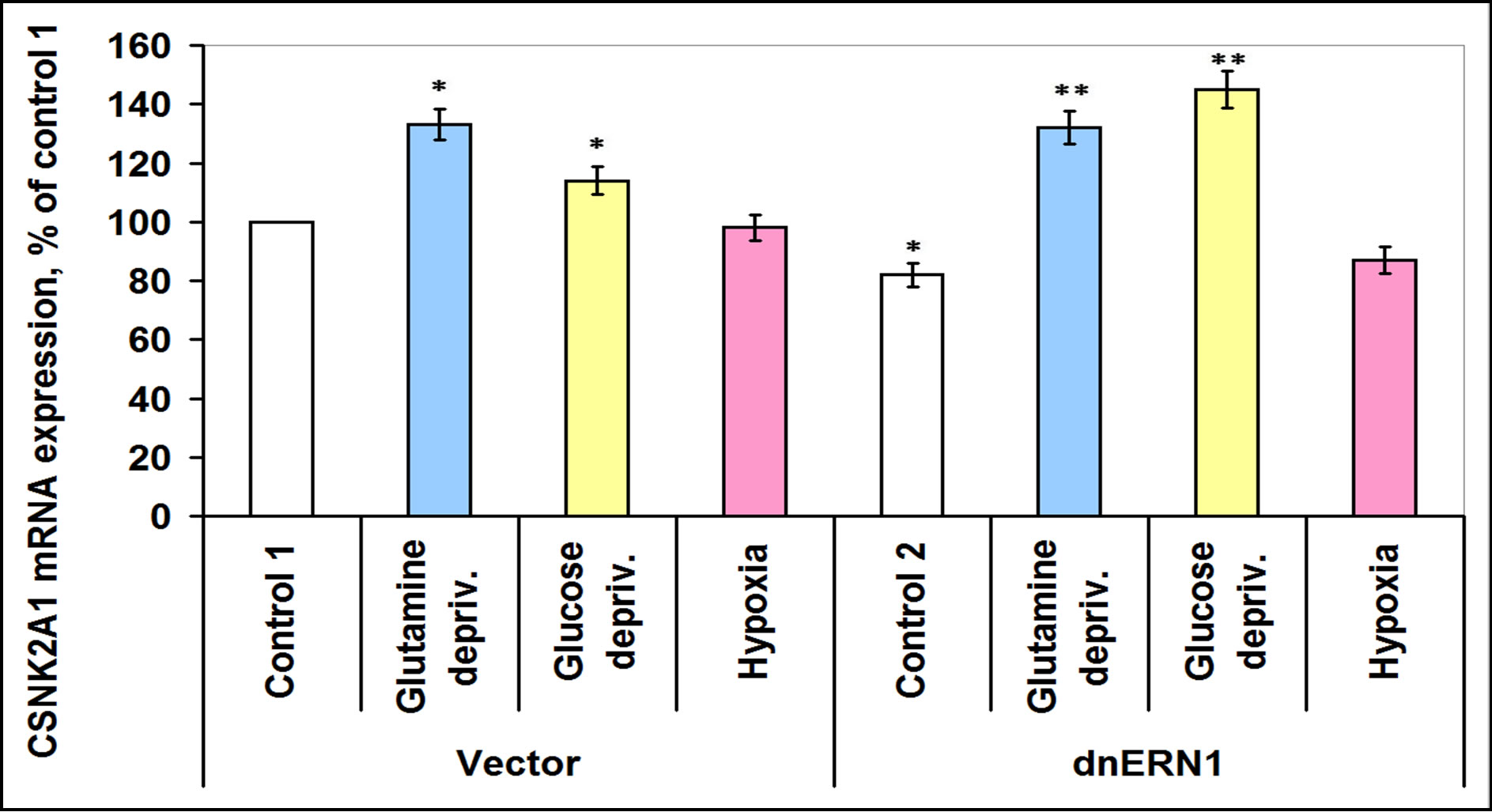
Figure 7. Effect of hypoxia and glutamine or glucose deprivation on the expression of casein kinase-2A1 (CSNK2A1) mRNA in glioma cell line U87 (control 1) and its subline with blockade of signaling enzyme endoplasmic reticulum-nuclei-1 (dnERN1) activity (control 2) measured by quantitative polymerase chain reaction. Values of CSNK2A1 mRNA expression were normalized to beta-actin mRNA expression and represent as percent for control 1 (100 %); n = 4; *P < 0.05 as compared to control 1; **P < 0.05 as compared to control 2.
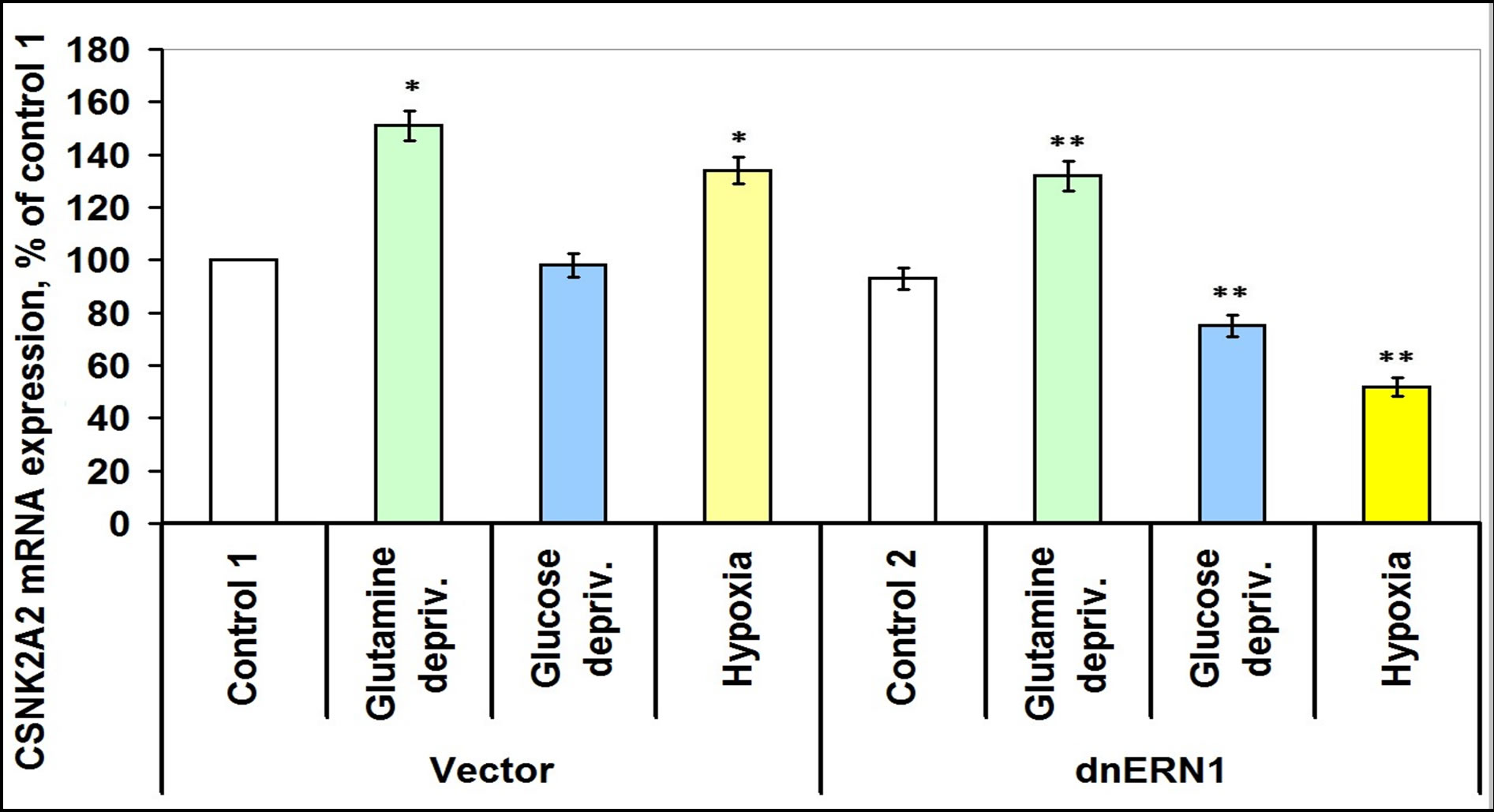
Figure 8. Effect of hypoxia and glutamine or glucose deprivation on the expression of casein kinase-2A2 (CSNK2A2) mRNA in glioma cell line U87 (control 1) and its subline with blockade of signaling enzyme endoplasmic reticulum-nuclei-1 (dnERN1) activity (control 2) measured by quantitative polymerase chain reaction. Values of CSNK2A2 mRNA expression were normalized to beta-actin mRNA expression and represent as percent for control 1 (100%); n = 4; *P < 0.05 as compared to control 1; **P < 0.05 as compared to control 2.
4. DISCUSSION
It has been known that the neovascularization process, tumor growth and cellular death processes are linked to the stress and its sensing and signal transduction pathways and to the ERN1 pathway, in particular, because the complete blockade of this signaling enzyme activity has anti-tumor effects [13-15] ADDIN EN.CITE ADDIN EN.CITE.DATA . Moreover, there is data that a growing tumor requires ischemia and hypoxia which initiate the endoplasmic reticulum stress for own neovascularization and growth, and for apoptosis inhibition [13,16,17]. It is known that many circadian factors and some casein kinases, in particular, are components of the endoplasmic reticulum stress system; they participate in the control of the cell cycle, in proliferation processes and in apoptosis [18,19,27].
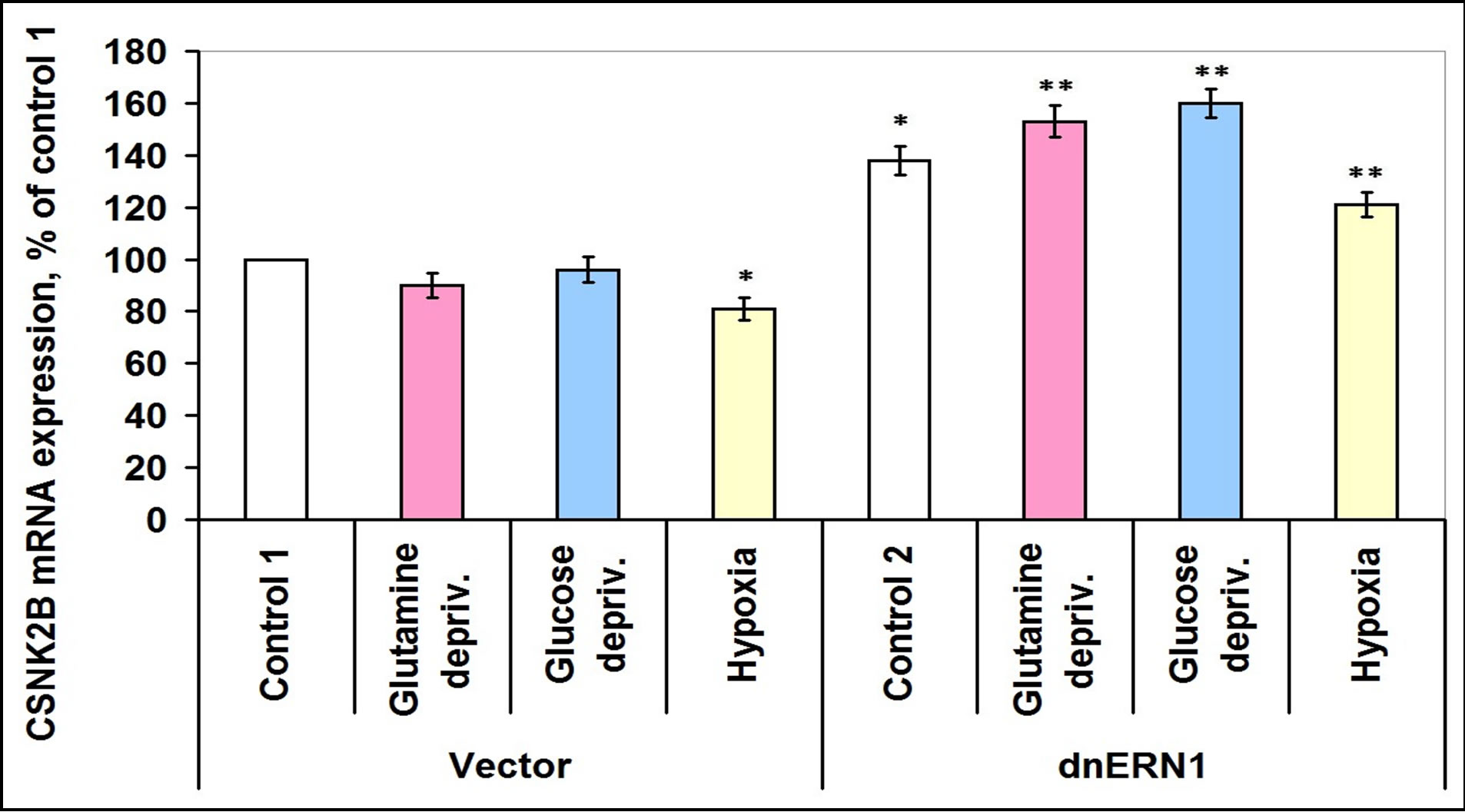
Figure 9. Effect of hypoxia and glutamine or glucose deprivation on the expression of casein kinase-2B (CSNK2B) mRNA in glioma cell line U87 (control 1) and its subline with blockade of signaling enzyme endoplasmic reticulum-nuclei-1 (dnERN1) activity (control 2) measured by quantitative polymerase chain reaction. Values of CSNK2B mRNA expression were normalized to beta-actin mRNA expression and represent as percent for control 1 (100%); n = 4; *P < 0.05 as compared to control 1; **P < 0.05 as compared to control 2.
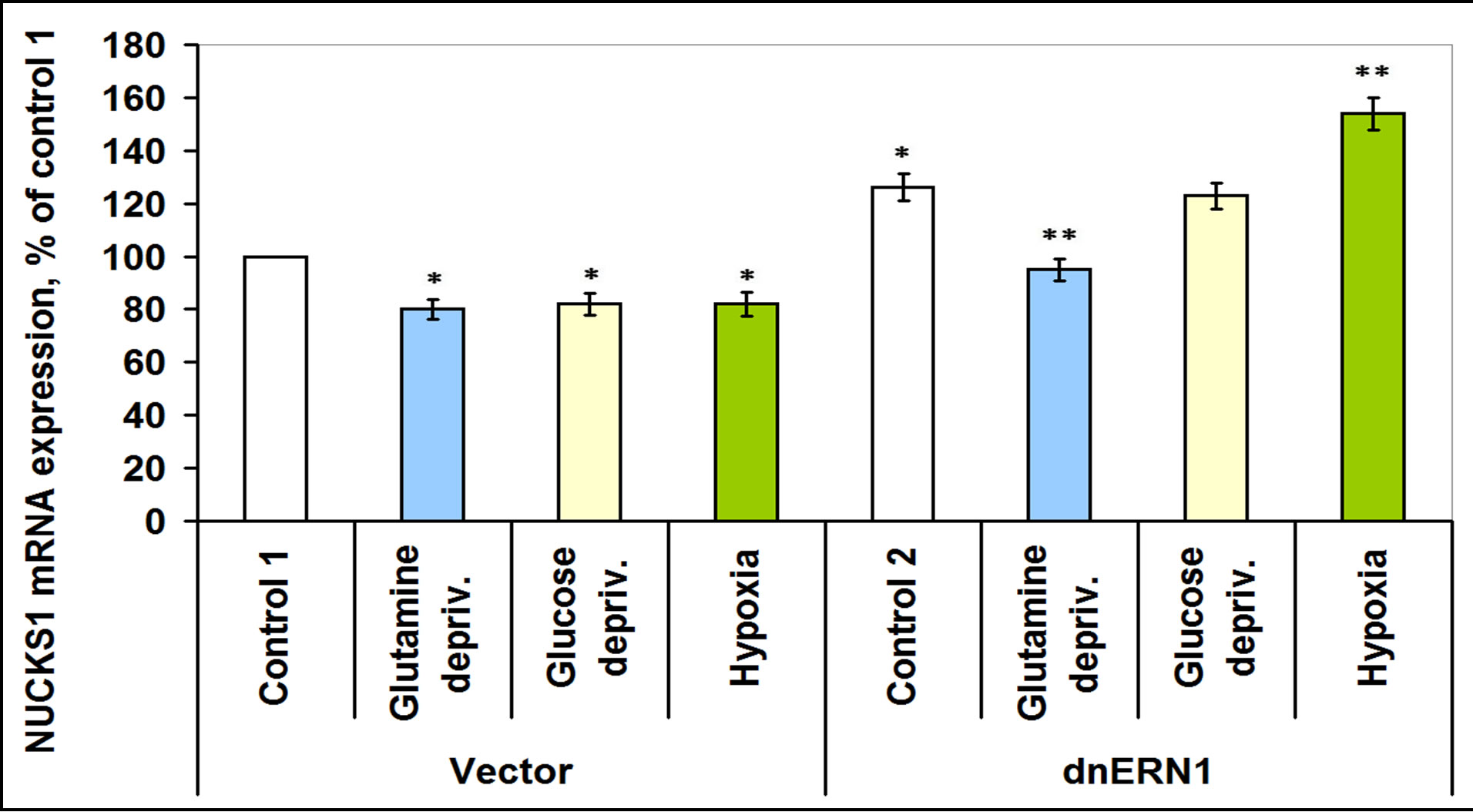
Figure 10. Effect of hypoxia and glutamine or glucose deprivation on the expression of nuclear ubiquitous casein kinase (NUCKS1) mRNA in glioma cell line U87 (control 1) and its subline with blockade of signaling enzyme endoplasmic reticulum-nuclei-1 (dnERN1) activity (control 2) measured by quantitative polymerase chain reaction. Values of NUCKS1 mRNA expression were normalized to beta-actin mRNA expression and represent as percent for control 1 (100 %); n = 5; *P < 0.05 as compared to control 1; **P < 0.05 as compared to control 2.
In this study we have shown that blockade of ERN1, the key endoplasmic reticulum stress sensor, changes the expression levels of most casein kinase-1 and -2 isoenzymes, which play an important role in the control of malignant tumor growth. This data agrees with the idea that some casein kinases participate in endoplasmic reticulum stress signaling via phosphorylation of different proteins which are important components of IRE-1 signaling [10,28,33]. We have found that the expression of different casein kinase-1 isoenzymes change in different ways in IRE-1α knockdown glioma cells and, it is quite possible, that this dysregulation is responsible for the suppression of proliferation of these cells [14], because different isoforms of casein kinase-1 (alpha, gamma-1, and delta) participate in the regulation of the Wnt and TGF-beta signaling cascades, in phosphorylation of TP53 [33,35,36]. Moreover, suppression of IRE-1α function in glioma cells also leads to a decrease of the catalytic subunit of casein kinase-2 (CSNK2A1) which has an important regulatory function in cell proliferation, cell differentiation, and in apoptosis via phosphorylation of a number of key intracellular signaling proteins implicated in tumorigenesis and tumor suppression [37].
Results of this investigation clearly demonstrate the regulation of the expression levels of different casein kinase-1 as well as casein kinase-2 isoenzymes by hypoxia and ischemic conditions and that this regulation in most cases is different in control glioma cells and in ERN1-deficient cells. Thus, hypoxia induces expression of casein kinase-1ε in glioma U87 cells and it is possible that this kinase participates in tumor growth via its effect on cell adhesion and migration [33,34]. Moreover, the effect of hypoxia and glutamine or glucose deprivation conditions on the expression levels of different casein kinases (CSNK1A1, CSNK1D and CSNK1G2 as well as for casein kinase-2A1) depends upon the functional activity of ERN1. This data also indicates the participation of these kinases in endoplasmic reticulum stress signaling. However, the molecular mechanisms underlying these seemingly mutually exclusive behaviors have not been elucidated. This provides a rationale for the molecular analysis of expression signatures of different related genes in glioma cells which control the neovascularization process, tumor growth, and cellular death processes for a comprehensive approach to the understanding of these complex mechanisms.
The major finding reported here is that the expression of most tested genes that encode casein kinases are dependent on the function of ERN1 signaling enzyme— both in normal, hypoxic and glutamine or glucose deprivation conditions and it is possible that they participate in glioma cell proliferation and tumor growth via regulation of various signaling pathways. However, the detailed molecular mechanisms of regulation of the genes encoding different casein kinase-1 and casein kinase-2 by endoplasmic reticulum—nuclei-1 signaling system under ischemic stress conditions is complex and warrants further study.
5. CONCLUSION
Results of this investigation clearly demonstrate that the expression of different casein kinase-1 and casein kinase-2 genes in glioma cells are regulated by hypoxia and glutamine or glucose deprivation and that this regulation depends on the functional activity of the signaling enzyme ERN1 for most isoenzymes of casein kinase and this is quite possible. Thus, casein kinase-1 and -2 isoenzymes participate in cell adaptive response to endoplasmic reticulum stress associated with hypoxia, glutamine or glucose deprivation.
6. ACKNOWLEDGEMENTS
This work was supported by a grant from the National Academy of Sciences of Ukraine (Grant No: 0111U002234). We thank Prof. Michel Moenner (INSERM U1029, France) for permanent support and interest in this work as well as the gift of IRE-1alpha knockdown U87 glioma cells.
REFERENCES
- Aragón, T., Van Anken, E., Pincus, D., Serafimova, I.M., Korennykh, A.V., Rubio, C.A. and Walter P. (2009) Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature, 457, 736-740. doi:10.1038/nature07641
- Bi, M., Naczki, C., Koritzinsky, M., Fels, D., Blais, J., Hu, N., Harding, H., Novoa, I., Varia, M., Raleigh, J., Scheuner, D., Kaufman, R.J., Bell, J., Ron, D., Wouters, B.G. and Koumenis, C. (2005) ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO Journal, 24, 3470-3481. doi:10.1038/sj.emboj.7600777
- Blais, J.D., Filipenko, V., Bi, M., Ron, D., Koumenis, C., Wouters, B.G. and Bell, J.C. (2004) Transcription factor 4 is translationally regulated by hypoxic stress. Molecular Cell Biology, 24, 7469-7482. doi:10.1128/MCB.24.17.7469-7482.2004
- Fels, D.R. and Koumenis, C. (2006) The PERK/eIF2a/ ATF4 module of the UPR in hypoxia resistance and tumor growth. Cancer Biology & Therapy, 5, 723-728. doi:10.4161/cbt.5.7.2967
- Luo, D., He, Y., Zhang, H., Yu, L., Chen, H., Xu, Z., Tang, S., Urano, F. and Min, W. (2008) AIP1 is critical in transducing IRE1-mediated endoplasmic reticulum stress response. The Journal of Biological Chemistry, 283, 11905-11912. doi:10.1074/jbc.M710557200
- Korennykh, A.V., Egea, P.F., Korostelev, A.A., Finer-Moore, J., Zhang, C., Shokat, K.M., Stroud, R.M. and Walter, P. (2009) The unfolded protein response signals through high-order assembly of Ire1. Nature, 457, 687-693. doi:10.1038/nature07661
- Romero-Ramirez, L., Cao, H., Nelson, D., Hammond, E., Lee, A.H., Yoshida, H., Mori, K., Glimcher, L.H., Denko, N.C., Giaccia, A.J., Le, Q.-T. and Koong, A.C. (2004) XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Research, 64, 5943-5947. doi:10.1158/0008-5472.CAN-04-1606
- Lin, J.H., Li, H., Yasumura, D. Cohen, H.R., Zhang, C., Panning, B., Shokat, K.M., Lavail, M.M. and Walter, P. (2007) IRE1 signaling affects cell fate during the unfolded protein response. Science, 318, 944-949. doi:10.1126/science.1146361
- Hollien, J., Lin, J.H., Li, H., Stevens, N., Walter, P. and Weissman, J.S. (2009) Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. The Journal of Cell Biology, 186, 323-331. doi:10.1083/jcb.200903014
- Acosta-Alvear, D., Zhou, Y., Blais, A., Tsikitis, M., Lents, N.H., Arias, C., Lennon, C.J., Kluger, Y. and Dynlacht D.D. (2007) XBP1 controls diverse cell typeand condition-specific transcriptional regulatory networks. Molecular Cell, 27, 53-66. doi:10.1016/j.molcel.2007.06.011
- Han, D., Upton, J.-P., Hagen, A., Callahan, J., Oakes, S.A. and Papa, F.R. (2008) A kinase inhibitor activates the IRE1alpha RNase to confer cytoprotection against ER stress. Biochemical and Biophysical Research Communications, 365, 777-783. doi:10.1016/j.bbrc.2007.11.040
- Greenman, C., Stephans, P., Smith, R. Dalgliesh, G.L., Hunter, C., Bignell, G., Davies, H., et al. (2007) Patterns of somatic mutation in human genomes. Nature, 446, 153-158. doi:10.1038/nature05610
- Auf, G., Jabouille, A., Guérit, S., Pineau, R., Delugin, M., Bouchecareilh, M., Favereaux, A., Maitre, M., Gaiser, T., Von Deimling, A., Czabanka, M., Vajkoczy, P., Chevet, E., Bikfalvi, A. and Moenner, M. (2010) A shift from an angiogenic to invasive phenotype induced in malignant glioma by inhibition of the unfolded protein response sensor IRE1. Proceedings of the National Academy of Sciences of the United States of America, 107, 15553- 15558. doi:10.1073/pnas.0914072107
- Drogat, B., Auguste, P., Nguyen, D.T., Bouchecareilh, M., Pineau, R., Nalbantoglu, J., Kaufman, R.J., Chevet, E., Bikfalvi, A. and Moenner, M. (2007) IRE1 signaling is essential for ischemia-induced vascular endothelial growth factor—A expression and contributes to angiogenesis and tumor growth in vivo. Cancer Research, 67, 6700-6707. doi:10.1158/0008-5472.CAN-06-3235
- Moenner, M., Pluquet, O., Bouchecareilh, M. and Chevet, E. (2007) Integrated endoplasmic reticulum stress responses in cancer. Cancer Research, 67, 10631-10634. doi:10.1158/0008-5472.CAN-07-1705
- Denko, N.C. (2008) Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nature Reviews Cancer, 8, 705-713. doi:10.1038/nrc2468
- Saito, A., Ochiai, K., Kondo, S., Tsumagari, K., Murakami, T., Cavener, D.R. and Imaizumi, K. (2011) Endoplasmic reticulum stress response mediated by the PERKeIF2-ATF4 pathway is involved in osteoblast differentiation induced by BMP2. The Journal of Biological Chemistry, 286, 4809-4818. doi:10.1074/jbc.M110.152900
- Hetz, C. and Glimcher, L.H. (2009) Fine-tuning of the unfolded protein response: Assembling the IRE1alpha interactome. Molecular Cell, 35, 551-561. doi:10.1016/j.molcel.2009.08.021
- Hunt, T. and Sassone-Corsi, P. (2007) Riding tandem: Circadian clocks and the cell cycle. Cell, 129, 461-464. doi:10.1016/j.cell.2007.04.015
- Hua, H., Wang, Y., Wan, C., Liu, Y., Zhu, B., Yang, C., Wang, X., Wang, Z., Cornelissen-Guillaume, G. and Halberg, F. (2006) Circadian gene mPer2 overexpression induces cancer cell apoptosis. Cancer Science, 97, 589-596. doi:10.1111/j.1349-7006.2006.00225.x
- Cao, Q., Gery, S., Dashti, A., Yin, D., Zhou, Y., Gu, J. and Koeffler, H.P. (2009) A role for the clock gene per1 in prostate cancer. Cancer Research, 69, 7619-7625. doi:10.1158/0008-5472.CAN-08-4199
- Chen, S.T., Choo, K.B., Hou, M.F., Yeh, K.T., Kuo, S.J. and Chang, J.G. (2005) Rhythmic PER abundance definesa critical nodal point for negative feedback within the circadian clock mechanism. Carcinogenesis, 26, 1241- 1246. doi:10.1093/carcin/bgi075
- Taniguchi, H., Fernandez, A.F. and Setien, F. (2009) Epigenetic inactivation of the circadian clock gene BMAL1 in hematologic malignancies. Cancer Research, 69, 8447- 8454. doi:10.1158/0008-5472.CAN-09-0551
- Climent, J., Perez-Losada, J., Quigley, D.A., Kim, I.J., Delrosario, R., Jen, K.Y., Bosch, A., Lluch, A., Mao, J.H. and Balmain, A. (2010) Deletion of the PER3 gene on chromosome 1p36 in recurrent ER-positive breast cancer. Journal of Clinical Oncology, 28, 3770-3778. doi:10.1200/JCO.2009.27.0215
- Huang, W., Ramsey, K.M. and Marcheva, B. (2011) Circadian rhythms, sleep, and metabolism. The Journal of Clinical Investigation, 121, 2133-2141. doi:10.1172/JCI46043
- Borgs, L., Beukelaers, P., Vandenbosch, R., Belachew, S., Nguyen, L. and Malgrange, B. (2009) Cell “circadian” cycle: New role for mammalian core clock genes. Cell Cycle, 8, 832-837. doi:10.4161/cc.8.6.7869
- Mizoguchi, T., Putterill, J. and Ohkoshi, Y. (2006) Kinase and phosphatase: The cog and spring of the circadian clock. International Review of Cytology, 250, 47-72. doi:10.1016/S0074-7696(06)50002-6
- Walton, K.M., Fisher, K., Rubitski, D., Marconi, M., Meng, Q.J., Sládek, M., Adams, J., Bass, M., Chandrasekaran, R., Butler, T., Griffor, M., Rajamohan, F., Serpa, M., Chen, Y., Claffey, M., Hastings, M., Loudon, A., Maywood, E., Ohren, J., Doran, A. and Wager, T.T. (2009) Selective inhibition of casein kinase 1 epsilon minimally alters circadian clock period. Journal of Pharmacology and Experimental Therapeutics, 330, 430-439. doi:10.1124/jpet.109.151415
- Meng, Q.J., Maywood, E.S., Bechtold, D.A., Lu, W.Q., Li, J., Gibbs, J.E., Dupré, S.M., Chesham, J.E., Rajamohan, F., Knafels, J., Sneed, B., Zawadzke, L.E., Ohren, J.F., Walton, K.M., Wager, T.T., Hastings, M.H. and Loudon, A.S. (2010) Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proceedings of the National Academy of Sciences of the United States of America, 107, 15240-15245. doi:10.1073/pnas.1005101107
- Sprouse, J., Reynolds, L., Swanson, T.A. and Engwall, M. (2009) Inhibition of casein kinase I epsilon/delta produces phase shifts in the circadian rhythms of Cynomolgus monkeys. Psychopharmacology (Berl), 204, 735-742. doi:10.1007/s00213-009-1503-x
- Takano, A., Hoe, H.S., Isojima, Y. and Nagai, K. (2004) Analysis of the expression, localization and activity of rat casein kinase 1epsilon-3. Neuro Report, 15, 1461-1464. doi:10.1097/01.wnr.0000133297.77278.81
- Rumpf, C., Cipak, L., Dudas, A., Benko, Z., Pozgajova, M., Riedel, C.G., Ammerer, G., Mechtler, K. and Gregan, J. (2010) Casein kinase 1 is required for efficient removal of Rec8 during meiosis I. Cell Cycle, 9, 2657-2662. doi:10.4161/cc.9.13.12146
- Foldynová-Trantírková, S., Sekyrová, P., Tmejová, K., Brumovská, E., Bernatík, O., Blankenfeldt, W., Krejcí, P., Kozubík, A., Dolezal, T., Trantírek, L. and Bryja, V. (2010) Breast cancer-specific mutations in CK1epsilon inhibit Wnt/beta-catenin and activate the Wnt/Rac1/JNK and NFAT pathways to decrease cell adhesion and promote cell migration. Breast Cancer Research, 12, R30. doi:10.1186/bcr2581
- Witte, F., Bernatik, O., Kirchner, K., Masek, J., Mahl, A., Krejci, P., Mundlos, S., Schambony, A., Bryja, V. and Stricker, S. (2010) Negative regulation of Wnt signaling mediated by CK1-phosphorylated Dishevelled via Ror2. The FASEB Journal, 24, 2417-2426. doi:10.1096/fj.09-150615
- Venerando, A., Marin, O., Cozza, G., Bustos, V.H., Sarno, S. and Pinna, L.A. (2010) Isoform specific phosphorylation of p53 by protein kinase CK1. Cellular and Molecular Life Sciences, 67, 1105-1118. doi:10.1007/s00018-009-0236-7
- Guo, X., Waddell, D.S., Wang, W., Wang, Z., Liberati, N.T., Yong, S., Liu, X. and Wang, X.F. (2008) Liganddependent ubiquitination of Smad3 is regulated by casein kinase 1 gamma 2, an inhibitor of TGF-beta signaling. Oncogene, 27, 7235-7247. doi:10.1038/onc.2008.337
- Hessenauer, A., Schneider, C.C., Gotz, C. and Montenarh, M. (2011) CK2 inhibition induces apoptosis via the ER stress response. Cellular Signalling, 23, 145-151. doi:10.1016/j.cellsig.2010.08.014
- Minchenko, D.M., Hubenya, O.V., Terletsky, B.M., Moenner, M. and Minchenko, O.H. (2011) Effect of hypoxia, glutamine and glucose deprivation on the expression of cyclin and cyclin-dependent kinase genes in glioma cell line U87 and its subline with suppressed activity of signaling enzyme endoplasmic reticulum-nuclei-1. Ukrainian Biochemical Journal, 83, 5-16.

