Advances in Biological Chemistry
Vol. 2 No. 3 (2012) , Article ID: 21808 , 8 pages DOI:10.4236/abc.2012.23031
Cloning of multicopper oxidase gene from Ochrobactrum sp. 531 and characterization of its alkaline laccase activity towards phenolic substrates
![]()
Faculty of Life Science, Hubei University, Wuhan, China
Email: *xgw569@hotmail.com
Received 22 March 2012; revised 29 April 2012; accepted 4 May 2012
Keywords: Ochrobactrum sp. 531; Multicopper Oxidase; Bacterial Laccase; Enzyme Kinetics
ABSTRACT
A 1602 bp fragment was cloned from a soil bacterium Ochrobactrum sp. 531. It contained an open reading frame (ORF) of 1092 bp which was identified as a multicopper oxidase (MCO) with potential laccase activity. After inserting the cloned gene into the expression vector pET23a, it was expressed in E. coli BL21(DE3)pLysS, and its product was purified to homogeneity through chromatography. The Ochrobactrum sp. 531 MCO, consisting of 533 amino acids with a molecular mass of 57.8 kDa, was quite stable in neutral pH and showed laccase-like activity oxidizing 2,6-dimethoxyphenol (DMP), 2,2’-azino-bis(3-ethylbenzthiazolinesulfonic acid) (ABTS), and syringaldazine (SGZ). The enzyme showed optimum activity towards DMP, ABTS, and SGZ at the pH 8.0, 3.6, and 7.5 respectively. Kinetic studies gave this enzyme Km, kcat and kcat//Km values of: 0.09 mM, 7.94 s–1, and 88.22 s–1·mM–1 for DMP; 0.072 mM, 2.95 s–1, and 40.97 s–1·mM–1 for ABTS; and 0.015 mM, 2.4 s–1, and 160 s–1·mM–1 for SGZ. Our results demonstrate that Ochrobactrum sp. 531 MCO is a bacterial laccase which oxidized phenolic substrates DMP and SGZ effectively under alkaline conditions. These unusual properties make the enzyme an interesting biocatalyst in applications for which classical laccases are unsuitable.
1. INTRODUCTION
Laccases (EC 1.10.3.2) are a family of multicopper oxidases which catalyze the oxidation of a wide range of inorganic and aromatic substrates by coupling four-electron reduction of dioxygen to water [1,2]. Laccases are widely found in plants, insects and fungi, and recent studies are increasing their number especially in prokaryotic sources [3]. Multicopper oxidases are found in Streptomycetes griseus [4,5], Streptomyces lavendulae [6], Sinorhizobium meliloti [7], Marinomonas mediterranea [8], Escherichia coli [9,10], Klebsiella sp. 601 [11], Bacillus subtilis [12], Bacillus sphaericus [13] and Bacillus halodurans [14]. Like fungal and plant laccases, bacterial laccases also contain four copper atoms: type I or blue copper, type II or normal copper, and type III or coupled binuclear copper center [15]. Compared to fungal laccases, bacterial laccases show little overall amino acid sequence similarity, low oxidation rate, and supplementation of Cu2+ [16]. Because of these differences, bacterial enzyme is often defined as “multicopper oxidase (MCO)”, and its activity as “laccase-like enzyme”.
Although fungal laccases are currently commercialized for industrial application, most of them have narrow substrate specificity and a limited effective pH range, especially in alkaline environments. The amount of fungal laccases does not meet current market requirements due to their expression difficulty in prokaryotic hosts in an active form, but bacterial laccases, unlike fungal ones, do not require any post-transcriptional modification, and are able to express in prokaryotic hosts as a soluble form. Bacterial laccases also act in a wide rang of pH, and have wide substrate specificities and good stability [16]. Its low activities can be improved, because the factors affecting enzyme activity can often be defined and optimized via enzymatic study and protein engineering [17-20].
Considering the potential applications of bacterial laccases in environment and green industry, we isolated a novel strain Ochrobactrum sp. 531 displaying laccase activity from soil, cloned a DNA fragment encoding a MCO gene, and studied its expression in E. coli BL21 (DE3) pLysS. The enzyme was purified to homogeneity, and its kinetic parameters for 2,6-dimethoxyphenol (DMP), 2,2’- azino-bis(3-ethylbenzthiazolinesulfonic acid) (ABTS), and syringaldazine (SGZ) were also determined through steadystate kinetics. Our results demonstrate that the cloned Ochrobactrum sp. 531 fragment encodes a bacterial laccase which can oxidize DMP and SGZ effectively under alkaline conditions.
2. MATERIALS AND METHODS
2.1. Strains, Plasmids and Chemicals
Ochrobactrum.531 was isolated from soil and stocked in our laboratory. E coli DH5α and E. coli BL21 (ED3) pLysS were used for DNA amplification and protein expression. All primers used in this study are listed in Table 1. PCR reagents, T-vector, ligase kits, restriction enzymes, and DNA markers were purchased from TaKaRa. The gel recovery kit was bought from V-Gene. ABTS, DMP, and SGZ were purchased from Sigma. The reagents for electrophoresis were bought from Ameresco. Inorganic salts, antibiotics, and culture medium were obtained from Zhong Ke (Shanghai, China).
2.2. Cloning the Core Sequence of Ochrobactrum sp. 531 MCO Gene by PCR
As the genomic DNA of Ochrobactrum sp. 531 is not sequenced yet, it was difficult to use PCR technique directly to clone its MCO gene. We adopted the methodology described in Li et al. [11] to clone the core segment of MCO from the genomic DNA of Ochrobactrum sp. 531. Sequence alignments revealed that amino acids at four copper-binding sites of both eukaryotic and prokaryotic laccases are highly conserved [21,22]. To reduce the degeneracy of DNA composition, Blast search in GenBank and sequence alignment found a conserved pattern WFHPHL(Q)H and HP(A)MF(A)HCH at copper-binding sites of the CueO-type multicopper oxidases of several bacteria (see Figure 1(a)). Based on these two conserved sequences, two degenerate primers, 531F1 and 531R1 (Table 1), were designed using the program CODEHOP [23]. 10 ng of genomic DNA extracted from Ochrobactrum sp. 531 was used as template in 100 ml reaction solution containing 10% 10 ´ PCR buffer, 0.2 mM dNTPs, 0.4 mM 531F1, and 531R1 primers. PCR was initiated by adding 2.5 U ExTaq DNA polymerase and performed at 94˚C for 5 min, followed by 30 cycles of denaturizing at 94˚C for 30 s, annealing at 54˚C for 45 s, and extension with ExTaq DNA polymerase at 72˚C for 90 s. The PCR product (1092 bp) was then recovered using gel extraction kits, inserted into pMD18-T vector by mixing DNA fragment and pMD18-T DNA (5:1) and incubated at 16˚C for 4 hours. 5 μl ligation mixture was used for transforming E. coli DH5α via calcium chloride method [24]. Positive transformants were screened using LB plates supplemented with 100 mg/ml Ampicillin, and further confirmed by PCR in which two primers, 531F1 and 531R1, were used. The cloned DNA segments were finally sent to Genomic Institute of Science and Technology Co., Ltd (Shanghai, China) for DNA sequencing. Finally a core DNA fragment (1092 bp) was obtained (see Figure 1(b)). Translation gave the cloned DNA 364 amino acids with Cu II, Cu III, and Cu IV sites.
2.3. Cloning Full Length of the Gene
BlastP search showed that amino acid sequence, derived from the cloned 1092 bp DNA, gave 98% similarity to Ochrobactrum anthropi (gi/153011149) and more than 75% to Brucella suis (gi/23500444). Phylogenetic analysis also showed the MCO of Ochrobactrum anthropi has a close evolutionary relationship with that of Brucella suis (data not shown). Results of BlastP and phylogenetic analysis suggested that the MCO of Ochrobactrum sp. 531 has perhaps strong homology with that of Ochrobactrum anthropi and Brucella suis. Multiple alignments revealed that 10 amino acids at both Nand C-termini (MTGITRRRLL and VGMMAQFVTV) of MCOs in Ochrobactrum anthropi and four Brucella species (B. suis, B. canis, B. abortus and B. ovis) are identical. Based on these 10 identical amino acids, we designed the rostrocaudal degenerate primers 531F2 and 531R2 (see Table 1). To clone the full length of Ochrobactrum sp. 531 MCO gene, PCR was performed by using two oligomers, 531F2 and 531R2, as a primer pair and Ochrobactrum sp. 531 genomic DNA as template. PCR amplification was executed at 94˚C for 5 min, followed by 30 cycles of denaturing at 94˚C for 30 s, annealing at 54˚C for 45 s, and extension at 72˚C for 90 s. 1.6 kb of PCR product was recovered, inserted into pMD18-T vector to form recombinant molecule and then transformed into E. coli DH5α as described previously. A 1602 bp DNA fragment was finally obtained via PCR. DNA sequence analysis showed that the 1602 bp DNA segment, enclosing the cloned core DNA fragment (1092 bp), contained intact information of an open reading frame (ORF). The gene sequence encoding Ochrobactrum sp. 531 MCO was deposited in NCBI database (Accession No: HM358156).
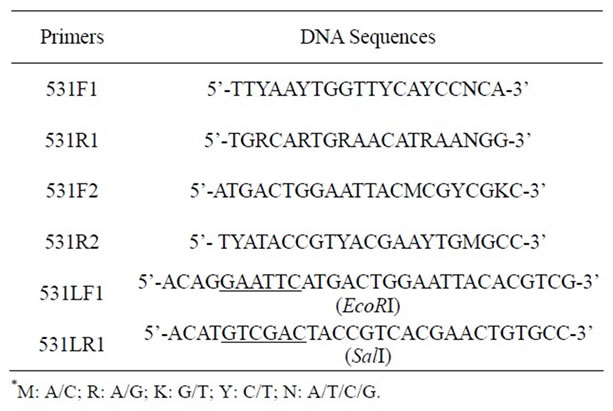
Table 1. DNA primers used for cloning the gene of Ochrobactrum sp. 531 MCO.
 (a)
(a) (b)
(b)
Figure 1. Amino acid sequence alignment of the two copper binding domains of bacterial multicopper oxidases and PCR product amplified from the genomic DNA of Ochrobactrum sp. 531. (a) alignment of the two copper binding domains of six bacterial multicopper oxidases (Ochrobactrum anthropi gi:153011149, Brucella suis gi:23500444, Salmonella enterica gi:194443616, Klebsiella pneumoniae gi:152968582, Escherichia coli gi:16128116, Shigella dysenteriae gi:194433448). Sequences in boxes are copper binding domains used for designing degenerate primers; (b) 1% agarose gel showing the DNA band amplified using the primers 531F1 and 531R1. Lane 1: PCR product, and Lane 2: DNA marker.
2.4. Enzyme Expression and Purification
Two primers 531LF1 and 531LR1 with a restriction site (see Table 1) were designed based on the DNA sequence of the cloned gene from Ochrobactrum sp. 531, and used to amplify the cloned gene via PCR from the recombinant plasmid pMD18-T carrying the cloned 1602 bp DNA segment in order to insert it into a pET23a vector at EcoR I and Sal I sites. As described above, similar PCR procedure was employed to subclone the gene encoding MCO. The amplified DNA was then digested with EcoR I and Sal I and inserted into pET23a vector. To eliminate possible contamination of E. coli CueO during purification, a 6 ´ his tag was designed at the C-terminus of Ochrobactrum sp. 531 MCO. The recombinant pET23a plasmid was finally introduced into E. coli BL21 (ED3) pLysS. Positive transformants were selected using LB plates plus 100 mg/ml Ampicillin, and confirmed by PCR. Single colony was inoculated into 5 ml LB containing 100 mg/ml Ampicillin and incubated at 37˚C overnight. 5ml overnight culture was then transferred into 500 ml fresh LB medium supplemented with 100 mg/ml Ampicillin, and bacterial culture was incubated at 23˚C. When OD600 approached to 0.6, protein expression was initiated by adding IPTG to the final concentration of 0.5 mM, and bacterial cells were grown continually for 6 hours at 23˚C. After harvested by centrifuging at 6000 rpm for 15 min at 4˚C, bacterial cells were re-suspended in 50 mM Tris-HCl (pH 8.0) and broken by sonication. The crude extract was collected and clarified by centrifugation at 12,000 rpm for 15 min at 4˚C. The enzyme was purified via Ni-affinity chromatography. The Ni-affinity column was equilibrated with buffer A (50 mM Tris-HCl (pH 8.0)), washed with buffer B (25 mM imidazole in 50 mM Tris-HCl (pH 8.0)), and eluted with buffer C (250 mM imidazole in 50 mM Tris-HCl (pH 8.0)). Protein concentration was estimated by measuring the absorbance at the wavelength of 280 nm and calculated by using its extinction coefficient of 40660 M–1·cm–1. Protein purity was routinely monitored on 10% SDS-PAGE. Enzyme activity was also monitored routinely by active staining on a 10% native gel as described below.
2.5. Active Staining
10% native gel was used to separate the crude extract as described previously [25]. After electrophoresis, the minigel was first soaked in 30 ml of 50 mM TrisHCl (pH 7.5 or 8.0) for 15 minutes, and then stained in 50 mM TrisHCl buffer (pH 7.5 or 8.0) containing 0.02 mM DMP (or SGZ) and 0.2 mM CuSO4 at room temperature for 15 minutes. All results were recorded by scanning or photography.
2.6. Enzyme Assay and Kinetic Parameter Determination
Enzyme activities towards substrates DMP, SGZ, and ABTS were examined at 37˚C according to the protocol described in Palmieri [26]. Absorbance was measured at 477 nm for DMP, 420 nm for ABTS and 530 nm for SGZ on a Shimadzu UV/visible spectrophotometer (UV-2550). The extinction coefficients used in active assays were 14.8 mM–1·cm–1 for DMP, 36 mM–1·cm–1 for ABTS, and 65 mM–1·cm–1 for SGZ. 30 mM HAc-NaAc buffer was used for pH 3.0 - 5.5, 30 mM K2HPO4-KH2PO4 buffer for pH 5.5 - 7.5, and 30 mM Tris-HCl for pH 7.5 - 9.5. In addition, 0.2 mM CuSO4 was routinely added into the reaction solution. Enzyme activity (U/ml) was defined as units of activity per milliliter of enzyme solution, where one unit (U) represents 1 μmol of substrate oxidized per min. Specific activity (U/mg) was expressed as units of activity per milligram of protein.
To determine kinetic parameters, rates were measured spectrophotometrically at 37˚C with 0.2 mM CuSO4 over a range of substrate concentrations (0.05 - 0.2 mM DMP, 0.03 - 0.2 mM ABTS, and 6 - 25 mM SGZ). Oxidation of DMP was assayed in 30 mM TrisHCl buffer (pH 8.0), of ABTS in 30 mM acetate buffer (pH 3.6), and of SGZ in 30 mM TrisHCl buffer (pH 7.5). Michaelis-Menten parameters were calculated using the UVProbe-[Kinetics] version 1.11a (SHIMADZU Corporation), and mainly determined by Lineweaver-Burk plot and also checked by Hanes-Woolf and Eadie-Hofstee plots. The deviation between the same parameters obtained from different plots was less than 5%.
3. RESULTS
3.1. Identification of Laccase Activity
The 1602 bp nucleotide sequence of intact MCO open reading frame cloned from Ochrobactrum sp. 531 is shown in Figure 2. 533 amino acids were deduced for Ochrobactrum sp. 531 MCO based on its DNA sequence. As shown in Figure 2, Ochrobactrum sp. 531 MCO contains four His-rich copper binding domains, which is a typical feature for multicopper oxidase family. Analysis of the primary structure suggested that Ochrobactrum sp. 531 MCO could be a multicopper oxidase that might act as a laccase. Furthermore, Ochrobactrum sp. 531 grew quite well at pH 9.0 (data not shown), suggesting that the enzyme could be active at higher pH.
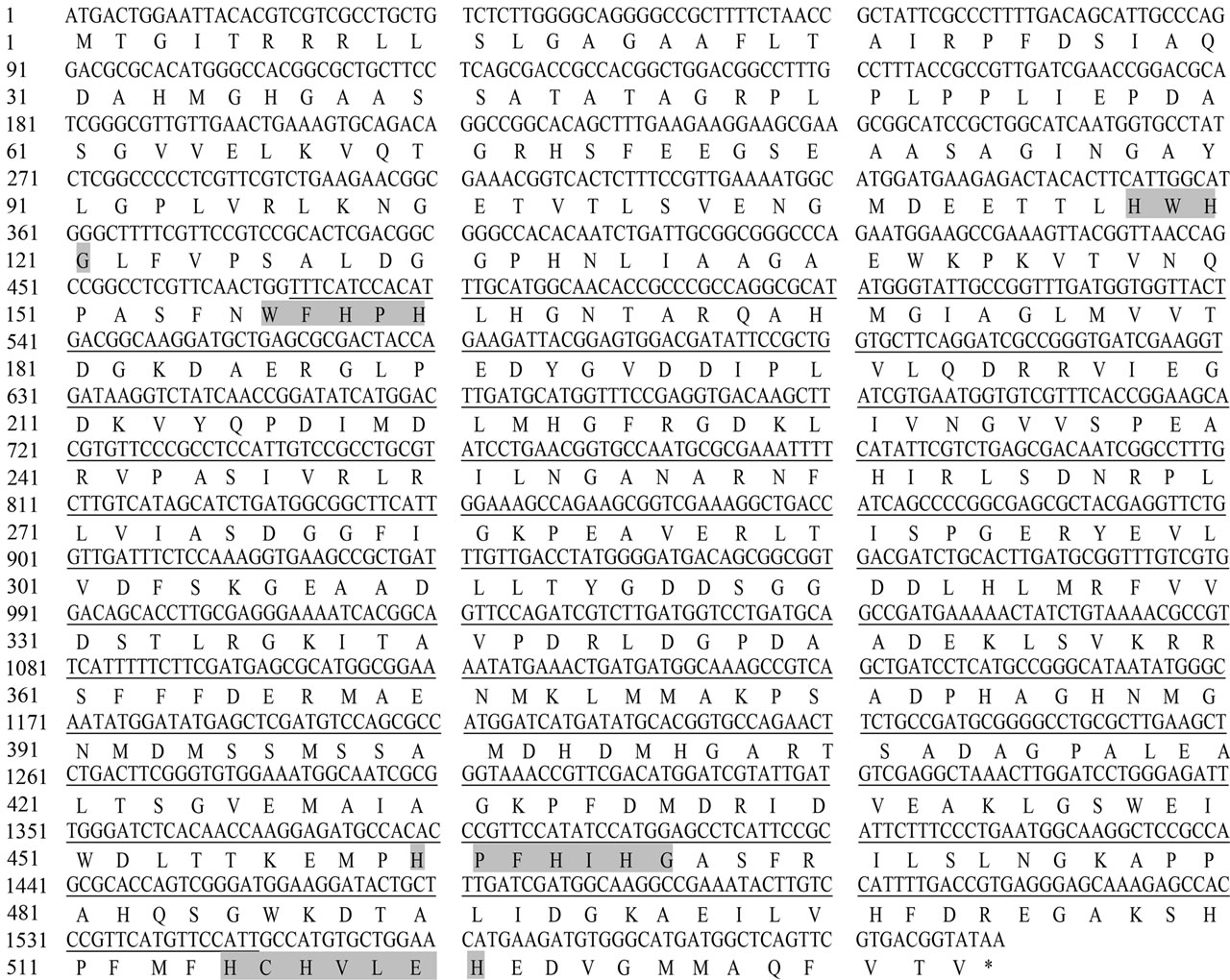
Figure 2. Nucleotide sequence of Ochrobactrum sp. 531 MCO gene and its corresponding amino acids. The core sequence amplified initially by PCR was underlined, and the amino acid sequences for four copper-binding domains were shaded. Both DNA and amino acid sequences were deposited in NCBI database (Accession No: HM358156).
To further confirm whether the cloned gene actually encodes an enzyme acting as a multicopper oxidase, the ORF cloned from Ochrobactrum sp. 531 was inserted into the expression vector pET23a and controlled under a T7 promoter. The recombinant plasmid was transformed into E. coli BL21(DE3) pLysS, and positive transformants were then used for protein expression. After induction by IPTG, about 60 kDa protein was detected on 10% SDS-PAGE gel (see Figure 3(a)), which is very close to 57.8 kDa calculated theoretically from the amino acid composition of Ochrobactrum sp. 531 MCO by ProtParam. Slightly larger molecular weight appearing on SDS-PAGE gel could be because of the presence of 6 ´ his tag at the carboxyl terminus of the enzyme. Activity staining on 10% native gel showed both purified enzyme and crude extract catalyzed oxidation of three laccase substrates DMP at pH 8.0, SGZ at pH 7.5) (see Figures 3(b) and (c)), and ABTS at pH 4.0 (data not shown). Addition of Cu2+ into the reaction system markedly enhanced oxidation of all three substrates in activity staining (data not shown), indicating that copper ion is essential for the enzyme activity. All these results clearly demonstrate that the cloned Ochrobactrum sp. 531 MCO indeed is a multiple oxidase acting as a laccase.
3.2. Properties of Ochrobactrum sp. 531 MCO
Theoretical calculation by ProtParam gave Ochrobactrum sp. 531 MCO a molecular mass of 57.8 kDa with a pI
 (a)
(a) (b)
(b)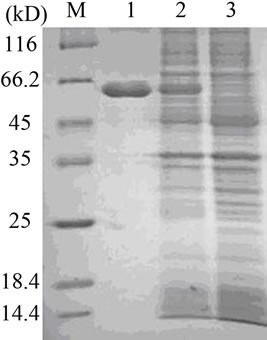 (c)
(c)
Figure 3. SDS-PAGE and native PAGE analysis. (a) 12% SDSPAGE analysis of Ochrobactrum sp. 531 MCO from supernatant of E. coli cell lysates and purified enzyme by Ni-affinity chromatography. M: protein marker; Lane 1: the purified enzyme via Ni-affinity chromatography; Lane 2: the crude extract from the bacterial cells transformed with the recombinant pET23a plasmid; and Lane 3: the crude extract from the bacterial cells transformed with pET23a plasmid as control; (b) 10% native gel showing active staining (at pH 8.0) of DMP oxidation by the purified enzyme in lane 1 and by the crude extract of the bacterial cells transformed with the recombinant pET23a plasmid in lane 2; (c) 10% native gel showing active staining (at pH 7.5) of SGZ oxidation by the purified enzyme in lane 1 and by the crude extract of the bacterial cells transformed with the recombinant pET23a plasmid in lane 2.
value of 5.58. This enzyme is rich in alanine (10.51%), glycine (9.76%) and leucine (8.82%), and contains 9.57% positively charged and 13.7% negatively charged amino acids. SingalP predicted a singal peptid located at N-terminus of the enzyme and a cleaving site of singal peptidase between the 29th and 30th amino acid, suggesting that this enzyme could be translocated from cytoplasm into periplasm as a secreatory protein in Ochrobactrum sp. 531. Predication of protein subcelluar location by PSORT suggested the enzyme might be located in both cytoplasm and periplasm with localization scores of 5.4 and 4.5. Hydropathic calculation (Kyte and Doolittle’s method) for the enzyme gave a grand average of hydropathicity (GRAVY) of –0.223, indicating that Ochrobactrum sp. 531 MCO could be a hydrophilic protein. Analysis by Dense Alignment Surface (DAS) method found 2 potential transmembrane segments located at positions 11 - 21 and 16 - 17 with the cutoff of 1.7. 13 amino acids of two transmembrane segments are 2.4% of total amino acids, suggesting the MCO is not strongly associated with bacterial membranes proteins. Protein hydrophobicity and transmembrane analysis indicated high solubility of the enzyme.
Enzyme activities were spectrophotometrically examined with a fixed concentration of DMP (1 mM), ABTS (0.5 mM), and SGZ (0.02 mM) over a range of pH values. Figure 4(a) showed that Ochrobactrum sp. 531 MCO acted over a wide range of pH (from 3.0 - 9.5). The optimal pH range for enzyme catalysis was found to be 8.0 for DMP, 3.6 for ABTS, and 7.5 for SGZ. This enzyme showed its activity towards DMP from pH 3.0 to 9.5, but strong oxidation activity occurred in the pH range of 7.5 to 9.0. For SGZ, enzyme activity was observed in the pH range of 6.0 to 9.0, with optimum activity at pH 7.5. These bell-shaped pH-dependent activity profiles are similar to those found for the oxidation of phenolic substrates by laccases. The redox potential difference between the phenol and the T1 copper of laccase, i.e. the driving force for electron transfer, increases with pH, but anions such as CN–, F–, Cl–, and OH- inhibit laccase activity by binding to the T2/T3 coppers, which interrupts the internal electron transfer from T1 to the T2/T3 centre [27,28]. With the substrate ABTS, the enzyme activity was detectable only at low pH range (pH 3.0 - 5.0). This may be explained by the non-phenolic nature of ABTS because only inhibittory effect of OH– is observed in this case.
Enzyme activities were also measured over a range of temperatures (20˚C - 50˚C) under optimal pH conditions. The results show that optimal temperature range for enzyme activity towards three substrates DMP, ABTS, and SGZ is 37˚C - 40˚C (data not shown). In addition, the effect of divalent ions on enzyme activity at 37˚C was also examined. The rates were measured in the assay solution containing 1 mM DMP and 30 mM Tris-HCl (pH 8.0) and 0.2 mM Cu2+, Ca2+, Mn2+, Mg2+, Ni2+, Cd2+, or Fe2+ by recording the increase at A477. Cu2+ was the only a divalent ion required by the enzyme for its high activity. Mn2+, Ni2+, or Cd2+ gave the enzyme less than 5% activity as compared to Cu2+, whereas Ca2+, Mg2+, and Fe2+ did not give the enzyme any detectable activity (see Figure 4(b)). Enzyme activity was examined at 37˚C and pH 8.0 by a fixed concentration of 1 mM DMP and varying Cu2+ concentrations (0 - 5 mM), the activity rapidly increased with the increase of Cu2+ concentrations from 0 to 0.2 mM, and then stayed constant up to at least 3 mM. When Cu2+ concentration was approached to 5 mM, the enzyme decreased 20% activity (data not shown).
Impact of pH on enzyme stability was also investigated. 50 ml of the purified enzyme (0.45 mg/ml) was added into 950 ml of different buffers in the pH range of 3.0 to 9.5, and stayed at 4˚C for 0 - 20 hours. Rates were measured at 37˚C in 20 mM Tris-HCl buffer (pH 7.5) containing 0.02 mM SGZ and 0.2 mM CuSO4. As shown in Figure 4(c), the activity of the enzyme kept in pH 6.5 remained almost constant for 20 hours. On pH 7.5, the enzyme still showed more than 80% activity after 20 hours. In contrast, enzyme activities decreased to less than 40% and 50% when the enzyme was stored at pH 3.0 and 9.5 respectively, for 20 hours. Our results demonstrate that Ochrobactrum.531 MCO kept in neutral pH buffers is quite stable. Figure 4(c) also shows that the enzyme kept in alkaline pH is more stable than in acidic conditions.
3.3. Kinetic Parameters of Ochrobactrum.531 MCO
Kinetic parameters were estimated under the conditions described in the experimental section in the varying concentrations of DMP (0.05 - 0.2 mM), ABTS (0.03 - 0.15 mM), and SGZ (6 - 25 mM). As shown in Table 2, kinetic studies gave Ochrobactrum sp.531 MCO Km, kcat, and kcat//Km values of: 0.09 mM, 7.94 s–1, and 88.22 s–1·mM–1 for DMP; 0.072 mM, 2.95 s–1, and 40.97 s–1·mM–1 for ABTS; and 0.015 mM, 2.4 s–1, and 160 s–1·mM–1 for SGZ. As compared with that for ABTS, 2 - 4 fold larger kcat//Km values for DMP and SGZ indicate that this enzyme favors catalyzing oxidation of phenolic substrates. The largest kcat//Km value for SGZ is because of its low Km value. According to kcat//Km values, catalytic efficiency of Ochrobactrum sp. 531 MCO towards three substrates is in the order: SGZ > DMP > ABTS.
4. DISCUSSION
Ochrobactrum anthropi MCO gene sequence was recently submitted in the Genebank (gi: 153011149; gi: 153010558), but its enzyme has not been purified and studied yet. The cloned Ochrobactrum sp. 531 MCO, like other MCOs, contains four copper-binding domains, a typical characteristic of multicopper oxidases, and requires Cu2+ for
 (a)
(a)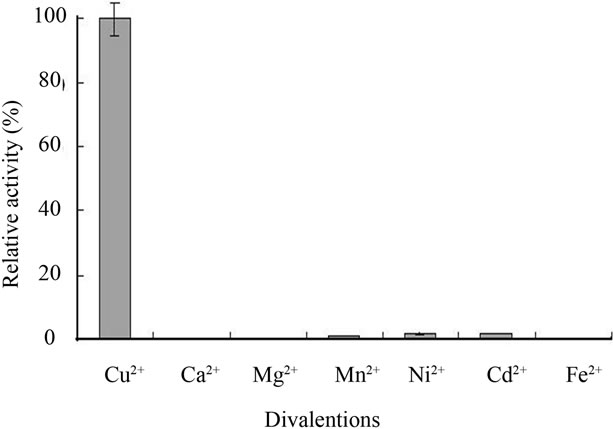 (b)
(b) (c)
(c)
Figure 4. Diagrams showing pH-dependent activity profiles of Ochrobactrum sp. 531 MCO, effects of divalent ions on enzyme activity and enzyme stability in different pH conditions. (a) pH-dependent activity profiles of Ochrobactrum sp. 531 MCO; (b) Effects of different divalent ions on the activity of Ochrobactrum sp. 531 MCO; (c) The stability of Ochrobactrum sp. 531 MCO stocked in different pH buffers at 4˚C for 20 hours. Activities were measured at 37˚C under optimal conditions. The detailed methods were described in the experimental section.
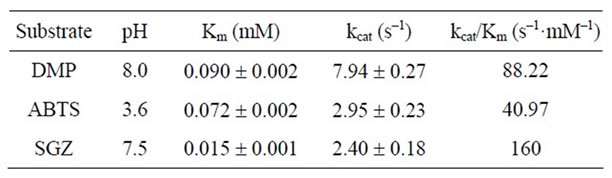
Table 2. Kinetic parameters of Ochrobactrum sp. 531 MCO for three laccase substrates.
catalytic activity. However, this Cu2+-dependent enzyme did not show any detectable Fe2+-oxidating activity that other multicopper oxidases do [10-12]. Further investigation is required to work out such differences between Ochrobactrum and other bacterial MCOs.
Bioinformatics programs theoretically predicated that Ochrobactrum sp. 531 MCO is a soluble enzame. However, like Klebsiella sp. 601 MCO [11], 85% enzyme expressed in E. coli precipitated together with cell debris as inclusion bodies into the pellet even when expressed at 20˚C (data not shown). Obviously, its insoluble behavior is contradictory to the results predicted by hydrophobicity analysis. The vital transmembrane segment (at position 11 - 21 and 16 - 17) is located inside the singal peptide. If the singal peptide were not cleaved by singal peptidase in E. coli, the enzyme containing the singal peptide might have anchor to the bacterial membranes via the transmembrane segment of the singal peptide. Without cleavage, the singal peptide part might also cause improper folding of the enzyme molecules. Either protein adherence to bacterial membranes or improper enzyme-folding could result in the formation of preciptation during crude extract preparation. Although we did not examine the cleavage of singal peptide in vivo, SDS-PAGE gave only one band of the purified enzyme with a molecular mass of about 60 kDa, suggesting that the singal peptid at N-terminus of the enzyme was not cleaved in E. coli. Given that the signal peptide was removed by E. coli singal peptidase, about 54 kDa protein band could emerge on SDS-PAGE gel. Obviously, the detailed study on the protein solubiliy of Ochrobactrum sp. 531 MCO is also required.
Laccases are very attractive enzymes for application in green chemistry, biotechnology, and environment engineering. Most bacterial MCOs display laccase activities, which make them a good potential candidate for Industrial applications. Different from fungal laccases, bacterial MCOs have a notable advantage that they can be easily expressed in E. coli or yeast in an active form of the enzyme without any glycosylation. Nevertheless, more in-depth study is needed about the detailed molecular mechanisms for substrate specificity, catalysis in different conditions, enzyme folding, and secretion, to improve catalytic activity of bacterial MCOs. Ochrobactrum sp. 531 MCO has unusual characteristics of bacterial laccases. For the phenolic substrates DMP and SGZ, enzyme shows optimum activity at alkaline pH. Our finding, together with those reported in other bacteria [14], underlines the potential of bacteria as a source of unusual alkaline laccases. Although Ochrobactrum sp. 531 MCO, like other bacterial laccases, displays lower activity, factors affecting enzyme activity can be optimized through protein engineering as reported recently [19,20]. Its expression in E. coli with low solubility will be improved in further studies by periplasmic protein expression, mutagenesis, incubating under low temperature plus microaerobic conditions, or secretion in yeast. It is expected that many more novel bacterial alkaline laccases, like Ochrobactrum sp. 531 MCO, B. halodurans Lbh1 [14], and the metagenome-derived Lac591 [29], will emerge from prokaryotic sources in near future. Unusual properties of bacterial laccases will draw more attention by industry for large scale production, green chemistry, biotechnology, and environment engineering.
5. ACKNOWLEDGEMENTS
This work was supported by the grant 201160823257 from Wuhan Science and Technology Bureau.
![]()
![]()
REFERENCES
- Xu, F. (1996) Oxidation of phenols, anilines, and benzenethiols by fungal laccases: Correlation between activity and redox potentials as well as halide inhibition. Biochemistry, 35, 7608-7614. doi:10.1021/bi952971a
- Solomon, E.I., Sundaram, U.M. and Machonkin, T.E. (1996) Multicopper oxidases and oxygenases. Chemical Reviews, 96, 2563-2605. doi:10.1021/cr950046o
- Claus, H. (2003) Laccases and their occurrence in prokaryotes. Archives of Microbiology, 179, 145-150.
- Endo, K., Hosono, K., Beppu, T. and Ueda, K. (2002) A novel extracytoplasmic phenol oxidase of Streptomyces: Its possible involvement in the onset of morphogenesis. Microbiology, 148, 1767-1776.
- Endo, K., Hayashi, Y., Hibi, T., Hosono, K., Beppu, T. and Ueda, K. (2003) Enzymological characterization of EpoA, a laccase-like phenol oxidase produced by Streptomyces griseus. Journal of Biochemistry, 133, 671-677. doi:10.1093/jb/mvg086
- Suzuki, T., Endo, K., Tsujubo, H., Miyamoto, K. and Inamori, Y. (2003) A thermostable laccsase from Streptomyces griseus REN-7: Purification, characterization, mucleotide sequence and expression. Bioscience Biochemistry, 67, 2167-2175. doi:10.1271/bbb.67.2167
- Castro, S.S, Martine, D.G. and Okon, Y. (2002) Laccase activity in melanin-producing strain of Sinorhzobium meliloti. FEMS Microbiology Letters, 209, 119-125. doi:10.1111/j.1574-6968.2002.tb11119.x
- Sanchez, A.A. and Solano, F. (1997) A pluripotent polyphenol oxidase from the melanogenic marine Alteromonas sp. share catalytic capabilities of tyrosinases and laccases. Biochemical and Biophysical Research Communications, 240, 787-792. doi:10.1006/bbrc.1997.7748
- Grass, G. and Rensing, C. (2001) CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochemical and Biophysical Research Communications, 286, 902-908. doi:10.1006/bbrc.2001.5474
- Kim, C., Lorenz, W.W., Hoopes, J.T. and Dean, J.F. (2001) Oxidation of phenolate siderophores by the multicopper oxidase encoded by the Escherichia coli yacK gene. Journal of Bacteriology, 183, 4866-4875. doi:10.1128/JB.183.16.4866-4875.2001
- Li, Y., et al. (2008) Gene cloning, protein purification and enzymatic properties of multicopper oxidase (MCO) from Klebsiella sp. 601. Canadian Journal of Microbiology, 54, 725-733. doi:10.1139/W08-063
- Hullo, M.F., Moszer, I., Danchin, A. and Martin, V.I. (2001) CotA of Bacillus subtilis is a copper-dependent laccase. Journal of Bacteriology, 83, 5426-5430. doi:10.1128/JB.183.18.5426-5430.2001
- Claus, H. and Filip, Z. (1997) The evidence of a laccase-like enzyme activity in a Bacillus sphaericus strain. Microbiological Research, 152, 209-216. doi:10.1016/S0944-5013(97)80014-6
- Ruijssenaars, H.J. and Hartmans, S. (2004) A cloned Bacillus halodruans multicopper oxidase exhibiting alkaline laccase activity. Applied Microbiology and Biotechnology, 65, 177-182. doi:10.1007/s00253-004-1571-0
- Solomon, E.I., Baldwin, M.J. and Lowery, M.D. (1992) Electronic structures of active sites in copper proteins: Contributions to reactivity. Chemical Reviews, 92, 521- 542. doi:10.1021/cr00012a003
- Solano, F., Lucas-Elio, P., Lopez-Serrano, D., Fernandez, E. and Sanchez-Amat, A. (2001) Dimethoxyphenol oxidase activity of different microbial blue multicopper proteins. FEMS Microbiology Letters, 204, 175-181. doi:10.1111/j.1574-6968.2001.tb10882.x
- Li, X., et al. (2007) Crystal structures of E. coli laccase CueO at different copper concentrations. Biochemical and Biophysical Research Communications, 354, 21-26. doi:10.1016/j.bbrc.2006.12.116
- Roberts, S.A., et al. (2002) Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli. Proceedings of the National Academy of Sciences USA, 99, 2766- 2771. doi:10.1073/pnas.052710499
- Kataoka, K., et al. (2007) Structure and function of the engineered multicopper oxidase CueO from Escherichia coli—Deletion of the methionine-rich helical region covering the substrate-binding site. Journal of Molecular Biology, 373, 141-152. doi:10.1016/j.jmb.2007.07.041
- Li, Y.D., Gong, Z.J., Li, X., Li, Y. and Wang, X.G. (2011) Engineering Klebsiella sp. 601 multicopper oxidase enhances the catalytic efficiency towards phenolic substrates. BMC Biochemistry, 12, 30. doi:10.1186/1471-2091-12-30
- Alexandre, G. and Zhulin, I.B. (2000) Laccases are widespread in bacteria. Trends in Biotechnology, 18, 41-42. doi:10.1016/S0167-7799(99)01406-7
- Claus, H. (2004) Laccases: Structure, reactions, distribution. Micron, 35, 93-96. doi:10.1016/j.micron.2003.10.029
- Rose, T.M., et al. (1998) Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Research, 26, 628-1635. doi:10.1093/nar/26.7.1628
- Sambrook, J., Fritsch, E.J. and Maniatis, T. (2001) Molecular cloning: A laboratory manual. 3 rd Edition, Cold Spring Harbor Laboratory Press, New York.
- Solano, F., Lucas-Elío, P., López-Serrano, D., Fernández, E. and Sanchez-Amat, A. (2001) Dimethoxyphenol oxidase activity of different microbial blue multicopper proteins. FEMS Microbiology Letters, 204, 175-181. doi:10.1111/j.1574-6968.2001.tb10882.x
- Palmieri, G., et al. (1997) A novel white laccase from Pleurotus ostreatus. Journal of Biology Chemistry, 272, 31301-31307. doi:10.1074/jbc.272.50.31301
- Xu, F. (1997) Effect of redox potential and hydroxide inhibition on the pH activity profile of fungal laccases. Journal of Biology Chemistry, 272, 924-928.
- Xu, F., et al. (1998) Site-directed mutagenesis in fungal laccase: Effect on redox potential, activity and pH profile. The Biochemical Journal, 334, 63-70.
- Ye, M., Li, G., Liang, W.Q. and Liu, Y.H. (2010) Molecular cloning and characterization of a novel metagenome-derived multicopper oxidase with alkaline laccase activity and high soluble expression. Applied Microbiology and Biotechnology, 87, 1023-1031. doi:10.1007/s00253-010-2507-5
NOTES
*Corresponding author.

