American Journal of Plant Sciences
Vol.5 No.5(2014), Article ID:43606,19 pages DOI:10.4236/ajps.2014.55071
RNAi-Mediated Resistance to Cucumber Mosaic Virus (CMV) in Genetically Engineered Tomato
Valentine Otang Ntui1,2, Kynet Kong1,3, Pejman Azadi1,4, Raham Sher Khan1, Dong Poh Chin1, Tomoko Igawa1, Masahiro Mii1, Ikuo Nakamura1
1Laboratory of Plant Cell Technology, Graduate School of Horticulture, Chiba University, Chiba, Japan
2Department of Genetics/Biotechnology, Faculty of Science, University of Calabar, Calabar, Nigeria
3Cambodia Agricultural Research and Development Institute, Phnom Penh, Cambodia
4Tissue Culture and Gene Transformation Department, Agricultural Biotechnology Research Institute of Iran (ABRII), Karaj, Iran
Email: ntuival@yahoo.com, inakamur@faculty.chiba-u.jp
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/


Received 26 December 2013; revised 2 February 2014; accepted 21 February 2014
ABSTRACT
Cucumber mosaic virus is one of the most constraints to the production of tomato and other vegetable crops worldwide. Here, we generated an RNAi construct containing inverted repeat of 1138 bp fragment of a partial replicase gene of CMV-O and used it to produce transgenic tomato plants expressing CMV-specific dsRNA of the replicase gene. Inoculation of transgenic plants with CMV strain O discriminated three categories of plants: plants that showed complete resistance, which were free of symptoms; highly resistant plants, which had mild symptoms, but later recovered because new leaves that emerged were free of symptoms; and susceptible plants, which showed severe symptoms similar to wild-type plants. The completely resistant lines were selected and challenged with a closely related strain, CMV-Y. Interestingly, the transgenic plant lines either remained immune or showed high levels of resistance to the strain. No virus could be detected in uninoculated new leaves of the resistant lines after RT-PCR and Dot immunobinding assay (DIBA) analyses. We could show that the resistance is correlated with post-transcriptional gene silencing because of the production of transgenic specific siRNA.
Keywords:CMV; dsRNA; PTGS; Replicase Gene; RNAi; Tomato

1. Introduction
RNA interference (RNAi) or RNA silencing was first discovered in plants as a mechanism for post-transcriptional gene silencing [1] . It is a conserved mechanism that recognizes double-stranded RNA (dsRNA) as a signal to trigger sequence-specific degradation of homologous mRNA. It is involved in regulating developmental processes, endogenous gene expression and also as a defense against viruses, transposons and foreign nucleic acids. The key feature of RNAi is short dsRNA fragments known as “short interfering RNAs (siRNAs)” of 21 - 25 bp in length, which are produced by the cleavage of dsRNA by a ds-specific ribonuclease termed “Dicer”, which occurs in plants as a family of four enzymes with different functions [2] . Once generated, the siRNAs are then recognized by a ribonuclease complex known as the RNA-induced silencing complex (RISC) and use as a guide for the recognition and sequence-specific degradation of homologous mRNAs [3] -[6] , resulting in post-transcriptional gene silencing (PTGS). It has been shown that RISC is present as a precursor complex that can be activated by ATP to form a complex with endonuclease activity that can cleave endogenous mRNAs [7] . In plants, dsRNA expressed from a transgene can induce not only PTGS, but also transcriptional gene silencing (TGS). siRNAs homologous to coding regions induce PTGS, which results in sequence-specific RNA degradation, whereas siRNAs homologous to promoter regions of target genes induce transcriptional gene silencing, which results in promoter methylation and consequent inhibition of transcription.
RNAi can be used to engineer resistance in plants against viruses. Plants expressing a copy of a viral gene in sense and/or antisense orientation have shown resistance upon infection with the virus (or other virus containing identical sequences) through RNAi [8] . Conventionally, antisense-mediated gene silencing has been widely used in the analysis of gene function in plants. Although antisense-mediated gene silencing is an RNAi-related phenomenon [9] , hpRNA-induced RNAi has been shown to be much more efficient [10] [11] . The hpRNA transgene is simply composed of a plant promoter and terminator between which an inverted repeat sequence of the target gene (sense and anti-sense) is inserted with a spacer region or intron, between the repeats. The RNA transcribed from such a transgene hybridizes with itself to form a hairpin structure comprising a single-stranded loop region, encoded by the spacer region/intron, and a base-paired stem encoded by the inverted repeats, which mimics the dsRNA structure that induces RNAi. The whole length of the stem acts as a substrate for the generation of siRNAs, whereas the spacer region/intron is not involved in siRNA production but is required for the stability of the inverted repeat in Escherichia coli, and appears to enhance the efficiency of silencing when directed. When an intron is used as the spacer, the efficiency becomes very high: almost 100% of transgenic plants show gene silencing [12] .
Cucumber mosaic virus (CMV) is one of the five most important viruses affecting production and causing significant economic losses in many vegetable and horticultural crops. It infects about 1200 species in over 100 plant families. CMV is a member of the cucumovirus group, having a tripartite single stranded RNA plus-sense genome [13] RNAs 1 and 2 (3.4 and 3.0 kb, respectively) encode the 1a and 2a proteins (111 and 97 kDa), which are components of the CMV replicase or RNA-dependent RNA polymerase (RdRP). The 2b protein, encoded by RNA 2, has a long distance movement function [14] . RNA 3 encodes two proteins, 3a movement protein and the viral coat protein (CP), which is expressed from subgenomic RNA 4. The 3a protein is known to facilitate the movement of CMV RNA from cell-to-cell [15] . Some CMV strains contain the CMV associated RNA 5 (CARNA-5) [16] satellite which modulates the symptoms of CMV. There are many isolates of CMV which differ in host range, pathogenicity, and biological characteristics. From nucleic acid or protein sequence composition and based on serological studies more than 70 well characterized strains of CMV were divided into two subgroups, referred to as subgroup I and II [17] -[20] , with a majority having high degree of identity (85% - 95%) at the amino acid sequence level [13] .
Tomato (Lycopersicon esculentum Mill.) is a popular vegetable widely grown in many parts of the world for its fruits, which are an excellent source of vitamin A and C, minerals like iron and phosphorus, besides having outstanding processing qualities [21] . Serious CMV infections have occurred in tomato, causing significant yield losses by reducing fruit production and quality [22] . In the late 90s, in the north-central tomato-growing region of Alabama, a CMV epidemic resulted in a 25% yield loss [23] . CMV epidemics have also been reported in tomato growing regions of Italy [16] , Spain [22] and China [24] . CMV symptoms in infected tomato include stunting of vegetative growth, distortion and mottling of new growth, and a characteristic shoestring-like leaf appearance [25] . Strains from both subgroups have been observed to infect tomatoes grown in the same field and sometimes mixed infection in the same plant have occurred [26] .
CMV is difficult to control because of its extremely broad natural host range and there are no sources of genetic resistance to CMV available in commercial fresh-market tomato cultivars [23] . Different approaches are currently under way to find suitable control measures for CMV. Efforts using traditional breeding for CMV resistance have mostly not been successful [27] . Also, pathogen-derived resistance to CMV often shows only partial resistance or very narrow spectrum resistance to the virus. Various levels of resistance were achieved via replicase-mediated resistance [28] . Very high levels of resistance and in some cases complete resistance were observed by expression of dsRNA of CMV in transgenic tobacco [29] [30] . Therefore, RNAi-mediated gene silencing especially the expression of dsRNA remains an important tool to obtaining complete resistance to CMV.
In this study, we report the expression of dsRNA of 1138 bp fragment of a partial replicase gene in transgenic tomato. Transgenic lines obtained showed high levels resistance to both CMV strains O and Y. siRNA were detected in transgenic lines prior to-and-after inoculation with the virus suggesting that the resistance was acquired through RNA silencing.
2. Materials and Methods
2.1. RNAi Vector Construction
A 1138 bp fragment spanning nucleotides 391 - 1528 of pBS+ SKCM2-50, which encodes a full length transcript of the replicase component of cucumber mosaic virus strain O, obtained from Dr. H. Takahashi, Tohoku University, Japan, was cloned by RT-PCR technique. The primers used for RT-PCR were CMVRep-5P and CMVRep-3P (Table 1). The PCR products were purified and subsequently cloned into Gateway entry vector, pCR®8/GW/TOPO® (Invitrogen, New Zealand), which contains attL1 and attL2 recombination sites. The transformants were sequenced and analyzed. The transformant with the correct insert (1138 bp) was positioned in the sense orientation between the attB2 and attB1 recombination sites and in the antisense orientation between the attB1 and attB2 recombination sites on either end of 278 bp fragment of cat1-intron derived from caster bean, to ensure stability of the construct [31] . The RNAi cassette was subcloned to the binary vector pEKH2IN2 (Nakamura et al. unpublished), by eLR clonase™ (Invitrogen, New Zealand) recombination reaction. The product was transformed into TOP10 chemical competent cells (Invitrogen, New Zealand) and selected on kanamycin-containing LB plates. Clones were verified by digestion with EcoRI. The plasmid, which contains marker genes for neomycin phosphotransferase (nptII) and hygromycin phosphotransferase (hpt), the CMV gene being driven by cauliflower mosaic virus (CaMV) 35S promoter was introduced into Agrobacterium tumefaciens strain EHA105 by triparental mating.
2.2. Plant Material, Transformation and PCR Analysis
Tomato seeds Lycopersicom seculentum Mill. (CL5915) kindly provided by Dr. Peter Hanson of Asian Vegetable Research and Development Center (AVRDC), Taiwan, were surface sterilized in 5% sodium hypochlorite for 10 min and geminated on half strength Murashige and Skoog (MS) medium [32] . Young expanded cotyledonary leaves were excised from 7 - 10 days-old seedlings and pre-cultured on (MZI), which was MS medium containing 1.5 mg/l of trans-zeatin and 0.5 mg/l of Indole-3-acetic acid (IAA), for 2 days. The pre-cultured explants were inoculated with overnight culture of A. tumefaciens strain EHA105 containing pEKH2IN2CMVRep
Table 1. Oligonucleotide primers used for amplification.
for 15 min, blotted dry and co-cultured on MZA (MZI supplemented with 100 μM acetosyringone) for 3 days at RT in the dark. After co-cultivation, explants were transferred to MZKM [MZI containing 50 mg/l of Kanamycin (to select transformants) and 20 mg/l of Meropenon (to kill bacteria carry over)] for 2 weeks. In subsequent subcultures until shoot formation and rooting, the concentration of Kanamycin in MZKM was increased to 100 mg/l. Shoots produced from the explants were excised and transferred to rooting medium, which was half strength MS medium containing the same concentrations of antibiotics but without plant growth regulators. To detect the presence of foreign genes, one well rooted T0 plant from each explant was selected as an independent clone and screened by PCR using primers CMVRep-5P and CMVRep-3P for the CMV gene, and NPTII-5P and NPTII-3P for the selectable marker gene (Table 1).
2.3. Southern Hybridization
Fifteen µg of the genomic DNA was digested overnight with HindIII, separated on a 0.8% agarose gel, transferred to a nylon membrane (Immobilon-Ny + Transfer Membrane; Millipore Co, Billerica, MA, USA) and probed with CMVRep PCR-DIG labelled probe. The probe DNA fragment was generated from plasmid DNA pEKH2IN2CMVRep and labelled by PCR using DIG-2’-deoxyuridine 5’-triphosphate (DIG-dUTP) according to the supplier’s instruction (PCR DIG probe synthesis kit, Roche Diagnostic GmbH, Boehringer Mannheim, Germany). Prehybridization (3 h) and hybridization (overnight) were carried out at 38˚C and 64˚C respectively, using high-SDS hybridization buffer containing 50% deionized formamide, 5 × SSC, 50 mM sodium phosphate (pH 7.0), 2% blocking solution, 0.1% N-lauroylsarcosine and 10% SDS. Hybridization signals were detected by exposing the membrane to a detection film (Lumi-Film Chemiluminescent Detection Film; Roche Diagnostics, Mannheim, Germany) for 20 min. Plants showing integration of the transgene as confirmed by Southern blot analysis were maintained in vitro and subcultured every 2 months to fresh medium.
2.4. Northern Blot Analysis of Total RNA
Total RNA was isolated from tomato leaves using Guanidine thiocyanate, 15 µg of the total RNA was denatured, separated on 1.5% agarose gel, transferred to nylon membrane (Immobilon-Ny + Transfer Membrane; Millipore Co, Billerica, MA, USA) by capillary blotting and probed with a DNA probe of CMVRep following the same procedure used in Southern blot analysis, with slight modification where necessary. Pre-hybridization (2 h), hybridization (overnight) and posthybridization washes were performed at 50˚C. mRNA signals were visualized using CDP-star reagent (Roche Applied Science) as described in Southern blot.
2.5. Analysis of Plants for siRNA before and after Virus Challenge
Transgenic tomato lines together with the control plants were analyzed for siRNA accumulation before and after challenge with CMV. Total RNA was isolated, and small RNAs were enriched from the total RNA by polyethylene glycol (PEG; MW8000) as described by Smith and Eamens [33] . Thirty micrograms of small RNAs were electrophoresed on a 17% polyacrylamide (acrylamide:bisacrylamide 19:1) containing 7 M urea and 10 × TBE buffer (0.9 M Tris-HCl, 0.9 M Boric acid, 20 mM EDTA). The gel was stained with 0.5 × TBE containing ethidium bromide (0.5 µg/ml) and photographed. siRNAs were transferred to Immobilon-NY+ membrane (Millipore Corporation, Billerica, MA, USA) in a semi-dry cell (Semi-dry apparatus NA-1512, Nippon Eido, Tokyo, Japan) for 1h at 10V/400mA and subjected to northern hybridization with CMVRep probe obtained by in vitro transcription of the CMV-replicase gene in the antisense and sense orientation using T7 RNA polymerase from DIG RNA labelling Kit SP6/T7, Roche Diagnostic GmbH, Boehringer Mannheim, Germany. In order to improve the signals, each probe was hydrolyzed with carbonate buffer (60 mM sodium carbonate and 40 mM sodium bicarbonate), and incubating at 60˚C for 3 h using the formula of Hamilton and Baulcombe [34] . Prehybridization (30 min) and hybridization (overnight) were performed at 37˚C. Posthybridization washes were performed with 2 × SSC for 2 × 5 min at room temperature, followed by 0.1 × SSC/0.2%SDS for 2 × 15 min at 50˚C. siRNA signals were detected using CDP-star (Roche Applied Science), as described in the DIG system and DIG application manual (Roche Diagnostic GmbH, Boehringer Mannheim, Germany).
2.6. Evaluation of Transgenic Tomato Plants for Resistance to CMV-O—In Vitro Bioassays
In this bioassay, 3 week-old rooted T0 transgenic and wild-type plants were used for analysis. Plants were mock-inoculated with phosphate buffer or inoculated with leaves sap extract (diluted in 50 mM sodium phosphate buffer containing 0.4% sodium sulphite, pH 7.5) from tobacco plants infected with CMV-O by rubbing the phosphate buffer or sap extract gently onto two completely opened young leaves with the help of carborundum as an abrasive agent (Mandal et al. 2001; Mbanzibwa et al. 2009). The inoculated plants were maintained in a growth room at 25˚C ± 2˚C, 16 h photoperiod, 30 - 40 µmol m−2·s−1 cool white fluorescent light and 70% relative humidity and observed daily for virus symptoms; pictures were taken at 30 days post inoculation (dpi).
2.7. Evaluation of Transgenic Tomato Plants for Resistance to CMV-O—Ex Vivo Bioassays
Transgenic and wild plants were grown in the greenhouse for 4 weeks before virus inoculation. Plants were inoculated with CMV-O as described above and maintained in transgenic greenhouse at 24˚C. The number of plants showing mosaic symptoms was recorded until 100% of the wild-type plants were infected. Plants were photographed at 30 dpi. Also, the number of leaves per plant showing mosaic symptoms and the plant height was recorded at 30 days post-inoculation (dpi). A five point disease rating scale of 0 - 4 grades based on the amount of leaf area affected by the disease was used to ascertain the severity of CMV symptoms: 0, no symptom; 1, faint mosaic, <25% of leaf area; 2, yellow mosaic malformation, 26% - 5.0% of leaf area; 3, severe mosaic up to 75% leaf area and 4, severe mosaic, >76% of leaf area. Disease index (D.I) was calculated as described by [35] [36]
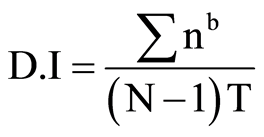
where n: number of leaves in each grade; b: grade; N: number of grades used in the scale; T: total number of leaves scored.
Plants with a disease index of 0.0 were considered as immune, those with a disease index <2.5 as having high resistance, those with a disease index of 2.6 - 5.0 as having moderate resistance, those with a disease index of 5.1 - 7.5 as being susceptible, and those with a disease index >7.6 as being highly susceptible.
2.8. Determination of Viral Load on Uninoculated New Leaves by RT-PCR
Four weeks after infection with CMV-O, total RNAs were extracted from new leaves that emerged from infected transgenic and wild-type plants using Guanidine thiocyanate as described above. The RNAs were treated with DNase and subjected to RT-PCR using the Superscript III RNase H reverse transcriptase (RT) kit (Invitrogen). The cDNAs were then used as templates for the amplification of CMV-CP using primers CMV-CP-5P and CMV-CP-3P (Table 1). Also, PCR analysis of rice actin gene (RAc1; X16280) was performed as a control to check the quality of cDNA synthesized in the RT-PCRs using the primers RAc1-5P and RAc1-3P (Table 1).
2.9. Determination of Viral Load on Uninoculated New Leaves by Dot-Immunobinding Assay (DIBA)
New leaves emerging from the inoculated plants were collected at 30 dpi. Ten mg of each leaf sample were ground in a centrifuge tube containing 100 µl TBS buffer (20 mM Tris/HCl, 500 mM NaCl, pH 7.5, and 0.5% Tween 20) to release the sap. The solution was centrifuged at 10,000 rpm for 10 min at 25˚C. Then, 10 µl each of the two concentrations; 1 and 10−1 (10 times dilution with TBS buffer) were gently dropped on separate positions on the surface of a PVDF membrane (Bio-Rad), air-dried for 5 min and submerged in Western blot blocking solution (ECL AdvancedTM Western Blotting detection Kit, Amersham, GE Healthcare UK Limited Little Chalfont, Buckinghamshire, UK.) for 30 min. Thereafter, the membrane was incubated in primary antibody raised against CP of CMV for 1 h, washed three times (10 min × 3) in TBS buffer and submerged in ECL anti-rabbit lgG immunoglobulin (Boehringer) as the secondary antibody for 1 h. To detect the presence of CMV, the membrane was stained with VEC SK-4800 kit (vector NovaRED substrate kit for peroxidase, Burlingame). All incubations were done at room temperature.
2.10. Statistical Analysis
Data on plant height were subjected to analysis of variance (ANOVA) test using a completely randomized design (CRD). Means were separated by least significant difference (LSD) test at the 5% probability level.
3. Results
3.1. Generation of the RNAi Construct and Tomato Transformation
To engineer tomato for resistance against CMV, we generated an RNAi construct containing inverted repeat of 1138 bp fragment spanning between nucleotides 391 - 1528 of a partial CMVRep gene of cucumber mosaic virus strain O. The RNAi cassette was cloned in the plant transformation vector, pEKH2IN2 under the control of CaMV 35S promoter, yielding pEKH2IN2CMVRep (Figure 1). To ensure stability of the inverted repeat in Escherichia coli, the two cDNA fragments were separated by a 278 bp unrelated spacer sequence.
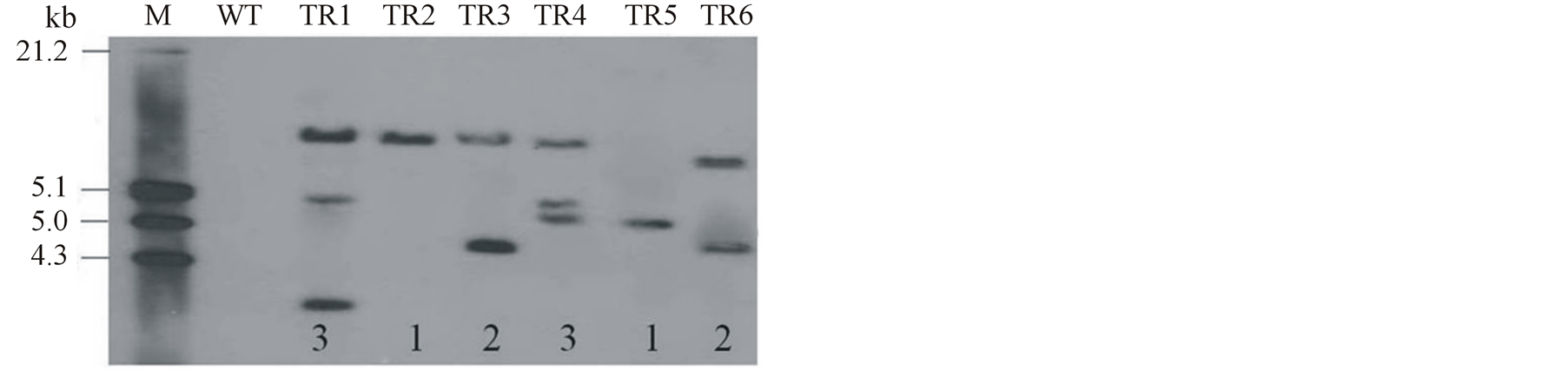
Transgenic tomato lines expressing dsRNA derived from CMVRep were generated via Agrobacterium tumefaciens-mediated transformation. A total of 15 T0 transgenic plant lines were regenerated by Kanamycin selection. Of these, 6 were selected at random and screened by PCR using CMVRep-5P and CMVRep-3P specific for CMVRep gene. The specific DNA fragment was amplified in all transgenic lines tested (data not shown). Also, 700 bp fragment of the nptII gene was amplified in all transgenic lines (data not shown). Since all the transgenic lines were phenotypically normal and no stunting was observed, the 6 lines were subjected to Southern and northern blot analyses to determine the copy number and expression of the transgene. Southern blot analysis indicated that the CMVRep gene was integrated into the DNA of the plants with 2 lines having a single locus, 2 with 2 loci and 2 with 3 loci (Figure 2(a)). Next, we determined the accumulation of transgene transcript in transgenic plants by northern hybridization. The Result shows that apart from line TR3 that produced intense mRNA, no hairpin transcript could be detected in the remainder of the transgenic lines (Figure 2(b)), indicating that engineered CMV-specific RNA molecules were being degraded to siRNA. The transgenic lines in which mRNA could not be detected showed increased levels of resistance to CMV, whereas transgenic line TR3 with significant level of mRNA was susceptible to CMV infection.
3.2. Challenging T0 Plants with CMV—In Vitro
Since no phenotypic abnormalities were observed in the transgenic plants carrying the CMVRep hairpin construct and in order to select the most promising CMV resistant plants for further propagation and testing, we first tested transgenic plants for resistance to CMV in vitro. For that purpose, T0 plants were inoculated with fresh inoculums of CMV strain O. Typical mosaic symptoms appeared on new emerging young leaves of the wild-type plants and plants of transgenic line TR3 in a few days, whereas the other transgenic lines showed no sign of disease development (data not shown). At 30 days post inoculation (dpi), all the leaves of the wild-type plants and transgenic line TR3 showed characteristic severe mosaic symptoms, which included mosaic bleaching of leaves, leaf deformation and decrease in leaf size and stunting (Figure 3). Among the other transgenic lines, line TR2 obtained from CMVRep reacted with severe symptoms in the older leaves and milder symptoms in the younger leaves (Figure 3). The remaining transgenic lines were immune to CMV infections (Figure 3). These plants remained asymptomic even after 45 dpi (data not shown) and grew taller.
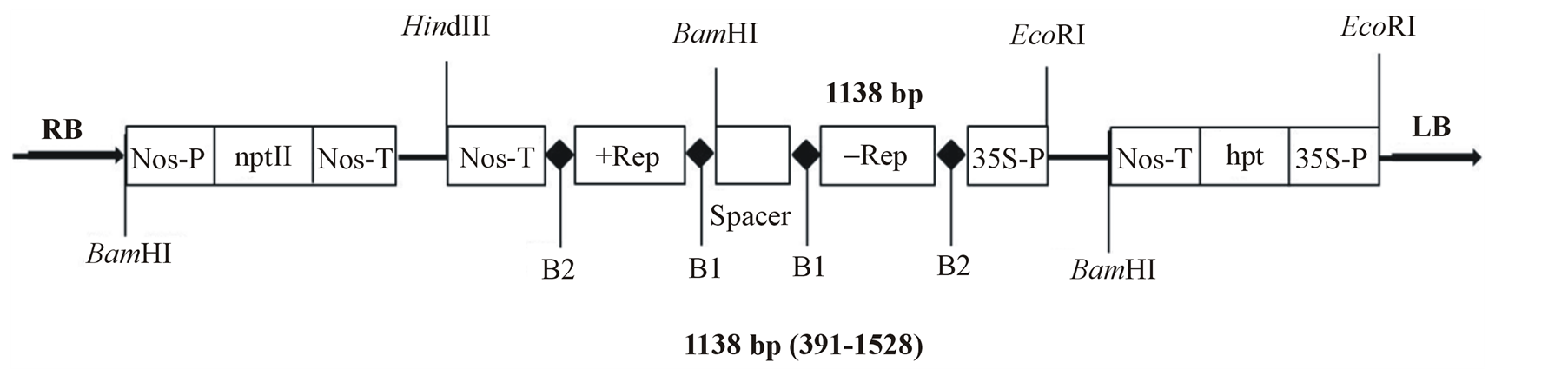
Figure 1. Map of T-DNA fragment of pEKH2IN2 carrying the partial replicase cucumber mosaic virus (CMV) inverted repeat. The CMV gene and hygromycin phosphotransferase (hpt) are driven by CaMV 35S promoter (35SP), and the gene for neomycin phosphotransferase (nptII) by nopaline synthase promoter (nos-p). B1 and B2 represent attB1 and attB2 recombination sites, respectively; LB and RB, left and right borders of T-DNA sequences, respectively; nos-T, terminator of the nopaline synthase gene. Recognition sites of restriction enzymes are also indicated.
 (a)
(a) (b)
(b)
Figure 2. Molecular analysis of transgenic tomato lines for transgene integration and expression. (a) Southern blot analysis of genomic DNA from transgenic and non-transformed control tomato plants digested with HindIII and hybridized with CMVRep probe. WT wild type control. TR1- TR6 transgenic lines. The transgene copy number is indicated in the bottom of each transgenic line. (b) Northern blot analysis for detection of transgene transcript levels. Apart from line TR3, which contains significant mRNA levels, transcript levels could not be detected in the remainder of the transgenic lines due to the specific degradation of the transgene product to siRNA soon after production.

Figure 3. In vitro evaluation of tomato lines infected with CMV-O. Photos were taken at 30 days post inoculation (dpi). WT non-transformed control plant. TR2, TR3 TR4 and TR6 transgenic tomato lines expressing hairpin CMVRep.
3.3. Challenging T0 Plants with CMV—Ex Vivo
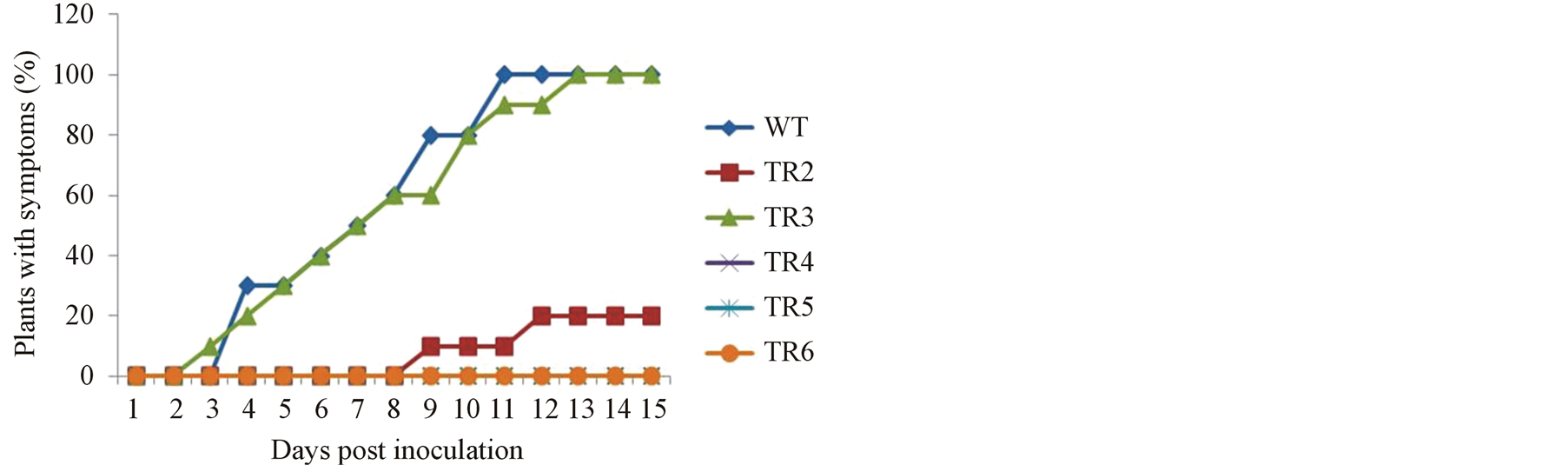
In order to further evaluate the resistance of transgenic tomato plants to CMV, ex vivo assay was carried out. Tests were performed with the same lines used for in vitro experiment. Plants were inoculated 4 weeks after transfer to the greenhouse and checked for CMV symptoms every day; the percentage of plants with symptoms was recorded. For each plant line, 10 plants were analyzed. Plants were considered infected if faint mosaic symptoms were spotted in the leaves. The result was not different from the in vitro experiment. Typical mosaic symptoms appeared on new emerging young leaves of the wild-type and plants of transgenic line TR3, 2 days post inoculation (dpi). At this period, about 40% of the plants were infected (Figure 4(a)). Mosaic symptoms continued to appear on every emerging new leaf of the wild-type and transgenic line TR3 and by 11 and 13 dpi, all the plants of wild-type and transgenic line TR3, respectively, had mosaic symptoms (Figure 4(a)). Infected wild-type and TR3 transgenic plants were stunted approximately two-third of the height of the mock inoculated control, the highly resistant or immune plants (Figure 4(b)). In transgenic lines TR1 and TR2, both susceptible
 (a)
(a) (b)
(b)
Figure 4. Ex vivo evaluation of resistance against CMV-O. (a) Symptom development in wild-type (WT) and representative T0 transgenic tomato lines after inoculation. (b) Representatives of different plant lines 30 dpi. WT Wild-type plant showing mosaic symptoms in emerging leaves, note that the height of the plant is reduced to almost a third of the resistant lines. TR2 Transgenic tomato line expressing hairpin CMVRep, showing mild/moderate symptoms after infection with CMV. TR4, TR5 and TR6 Transgenic tomato lines showing complete resistance to CMV under the period of study. WTM Mock inoculated control.
and resistant individuals were obtained. The susceptible individuals had symptoms similar to those of the wildtype plants (data not shown). In the resistant individuals, symptoms started appearing 8 dpi (Figure 4(a)). The symptoms were predominantly limited to the inoculated and emerging new leaf 1 - 3. After emerging new leaf 3, all other new leaves were free of symptoms. The reason for variation in these lines is not clear. However, it is possible that not all plants of these lines were homogenous in their transgenic content, and therefore may not represent a single “line”. It may also be possible that resistance in these lines may depend on the portion of the virus that is actually penetrating the plant.
Three of the transgenic lines tested, TR4, TR5 and TR6, were immune (i.e. showed complete resistance) to CMV infection throughout the observation period as no symptoms were observed in any of the plants. Moreover, the plants presented similar growth when compared with the mock inoculated control, indicating that the expression of CMVRep gene induced strong resistance in transgenic plants and did not affect the agronomic performance of the plants (Figures 4(a) and (b)).
We also measured the degree of disease spread per plant by evaluating the number emerging leaves showing mosaic symptom every day, but the final score was recorded at 60 dpi. Since different leaves that emerged usually showed a range of symptoms especially in the wild-type and susceptible plants, symptom severity was evaluated according to a standard scale of 0 (asymptomic) to 4 (severe mosaic, >76% of leaf area) (Figure 5(a)). Based on the above scale, disease indices were calculated according to the formula shown in materials and methods. Three categories of plants (highly susceptible, highly resistant and immune) were observed (Table 2).
The first category comprises the highly susceptible plants. Among the transgenic plants tested, one line, TR3 was grouped in this category as the severity of symptoms observed in this line was similar to that observed in the wild-type plants. As shown in Figure 5(b), all the emerging new leaves of the wild-type plants and transgenic line TR3 developed a ranged of symptoms, from mild (grade 1) to severe (grade 4). In the wild-type plants, 86.8% of the new leaves had severe disease symptoms, of this, 36.4% of the leaves were classified to be of grade 3 while 50.4% were classified to be of grade 4 severity score (Figure 5(b)). Similarly, in transgenic line TR3, 85% of the new leaves had severe disease symptoms, with 41.7 being of grade 4 and 33.3 being of grade 3. The severity of the disease in these plants produced high disease indices, which were 9.8 ± 2.7 for the wild-type plants and 8.8 ± 4.6 for TR3 (Table 2). Therefore, these plants were classified as being highly susceptible (HS) according to the severity score. It is worth noting that disease severity in these plants increased with age of the leaves i.e. lower leaves had symptoms which were mostly classified to be of grade 3 and 4 compared to upper leaves in which the symptoms were mostly of grade 2 or 1.
The second category comprises the resistant plants. The leaves of these plants reacted with mild to moderate symptoms and were classified in grade 1 and 2 severity score. Transgenic lines classified in this category were TR1 and TR2. In these lines, 66.6% of the leaves, which were from new leaf 4 and above, were asymptomic. The lower leaves, which included the inoculated leaves and new leaf 1-3, were either grouped under grade 1 (16.7) or grade 2 (16.7) severity score. Disease indices calculated for these line were very low (<2), hence the lines were classified as being highly resistant to CMV (Table 2), suggesting that the virus may move from cell to cell but do not invade or survive in the vascular system.
The last category of plants is the immune plants. All the leaves of these plants were grouped under grade 0 severity score (Figure 5(b)) as they were devoid of any disease symptom throughout the entire testing period. Consequently, their disease indices were 0 and the plants were classified as being immune to CMV (Table 2) according to the disease rating. Three plant lines fall under this category; these are TR4, TR5 and TTR6.
3.4. Detection of CMV Specific siRNA in Transgenic Plants
Virus resistance in plants can be induced by the expression of small RNAs. The presence of siRNAs would indicate whether the PTGS response had been activated. Therefore, prior to challenging the plants with CMV, northern blot analysis was carried out to detect CMVspecific siRNAs in transgenic and wild-type plants. Plants were hardened in the greenhouse for six weeks and fresh leaves were used for siRNA analysis.
Table 2. Evaluation of transgenic and wild type plants after inoculation with CMV-O.
aData were recorded at 30 days post inoculation (dpi). bDisease rating is based on calculated disease indices. The copy number of the transgene is as determined by Southern blot analysis. Mean with the different case letters are significantly different at 5% probability level. WT inoculated wild-type plant; HS Highly susceptible; HR Highly resistant; NA Not applicable; WTM Mock inoculated control plant; TR1-TR6 Transgenic lines expressing hairpin CMVRep.
 (a)
(a)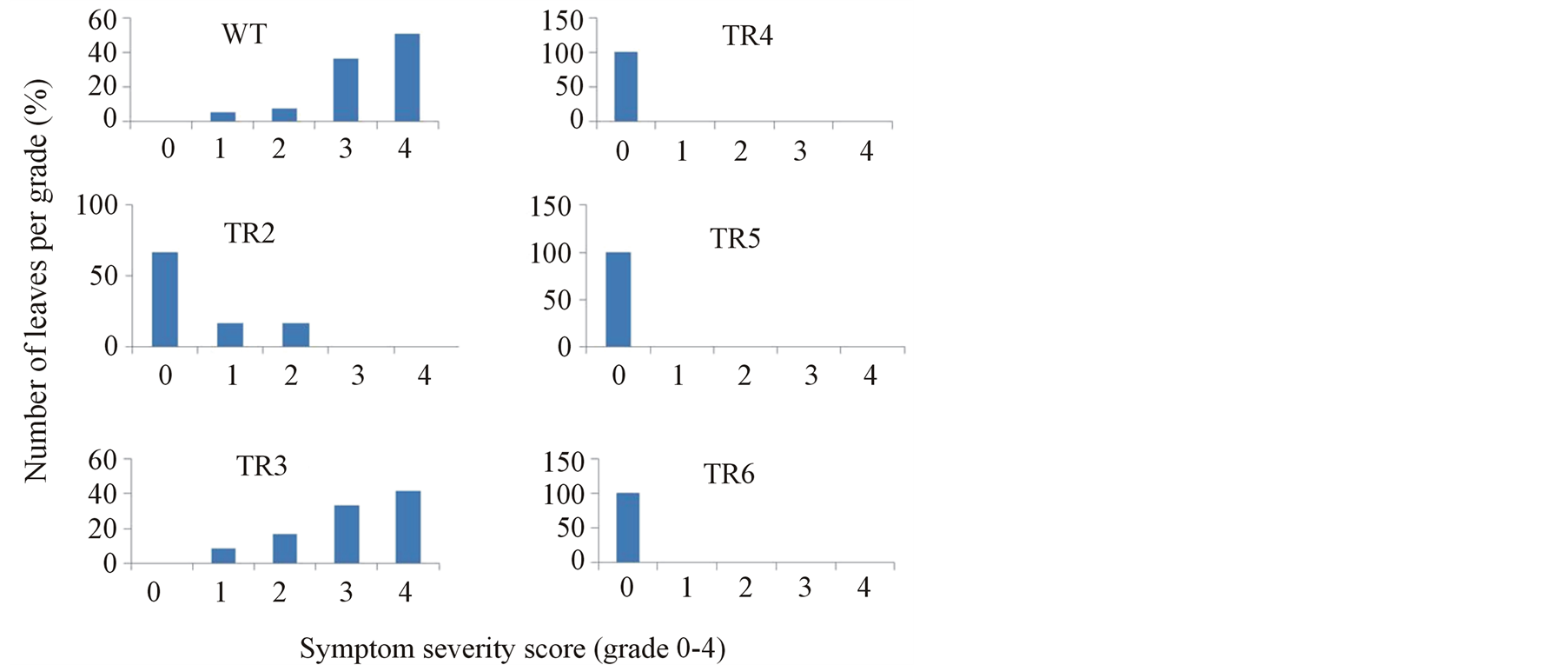 (b)
(b)
Figure 5. Disease severity score of emerging new leaves of tomato plants after inoculation with CMV. (a) Representative leaves showing different degree of symptoms for evaluation of symptom severity score (0, no symptoms; 4, severe symptoms). (b) Classification of leaves with respect to symptom severity at 60 dpi. WT Inoculated wild-type plant. TR2-TR6 transgenic lines expressing hairpin CMVRep.
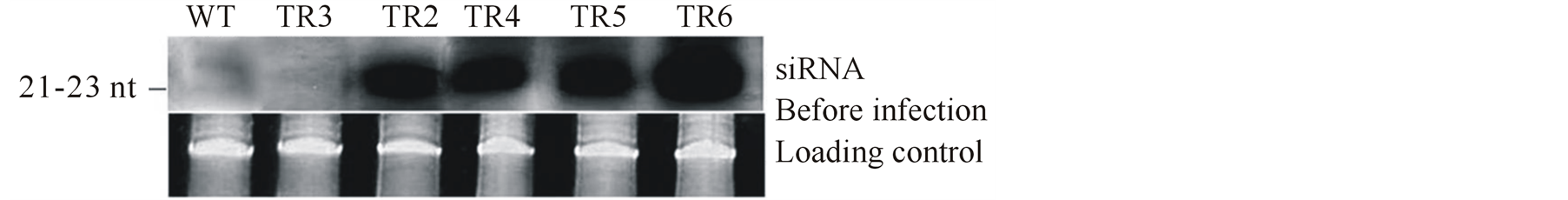
To exclude a potential latent infection with CMV, which might result in virus-derived siRNA that are not of transgene origin, the plants were subjected to RT-PCR and DIBA. As expected, no CMV signal was detected in any of the lines studied (data not shown). Northern analysis showed accumulation of siRNA with slightly different intensities in the resistant plants TR2, TR4, TR5 and TR6 (Figure 6(a)). In the wild-type plants and the susceptible transgenic line TR3, no siRNAs were detected (Figure 6(a)). It is interesting to note that although transgenic line TR3 accumulated high levels of CMV-specific transcript, siRNA could not be detected in this line. Since northern blot represent the steady state of RNA concentration, but not the transcription rate [30] , it could be that mRNA in TR3 were not degraded to siRNA resulting to inactivation of PTGS mechanism. This probably explains why this line is susceptible to CMV infection.
We examined the presence of siRNA accumulation in transgenic and wild-type plants after challenge with CMV. Four weeks after inoculation with the virus, the wild-type plant, mock inoculated control, and four transgenic lines TR2, TR4, TR5 and TR6 were selected and analyzed for the presence of siRNA. The analysis showed detectable levels of siRNAs in all the transgenic lines tested including the susceptible wild-type plant.
However, the level of siRNA signal did not correlate with severity of the symptoms; for example, plants that were immune to CMV infections produced intense siRNA signals than the susceptible plants (Figure 6(b)). This result suggests that plant virus infection causes accumulation of siRNA in both susceptible and resistant phenotypes [30] [34] . No signals were detected in the mock inoculated control plants.
3.5. Effect of Temperature on Resistance of Transgenic Lines
In view of the observation that some transgenic lines were either highly resistant or immune to CMV infection at 24˚C, we tested whether increased or reduced temperature will affect the resistance of these transgenic lines. It
 (a)
(a) (b)
(b)
Figure 6. Northern blot analysis of CMV replicase specific siRNA accumulation in tomato plants before (a) and after (b) inoculation with CMV. The inoculated plants were analyzed for siRNA accumulation at 30 days post inoculation (dpi). The lower panel shows the loading level of each sample after ethidium bromide staining. WT Inoculated wild-type plant. WTM Mock inoculated control plant. TR2-TR6 Transgenic lines expressing hairpin CMVRep.
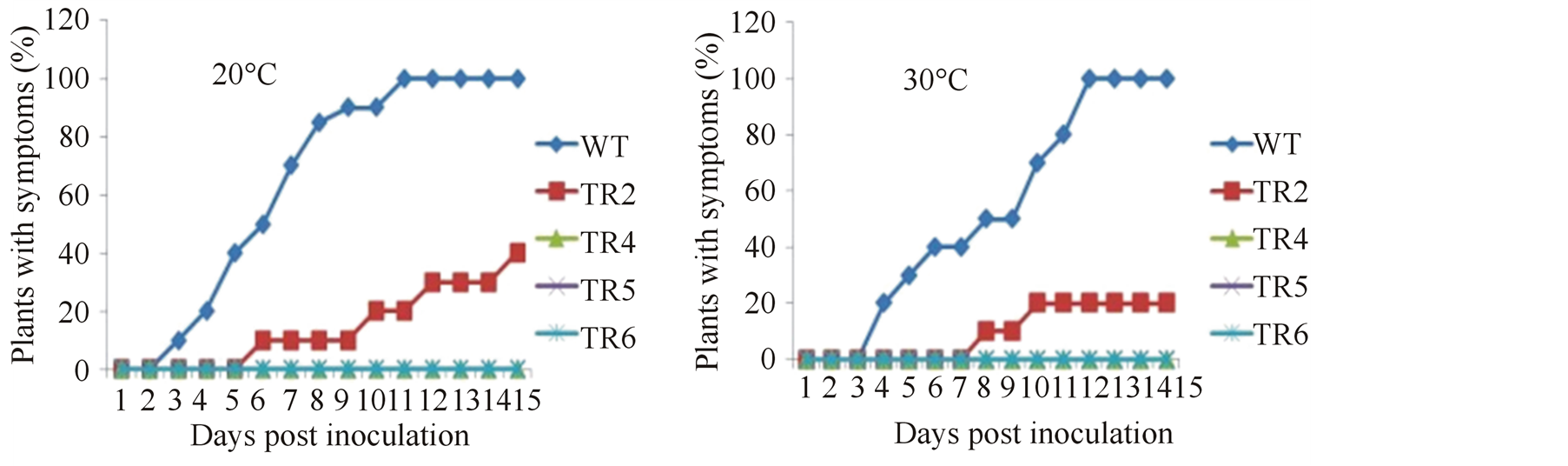
has been reported that environmental conditions such as temperature influence the response of plants to virus infection [37] . In this regard, four transgenic lines TR2, TR4, TR5 and TR6 were inoculated with CMV and exposed to either 20˚C or 30˚C ex vivo. Plants were monitored for disease development for a period of 30 days. As shown in Figures 7(a) and (b), transgenic lines TR4, TR5 and TR6 remained without symptoms at both temperatures throughout the observation period. At 20˚C and 30˚, the progeny of transgenic line TR2 showed a range of symptoms similar to those observed for the same line at 24˚C. In this line, 40% of the plants showed CMV symptoms when grown at 20˚C (Figure 7(a)), whereas only 20% had CMV symptoms when maintained at 30˚C (Figure 7(b)). Furthermore, at 20˚C symptoms appeared earlier (6 dpi) than at 30˚C (8 dpi) (Figure 7(b)). The result observed at 30˚C was not different from that obtained at 24˚C for this same line. Compared to the transgenic plants, all the wild-type plants developed symptoms at both temperatures within the period of study. Comparing temperatures, attenuation of symptom development and a decrease in symptom severity was observed when the plants were maintained at 30˚C (data not shown). From this result, if we disregard the slight difference in the percentage of plants with symptoms and the early appearance of the disease in transgenic line TR2 at 20˚C, and since the general trend of resistance is same for all the lines tested, we could suggest that transgene-mediated virus resistance was not affected by temperature. Moreover, the virus siRNA was stable at both temperatures (Figure 7(c), only lines TR4 and TR6 are shown here).
3.6. Screening of Transgenic Plants for Detection of Virus
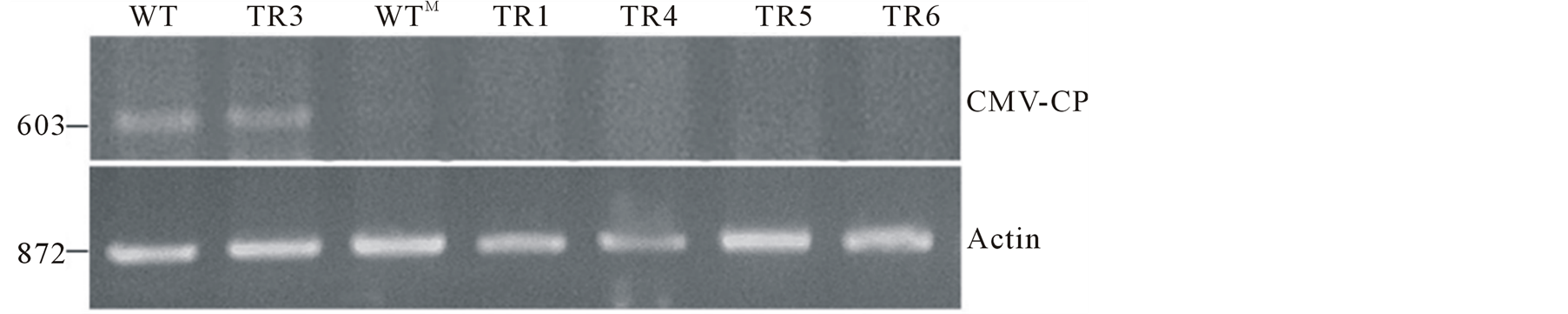
This analysis was performed to determine that there is no viral load in uninoculated new leaves of transgenic plants. Only plant lines that were immune to the virus were selected for the analysis. In parallel, leaves from a mock inoculated plant, wild-type and susceptible transgenic line TR3 were also tested. Two tests were performed, RT-PCR and DIBA. For RT-PCR, RNA was extracted from new leaves 30 days after challenge with the virus and analyzed for the presence of CMV-O coat protein using coat protein specific primers. The result indicates complete resistance to the virus in the immune plants as viral RNA was not detected in new leaves of all the transgenic lines tested (Figure 8(a)). On the contrary, clear signal of the CP messages were detected in new leaves of the inoculated wild-type plant and line TR3 (Figure 8(a)).
The result of DIBA was consistent with the RT-PCR analysis. No signal of the virus was detected in the new leaves of all the immune plants and mock inoculated plant at both concentration of plant sap. However, new leaves of the inoculated control plant and the susceptible transgenic line TR3 showed high intensity of CP signal of CMV-O even at low concentration (10−1, 10 times dilution) of plant sap (Figure 8(b)). From the above results it is evident that there is no viral load in these transgenic lines.
 (a)
(a) (b)
(b)
Figure 7. Response of transgenic plants to CMV at different temperature conditions. Four transgenic lines expressing the hairpin CMVRep gene were inoculated with CMV and exposed to either 20˚C (a) or 30˚C (b). (c) Effect of temperature on siRNAs accumulation of transgenic lines TR4 and TR6.
 (a)
(a)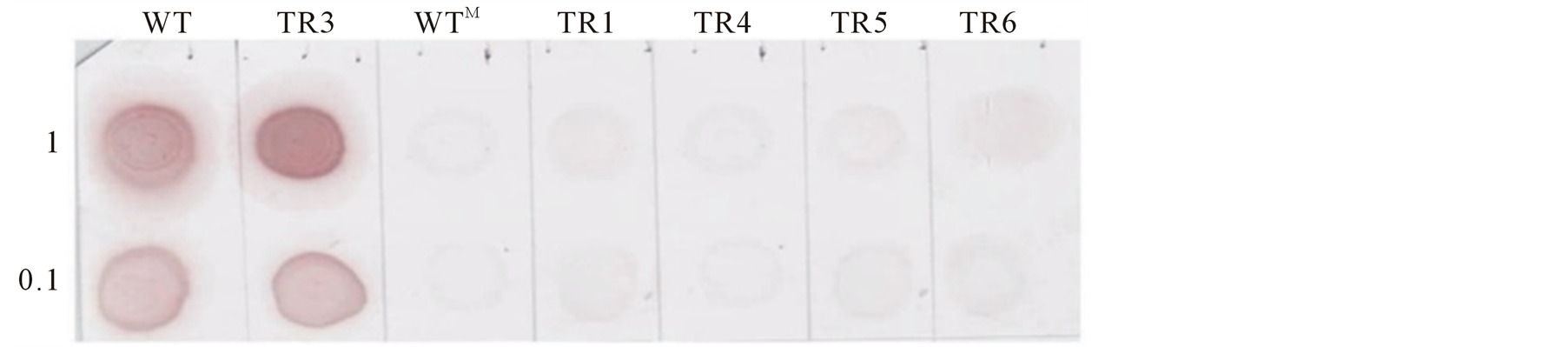 (b)
(b)
Figure 8. Screening of transgenic plants for detection of CMV virus after infection (A) RT-PCR analysis for the detection of coat protein (CP) of cucumber mosaic virus in emerging new leaves of transgenic lines infected with CMV-O (upper panel). Rice actin (RAc1) gene (lower panel) was used as an internal control for RNA input. (B) Dot-immunobinding assay for detection CP of cucumber mosaic virus in emerging new leaves of plants infected with CMV-O. 1 and 0.1 (10 times dilution) represent the concentration of plant sap used. Leaves were collected for RT-PCR and DIBA analysis at 30 dpi. WT inoculated wild-type plant. WTM Mock inoculated control plant. TR2-TR6 Transgenic tomato lines expressing hairpin CMVRep gene.
3.7. Relationship between Copy Number of Transgene and Disease Index
To determine whether copy number of the transgene has effect on the resistance of transgenic plants, we checked the relationship between copy number of the transgene and disease index. The results showed insignificant negative relationship (R = −0.146) (Figure 9), suggesting that resistance is not dependent upon the copy number. For example, transgenic lines, TR3 with 2 copies of the transgene was susceptible, whereas lines TR4, TR5 and TR6 with 3, 1, and 2 copies of the transgene, respectively, were immune to CMV. Furthermore, lines TR1 and TR2 with 3 and 1 copy, respectively had mild symptoms.
3.8. Resistance to another CMV Strain
Since some transgenic lines were immune to CMV-O, we were interested to see whether these transgenic lines will display the same level of resistance to a closely related CMV strain. Therefore, T0 plants of line TR4, TR5 and TR6 were inoculated with CMV-Y. In parallel, one Wild-type plant was also inoculated as well with this strain. CMV-O and Y belong to the same subgroup and show high degree of similarity in their nucleotide sequence [18] . CMV-Y, however, produces severe systemic yellow/white mosaic symptoms than CMV-O [38] . Two plants from each line were inoculated; the time (in days post inoculation) for disease to appear (Figure 10(a)) was recorded. Also, the total number of emerging new leaves (in percent) showing mosaic symptoms per plant line was recorded at 30 dpi (Figure 10(b)). Results revealed that both plants of line TR4 presented symptoms, which started appearing on 7 and 9 dpi in plant TR4-1 and TR4-2, respectively (Figure 10(a)). For each of the plants, greater than 50% of the new leaves had symptoms (Figure 10(b)). However, the symptoms were mostly of grade 2 severity score and did not significantly affect the growth of the plants (Figure 10(c)). The remainder of the tested plants, TR5-1, TR5-2, TR6-1 and TR6-2 were without symptoms throughout the observation period (Figures 10(a), (b) and (c)). Moreover, the viral DNA could not be detected by DIBA in plants that were free of symptoms (data not shown). However, as the number of plants per transgenic line used for inoculation with CMV-Y is few, further research is needed to confirm this result. Compared to the transgenic lines, symptoms were seen in the wild-type plant 2 dpi (Figure 10(a)) and all the new leaves that emerged were symptomic (Figure 10(b)).
4. Discussion
Cucumber mosaic virus is one of the most important constraints to the production of tomato and other vegetable crops worldwide. The virus is known to have the widest host range of any known plant virus. Over the years, engineering resistance to CMV has been approached using pathogen-derived resistance. However, pathogen-derived resistance often resulted to only partial or very narrow spectrum of resistance to the virus. The aim
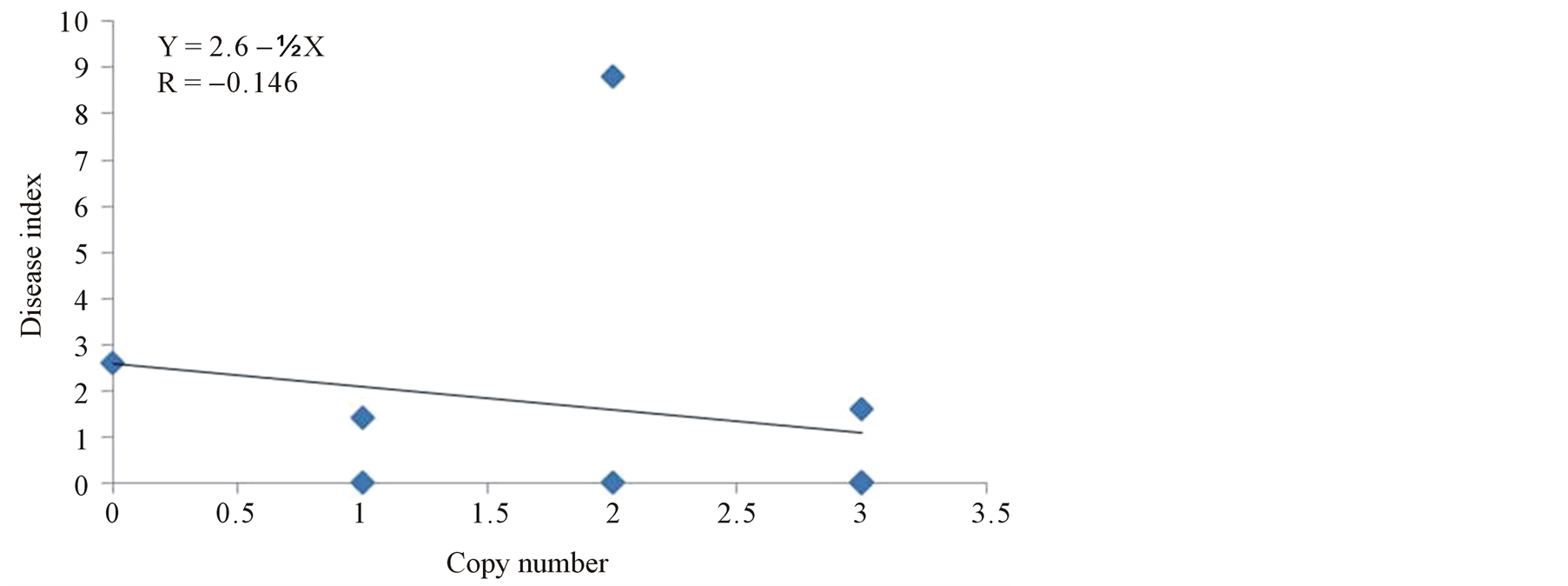
Figure 9. Linear regression analysis to establish the correlation between transgene copy number and disease index (infection index) of transgenic tomato lines inoculated with CMV-O. The transgene copy number as determined by Southern blot analysis is shown in the bottom of each line in Figure 2(b).

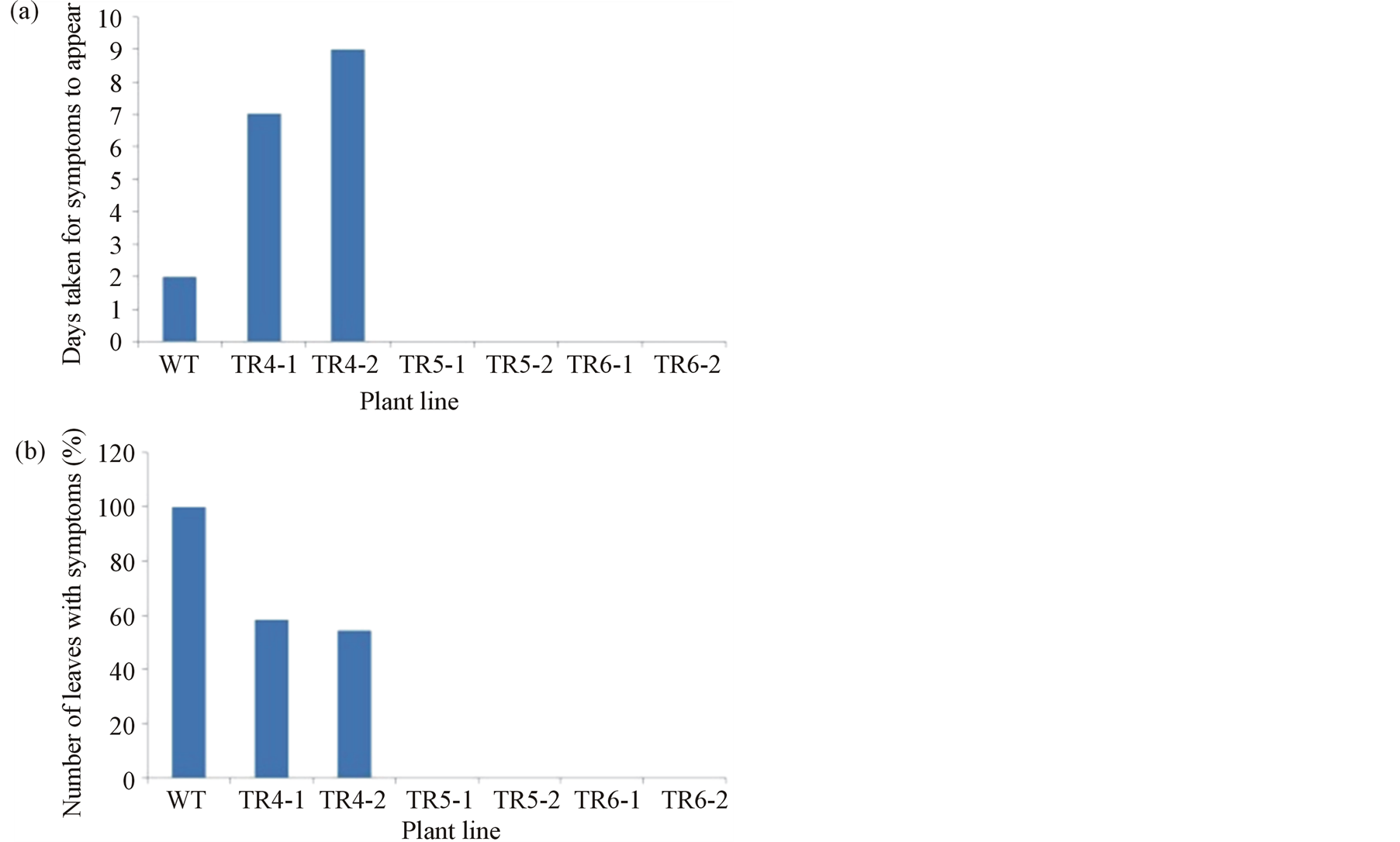
Figure 10. Ex vivo evaluation of resistance against CMV-Y. Three of the lines that showed complete resistance to CMV-O were used for this assay; two plants from each line were inoculated with the virus. (a) The time (in days post inoculation) for disease to appear. (b) The total number (in percent) of emerging new leaves per plant line showing mosaic symptoms at 30 dpi. (c) Representatives of different plant lines at 30 dpi. WT wild-type plant. TR4, TR5 and TR6 represent individual transgenic lines, 1 and 2 represent plant number.
of this work was to efficiently engineer resistance to CMV in tomato, an economically very important vegetable that is affected by the virus, through RNA silencing by the expression of dsRNA of CMV replicase gene. Engineering virus-resistance in transgenic crops through RNA silencing takes advantage of the natural defence mechanism of plants against viruses [39] , which is also active in wild-type plants. We have previously expressed dsRNA of a of defective replicase CMV gene in transgenic potato [40] and tobacco [41] . The replicase gene was modified by deleting a 9 bp region between nucleotides 1909-1918, which caused a shift in the active centre motif of polymerases, producing defective translated product 9 nucleotides shorter than the full length protein. Transgenic potato and tobacco plants obtained were immune to CMV infections. In this work, we decided to express dsRNA of a partial (none modified) replicase gene of CMV-O in tomato. We also set out to explore whether the resistance in tomato would be similar to that observed in potato and tobacco, which both contain the defective replicase gene. The introduction of CMV-specific dsRNA derived from the partial replicase gene into tomato discriminated three categories of plants: plants that showed complete resistance (three lines), these plants were free of symptoms; highly resistant plants, which had mild symptoms, but later recovered because new leaves that emerged were free of symptoms; and susceptible plants, which showed severe symptoms similar to wild-type plants.
In transgenic potato and tobacco containing the defective gene, no susceptible plants were observed [40] [41] . We could suggest that the two systems, both of which involve the expression of replicase derived dsRNA may operate by different mechanisms, one dealing with a mutant (defective) replicase while the other deals with the normal replicase gene.
In order to determine whether the transgenic tomato plants would be resistant to other cucumoviruses and many other plant viruses, we decided to challenge the completely resistant lines with a closely related CMV strain, CMV-Y. CMV-O and Y belong to the same subgroup and show high degree of similarity in their nucleotide sequence [18] ; however, infectivity of CMV-Y is severe than that of CMV-O. Sequence analysis show that the CMV-O nucleotide sequence used for hairpin in the constructs and the CMV strains (CMV-O and CMV-Y) used for infection share 99% identity. Furthermore, the nucleotide sequence of the majority of CMV strains found in database is highly conserved with greater than 95% identity within the part of the CMV sequence used in this work. Interestingly, two transgenic plant lines TR5 and TR6 remained immune, even after infection with CMV-Y, while one plant line TR4 showed mild to moderate symptoms upon inoculation with CMV-Y (Figure 9). Therefore, it is possible the resistance gained here might be effective against other cucumoviruses. As the number of plants used for inoculation with CMV-Y was quite small, further experiment is needed to assess fully the RNAi-mediated protection by CMVRep against CMV-Y and other cucumoviruses.
Regardless of the mechanistic approach used here, the expression of dsRNA resulting from replicase gene has advantages for environmental safety over the coat protein (CP)-mediated resistance. It has been reported that CPs expressed by transgenic plants can partially or completely encapsidate the genome of the challenged RNA viruses [42] [43] . We analyzed transgenic plants for viral coat protein 4 weeks after infection with CMV-O by RT-PCR and DIBA. Coat protein represents the stably packaged form of the viral genome, which is transmitted by aphids. The absence of CMV coat protein in new leaves of the highly and completely resistant transgenic lines suggests the absence of viral particles. As there is no virus CP load in these plants, the potential risks of heterologus encapsidation and recombination of virus are diminished. Also, it is likely that infected transgenic tomato plants would be a poor source of virus acquisition by aphids when tried under field conditions and therefore may reduce further spreading of CMV by the aphid vectors. The propagation of plants that display no symptoms but carry a high virus load would be undesirable with regard to virus control under field conditions as such plant would serve as a source of inoculum for subsequent dissemination via aphids [44] .
The key feature of RNAi-mediated resistance is the production of short dsRNA fragments known as “short interfering RNAs (siRNAs) of 21 - 25 bp in length. It is believed that the presence of artificially expressed dsRNA activates the plant’s PTGS machinery even in the absence of the virus. According to the model of PTGS, the strand of siRNA complementary to the target RNA becomes incorporated into the RNA-induced silencing complex (RISC), which is responsible for the actual RNA degradation of the homologous target RNA [7] . Of the transgenic lines analyzed only one (TR3) did not produce detectable levels of siRNA prior to challenge by the virus and was susceptible to CMV infection. We could thus suggest that the presence of siRNA prior to challenge by CMV plays a vital role in the resistance of transgenic lines to the virus. Plants that produced detectable levels of siRNA were resistant to viral infections. Therefore, the occurrence of siRNAs can be taken as a molecular marker to predict the degree of resistance which is an advantage of RNAi-mediated strategy [30] . After virus challenge, high levels CMV siRNA was detected in all transgenic lines tested including the susceptible wild-type plants. In the plants that produced detectable of siRNA but were susceptible to CMV infections, it is possible that CMV was actively replicating despite PTGS, resulting to a seemingly fragile balance between the plant and the virus: the plant suppressing the virus via PTGS (and possibly other mechanisms) and the virus responds by rapid replication and suppression of the host’s silencing mechanism [45] [46] . As a counterdefense, certain plant viruses encode proteins that can suppress the RNA silencing in order to overcome the defense mechanism. Cucumber mosaic virus (CMV)-contains the 2b protein [47] , which inhibits post-transcriptional gene silencing (PTGS). The 2b protein is a protein that contains a nuclear localization signal required for long distance viral movement in some hosts and it suppresses silencing only in newly emerged tissues that develop after infection [48] . It is possible that the 2b protein inhibited gene silencing by sequestrating siRNAs and preventing them from entering the RISC. Incomplete suppression of intracellular silencing, possibly due to CMV 2b not being expressed in every cell has been reported [49] .
Environmental conditions such as temperature have been reported to influence plant-virus interaction and the expression of siRNA. Szittya et al. [50] indicated that RNA silencing-mediated plant defense is temperature dependent. At low temperature, both virus and transgene silencing was inhibited leading to susceptibility. In our work, temperature difference did not affect the silencing mechanism of transgenic plants as all the lines remained immune or highly resistant to CMV (Figures 7(a) and (b)). Moreover, the virus siRNA signals were stable at both temperature, and no apparent decrease was observed at 20˚C (Figure 7(c)). Our results differ from that of Kalantidis and his associates [30] who found that elevated temperature favoured the formation of CMV derived siRNA in tobacco, resulting to increased resistance. The discrepancies in our result and that of Kalantidis et al. [30] may probably be because of the target gene used. Since Kalantidis et al. [30] used coat protein gene of CMV to confer resistance to the virus, it is possible that both replicase and coat protein sequences may operate by different mechanism, hence the discrepancy.
In most transformation experiments, expression of the transgene is dependent upon the copy number and abnormal morphology has often been reported [51] [52] . In some cases, increased copy number resulted to gene silencing and hence susceptible phenotypes [52] . In our experiment, increased copy number did not induce any phenotypic abnormality and resistance was not dependent upon the copy number of the transgene. In transgenic line TR3 having 2 copies of the transgene but was susceptible to CMV infection, we concluded that siRNA accumulation and not the transgene copy was responsible for the severe symptoms. As mentioned above, no siRNA was detected in this line prior to challenge with CMV.
Together, our results demonstrate that the resistance of tomato against CMV infection can be achieved by expression of dsRNA of the partial replicase gene. The transgenic lines generated here have not been yet tested in the field. To establish the economic viability of this system, extensive study of field performance of the transgenic lines to CMV infection will be necessary.
Acknowledgements
We thank Professor Hideki Takahashi of Tohoku University, Japan, for kindly donating the CMV-O and Y strains. This work was supported by Japan Society for the Promotion of Science (JSPS). V. O. Ntui is a recipient of JSPS fellowship.
References
- Baulcombe, D.C (2000) Unwinding RNA Silencing. Science, 290, 1108. http://dx.doi.org/10.1126/science.290.5494.1108
- Schauer, S.E., Jacobsen, S.E., Meinke, D.W. and Ray, A. (2002) DICER-LIKE1: Blind Men and Elephants in Arabidopsis Development. Trends in Plant Science, 7, 487-491. http://dx.doi.org/10.1016/S1360-1385(02)02355-5
- Nykanen, A., Haley, B. and Zamore, P.D. (2001) ATP Requirements and Small Interfering RNA Structure in the RNA Interference Pathway. Cell, 107, 309-321. http://dx.doi.org/10.1016/S0092-8674(01)00547-5
- Hammond, S.M., Bernstein, E., Beach, D. and Hannon, G.J. (2000) An RNA-Directed Nuclease Mediates PostTranscriptional Gene Silencing in Drosophila Cells. Nature, 404, 293-296. http://dx.doi.org/10.1038/35005107
- Zamorem P.D., Tuschlm T., Sharpm P.A. and Bartelm D.P. (2000) RNAi: Double-Stranded RNA Directs the ATP-Dependent Cleavage of mRNA at 21 to 23 Nucleotide Intervals. Cell, 101, 25-33. http://dx.doi.org/10.1016/S0092-8674(00)80620-0
- Tuschl, T., Zamore, P.D., Lehmann, R., Bartel, D.P. and Sharp, P.A. (1999) Targeted mRNA Degradation by DoubleStranded RNA in Vitro. Genes and Development, 13, 3191-3197. http://dx.doi.org/10.1101/gad.13.24.3191
- Hammond, S.M., Boettcher, S., Caudy, A.A., Kobayashi, R. and Hannon, G.J. (2001) Argonaute 2, a Link between Genetic and Biochemical Analyses of RNAi. Science, 293, 1146-1150. http://dx.doi.org/10.1126/science.1064023
- Waterhouse, P.M. and Helliwell, C.A. (2003) Exploring Plant Genomes by RNA-Induced Gene Silencing. Nature Reviews Genetics, 4, 29-38. http://dx.doi.org/10.1038/nrg982
- Serio, F.D., Schöb, H., Iglesias, A., Tarina, C., Bouldoires, E. and Meins, F.J. (2001) Senseand Antisense-Mediated Gene Silencing in Tobacco Is Inhibited by the Same Viral Suppressors and Is Associated with Accumulation of Small RNAs. Proceedings of the National Academy of Sciences of the United States of America, 98, 6506-6510. http://dx.doi.org/10.1073/pnas.111423098
- Chuang, C.F. and Meyerowitz, E.M (2000) Specific and Heritable Genetic Interference by Double-Stranded RNA in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America, 97, 4985- 4990. http://dx.doi.org/10.1073/pnas.060034297
- Smith, N.A., Wang, M.B., Stoutjesdijk, P.A., Green, A.G. and Waterhouse, P.M. (2000) Total Silencing by IntronSpliced Hairpin RNAs. Nature, 407, 319-320.
 http://dx.doi.org/10.1038/35036500
http://dx.doi.org/10.1038/35036500 - Wesley, S.V., Helliwell, C.A., Smith, N.A., Wang, M.B., Rouse, D.T., Liu, Q., Gooding, P.S., Singh, S.P., Abbott, D., Stoutjesdijk, P.A., Robinson, S.P., Gleave, A.P., Green, A.G. and Waterhouse, P.M. (2001) Construct Design for Efficient, Effective and High-Throughput Gene Silencing in Plants. The Plant Journal, 27, 581-590. http://dx.doi.org/10.1046/j.1365-313X.2001.01105.x
- Palukaitis, P., Roossinck, M.J., Dietzgen, R.G. and Francki, R.I.B. (1992) Cucumber Mosaic Virus. Advances in Virus Research, 41, 281-347. http://dx.doi.org/10.1016/S0065-3527(08)60039-1
- Ding, S.W., Li, W.X. and Symons, R.H. (1995) A Novel Naturally Occurring Hybrid Gene Encoded by a Plant RNA Virus Facilitates Long Distance Virus Movement. EMBO Journal, 14, 5762-5772.
- Kaplan, I.B., Gal-On, A. and Palukaitis, P. (1997) Characterization of Cucumber Mosaic Virus III. Localization of Sequences in the Movement Protein Controlling Systemic Infection in Cucurbits. Virology, 230, 343-349. http://dx.doi.org/10.1006/viro.1997.8468
- Kaper, J.M., Gallitelli, D. and Tousignant, M.E. (1990) dentification of a 334-Ribonucleotide Viral Satellite as Principal Aetiological Agent in Tomato Necrosis Epidemic. Research in Virology, 141, 81-95. http://dx.doi.org/10.1016/0923-2516(90)90058-Q
- Wahyundi, W.S., Dietzgen, R.G., Hanada, K. and Francki, R.I.B. (1992) Serological and Biological Variation between and within Subgroup I and II Strains of Cucumber Mosaic Virus. Plant Pathology, 41, 282-297. http://dx.doi.org/10.1111/j.1365-3059.1992.tb02350.x
- Hayakawa, T., Mizukami, M., Nakamura, I. and Suzuki, M. (1989) Cloning and Sequencing of RNA-1 cDNA from Cucumber Mosaic Virus Strain O. Gene, 85, 533-540. http://dx.doi.org/10.1016/0378-1119(89)90448-4
- Owen, J. and Palukaitis, P. (1988) Characterization of Cucumber Mosaic Virus. I. Molecular Heterogeneity Mapping of RNA3 in Eight CMV Strains. Virology, 166, 495-502. http://dx.doi.org/10.1016/0042-6822(88)90520-X
- Piazzolla, P., Diaz-Ruiz, J. R. and Kaper, J.M. (1979) Nucleic Acid Homologies of Eighteen Cucumber Mosaic Virus Isolates Determined by Competition Hybridization. Journal of General Virology, 45, 361-369. http://dx.doi.org/10.1099/0022-1317-45-2-361
- Christopher, D.J., Raj, T.S., Rani, S.U. and Udhayakumar, R. (2010) Role of Defense Enzymes Activity in Tomato as Induced by Trichoderma virens against Fusarium wilt Caused by Fusarium oxysporum f sp. Lycopersici. Journal of Biopesticides, 3, 158-162.
- Jorda, C. and Alfaro, A. (1992) Epidemic of Cucumber Mosaic Virus plus Satellite RNA in Tomatoes in Eastern Spain. Plant Disease, 76, 363-366. http://dx.doi.org/10.1094/PD-76-0363
- Sikora, E.J., Gudauskas, R.T., Murphy, J.F., Porch, D.W., Andrianifahanana, M., Zehnder, G.W., Bauske, E.M., Kemble, J.M. and Lester, D.F. (1998) A Multivirus Epidemic of Tomatoes in Alabama. Plant Disease, 82, 117-120. http://dx.doi.org/10.1094/PDIS.1998.82.1.117
- Kearney, C.M., Gonsalves, D. and Provvidenti, R. (1990) A Severe Strain of Cucumber Mosaic Virus from China and Its Associated Satellite RNA. Plant Disease, 74, 819-823. http://dx.doi.org/10.1094/PD-74-0819
- Zitter, T.A. (1991) Diseases Caused by Viruses. In: Jones, J.B., Jones, J.P., Stall, R.E. and Zitter, T.A., Eds., Compendium of Tomato Diseases, The American Phytopathological Society, St. Paul, 31-42.
- Ilardi, V., Mazzei, M., Loreti, S., Tomassoli, L. and Barba, M. (1995) Biomolecular and Serological Methods to Identify Strains of Cucumber Mosaic Cucumovirus on Tomato. OEPP Bulletin, 25, 321-327. http://dx.doi.org/10.1111/j.1365-2338.1995.tb01473.x
- Watterson, J.C. (1993) Development and Breeding of Resistance to Pepper and Tomato Viruses. In: Kyle, M.M., Ed., Resistance to Viral Disease of Vegetables, Timber Press, Portland, 80-110.
- Anderson, J.M., Palukatis, P. and Zaitlin, M. (1992) A Defective Replicase Gene Induces Resistance to Cucumber Mosaic Virus in Transgenic Tobacco Plants. Proceedings of the National Academy of Sciences of the United States of America, 89, 8759-8763. http://dx.doi.org/10.1073/pnas.89.18.8759
- Hu, Q., Niu, Y., Zhang, K., Liu, Y. and Zhou, X. (2011) Virus-Derived Transgenes Expressing Hairpin RNA Give Immunity to Tobacco Mosaic Virus and Cucumber Mosaic Virus. Virology Journal, 8, 41-51. http://dx.doi.org/10.1186/1743-422X-8-41
- Kalantidis, K., Psaradakis, S., Tabler, M. and Tsagris, M. (2002) The Occurrence of CMV-Specific Short RNAs in transgenic Tobacco Expressing Virus-Derived Double-Stranded RNA Is Indicative of Resistance to the Virus. Molecular Plant-Microbe Interactions, 15, 826-833. http://dx.doi.org/10.1094/MPMI.2002.15.8.826
- Tanaka, A., Mita, S., Ohta, S., Kyozuka, J., Shimamoto, K. and Nakamura, K. (1990) Enhancement of Foreign Gene Expression by a Dicot Intron in Rice but Not in Tobacco Is Correlated with an Increased Level of mRNA and an Efficient Splicing of the Intron. Nucleic Acid Research, 18, 6767-6770. http://dx.doi.org/10.1093/nar/18.23.6767
- Murashige, T. and Skoog, F. (1962) A Revised Medium for Rapid Growth and Bioassay with Tobacco Tissue Cultures. Physiologia Plantarum, 15, 473-497. http://dx.doi.org/10.1111/j.1399-3054.1962.tb08052.x
- Smith, N.A. and Eamens, A. (2012) Isolation and Detection of Small RNAs from Plant Tissues. Methods in Molecular Biology, 894, 155-172. http://dx.doi.org/10.1007/978-1-61779-882-5_11
- Hamilton, A.J. and Baulcombe, D.C. (1999) A Species of Small Antisense RNA in Post Transcriptional Gene Silencing. Science, 286, 950-995. http://dx.doi.org/10.1126/science.286.5441.950
- Ntui, V.O., Thirukkumaran, G., Azadi, P, Khan, R.S., Nakamura, I. and Mii, M. (2010) Stable Integration and Expression of Wasabi Defensin Gene in “Egusi” Melon (Colocynthis citrullus L.) Confers Resistance to Fusarium wilt and Alternaria Leaf Spot. Plant Cell Reports, 29, 943-954. http://dx.doi.org/10.1007/s00299-010-0880-2
- Krishnamoorthy, V., Kumar, N., Angappan, K. and Soorianathasundaram, K. (2004) Evaluation of New Banana Hybrids against Black Leaf Streak Disease. Infomusa, 13, 25-27.
- Bonfim, K., Faria, J.C., Nogueira, E.O.P.L., Mendes, E.A. and Aragao, J.L. (2007) RNAi-Mediated Resistance to Bean Golden Mosaic Virus in Genetically Engineered Common Bean Phaseolus vulgaris. Molecular Plant-Microbe Interactions, 20, 717-726. http://dx.doi.org/10.1094/MPMI-20-6-0717
- Takahashi, H. and Ehara, Y. (1993) Severe Chlorotic Spot Symptoms in Cucumber Mosaic Virus Strain Y-Infected Tobaccos Are Induced by a Combination of the Virus Coat Protein Gene and Two Recessive Genes. Molecular PlantMicrobe Interactions, 6, 182-189. http://dx.doi.org/10.1094/MPMI-6-182
- Watterhouse, P.M., Wang, M.B. and Lough, T. (2001) Gene Silencing as an Adaptive Defense against Viruses. Nature, 411, 834-842. http://dx.doi.org/10.1038/35081168
- Ntui, V.O., Kong, K., Azadi, P., Khan, R.S., Chin, D.P., Nakamura, I. and Mii, M. (2013) Transgenic Accumulation of a Defective Cucumber Mosaic Virus (CMV) Replicase Derived Double Stranded RNA Modulates Plant Defence against CMV Strains O and Y in Potato. Transgenic Research, 22, 1191-1205. http://dx.doi.org/10.1007/s11248-013-9721-8
- Ntui, V.O., Kong, K., Khan, R.S., Ohara, M., Goto, Y., Watanabe, M., Fukami, M., Nakamura, I. and Mii, M. (2013) Transgenic Tobacco Lines Expressing Defective CMV Replicase-Derived dsRNA Are Resistant to CMV-O and CMV-Y. Molecular Biotechnology, 56, 50-63. http://dx.doi.org/10.1007/s12033-013-9681-5
- Hammond, J. and Dienelt, M.M. (1997) Encapsidation of Potyviral RNA in Various Forms of Transgene Coat Protein Is not Correlated with Resistance in Transgenic Plants. Molecular Plant-Microbe Interactions, 10, 1023-1027. http://dx.doi.org/10.1094/MPMI.1997.10.8.1023
- Lecoq, H., Ravelonandro, M., Wipf-Scheibel, C., Monsion, M., Raccah, B. and Dunez, J. (1993) Aphid Transmission of a Non-Aphid-Transmissible Strain of Zucchini Yellow Mosaic Virus Potyvirus from Transgenic Plants Expressing the Capsid Protein of Plum Pox Potyvirus. Molecular Plant-Microbe Interactions, 6, 403-406. http://dx.doi.org/10.1094/MPMI-6-403
- Patil, B.L., Ogwok, E., Wagaba, H., Mohammed, I., Ydav, S.J., Bagewadi, B., Taylor, N.J., Kreuze, J.F., Maruthi, M.N., Alicai, T. and Fauquet, C.M. (2011) RNAi-Mediated Resistance to Diverse Isolates Belonging to Two Virus Species Involved in Cassava Brown Streak Disease. Molecular Plant Pathology, 12, 31-41. http://dx.doi.org/10.1111/j.1364-3703.2010.00650.x
- Kasschau, K.D. and Carrington, J.C. (1998) A Counter-Defensive Strategy of Plant Viruses: Suppression of Posttranscriptional Gene Silencing. Cell, 95, 461-470. http://dx.doi.org/10.1016/S0092-8674(00)81614-1
- Voinnet, O., Pinto, Y.M. and Baulcombe, D.C. (1999) Suppression of Gene Silencing: A General Strategy Used by Diverse and RNA Viruses of Plants. Proceedings of the National Academy of Sciences of the United States of America, 96, 14147-14152. http://dx.doi.org/10.1073/pnas.96.24.14147
- Beclin, C., Berthome, R., Palauqui, J.C., Tepfer, M. and Vaucheret, H. (1998) Infection of Tobacco or Arabidopsis Plants by CMV Counteracts Systemic Post Transcriptional Silencing of Nonviral (trans) Genes. Virology, 252, 313-317. http://dx.doi.org/10.1006/viro.1998.9457
- Brigneti, G., Voinnet, O., Li, W.X., Ji, L.H., Ding, S.W. and Baulcombe, D.C. (1998) Viral Pathenogenicty Determinants Are Suppressor of Transgene Silencing in Nicotiana benthamiana. EMBO Journal, 17, 6739-6746. http://dx.doi.org/10.1093/emboj/17.22.6739
- Guo, H.S. and Ding, S.W. (2002) A Viral Protein Inhibits the Long Range Signalling Activity of Gene Silencing Signal. EMBO Journal, 21, 398-407. http://dx.doi.org/10.1093/emboj/21.3.398
- Szittya, G., Silhavy, D., Molnar, A., Havelda, Z., Lovas, A., Lakatos, L., Banfalvi, Z. and Burgyan, J. (2003) Low Temperature Inhibits RNA Silencing-Mediated Defense by the Control of siRNA Generation. EMBO Journal, 22, 633- 640. http://dx.doi.org/10.1093/emboj/cdg74
- Matzke, M., Matzke, A.J.M. and Scheid, O.M. (1994) Inactivation of Repeated Genes: DNA-DNA Interaction? In: Paszkowaski, J., Ed., Homologous Recombination and Gene Silencing in Plants, Kluwer Academic Publishers, Dordrecht, 271-307.
- Finnegan, J. and McElroy, D. (1994) Transgene Inactivation: Plants Fight Back. Nature Biotechnology, 12, 883-888. http://dx.doi.org/10.1038/nbt0994-883
Abbreviations
CMV: Cucumber mosaic virus; CMV-O: Cucumber mosaic virus strain O; CMV-Y: Cucumber mosaic virus strain Y; DIBA: Dot-immunobinding assay; dsRNA: Double-stranded RNA; PTGS: Post-transcriptional gene silencing; RISC: RNA-induced silencing complex; RNAi: RNA interference; siRNA: Small interfering RNA; HS: Highly susceptible; HR: Highly resistant; NA: Not applicable; WTM: Mock inoculated control plant.


