American Journal of Plant Sciences
Vol. 4 No. 2A (2013) , Article ID: 28459 , 9 pages DOI:10.4236/ajps.2013.42A053
Characterization of a Highly Potent Insecticidal Lectin from Colocasia esculenta Tuber and Cloning of Its Coding Sequence
![]()
1Division of Plant Biology, Centenary Campus, Bose Institute, Kolkata, India; 2The Protein Analysis Facility, Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland.
Email: ayandas80@gmail.com, amitroy_81@yahoo.co.in, #sampa@bic.boseinst.ernet.in
Received December 20th, 2012; revised January 28th, 2013; accepted February 6th, 2013
Keywords: Colocasia esculenta Tuber Agglutinin (CEA); Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS); Genome Walk; Hemipteran Insect; Insect Bioassay
ABSTRACT
Hemipteran insects are the most devastating pest for different crops of high economic value. Colocasia esculenta tuber agglutinin (CEA), a mannose binding monocot lectin from araceae family was previously reported by the present group to be effective against some members of this class of pests. In the present study, efficacy of this potent lectin has been extended to cotton aphid (Aphis gossypii) which is becoming a highly damaging pest of cotton in recent days. Because, like other aphids, A. gossypii not only extracts the phloem fluid but also transmit disease causing viruses and add to the high degree of yield loss. Efficacy of the lectin on cotton aphid as well as other hemipteran insects prompted us further to clone the protein coding gene. Very little sequence information of this gene was available in the database. Hence, attempt had been made to study the protein through liquid chromatography-tandem mass spectrometry (LC-MS/MS) to have the detailed peptide information. On the basis of the peptide homology information obtained from LC-MS/MS the complete coding sequence of CEA was determined. The coding sequence corresponding to CEA was cloned further using primers designed on the basis of above information and genome walk technology for its potential utilisation in insect management programme.
1. Introduction
Hemipteran insects are one of the most damaging pests affecting a wide array of economically important plants. Amongst all hemipterans, aphids alone account for 13% of the total crop loss in the world [1]. Use of chemical insecticides is not at all acceptable option for controlling them not only for the sake of environmental safety but they often provoke the emergence of resistant biotypes of various insect pests. Bt toxin, the most commonly used bio-control agent has found to be ineffective against this group of pests [2,3].
Plant lectins are a class of non-immugenic carbohydrate binding proteins often found to be effective against this group of insect pests. Earlier reports indicated about the efficacy of different plant lectins, namely, Galanthus nivalis agglutinin (GNA), Allium sativum leaf agglutinin (ASAL), Arum maculatum tuber lectin (ATL), Amorphophallus paeonifolius tuber agglutinin (AMTL) against various insect pests [4-9]. The mode of action of one such mannose-binding lectin Amorphophallus paeonifolius tuber agglutinin (AMTL) on sucking type of hemipteran pest described in detail by Mondal et al. [9] with additional information on interaction of lectin with insect midgut receptor and predicted the cause of toxicity either through nutrient leaching or causing anti-absorption detrimental effect due to lectin binding.
A single insecticidal agent, in due course of time might become ineffective against its target insects due to its behavioural reorientation [9,10]. Keeping this in mind the present study was conducted to investigate extensively the efficacy of another mannose binding lectin Colocasia esculenta tuber agglutinin (CEA) which was previously reported by the same group [11,12]. After purifying the protein at homogeneity detailed bioassay of CEA was performed on three different hemipteran insects, namely, Red Cotton bug (Dysdercus cingulatus), Cowpea aphid (Aphis craccivora) and Cotton aphid (Aphis gossypii)—the last one has emerged as a devastating pest for cotton due to its ability to damage the crop as a sucking pest and as a vector for a number of disease causing viruses. Significant performance of this lectin on all the target insects encouraged its further utilisation in the plant biotechnological programme. The protein was studied further by mass spectrometry and on the basis of the peptide sequence information obtained from LC MS/MS analysis, the complete coding sequence of the protein was determined. This is now being studied for further utilisation.
2. Materials and Methods
2.1. Plant Materials and Insects Used
Fresh tubers of Colocasia esculenta and nymphs of Aphis gossypii, Dysdercus cingulatus & Aphis craccivora were collected from institutional farm as per requirement.
2.2. Extraction and Isolation of the Lectin from Colocasia esculenta Tuber
Lectin was isolated from tuber tissue extracts of Colocasia esculenta by affinity chromatography using affinity matrix [α-D-Mannose, immobilized on cross linked 4% beaded agarose, Sigma-Aldrich], pre-equilibriated with 20 mM Tris-HCl (pH 7.2) following the protocol of Van Damme et al. [13] with some modification as described by Roy et al. [11]. The matrix bound lectin was desorbed with 20 mM unbuffered 1,3-diaminopropane [DAP] and lyophilised.
2.3. Ion Exchange Chromatography of CEA
The lyophilised semi-purified sample from affinity purification was dialyzed against phosphate-buffered saline [PBS], pH 7.2 and loaded onto a DEAE-Sephadex ion exchange column equilibrated with PBS, pH 7.2. A gradient of 0 - 500 mM NaCl solution was passed through and the eluted fractions were collected, pooled & lyophilised. Bradford assay was carried out to quantify the purified protein [14].
2.4. SDS-PAGE and Western Analysis
Lectin preparations after chromatographic separation were resolved in 15% SDS-PAGE according to Laemmli [15]. Western blot analysis was carried out by using an antiCEA polyclonal primary antibody developed in our laboratory following the protocol of Harlow et al. [16] and anti-rabbit IgG-horse radish peroxidase conjugates as the secondary antibody obtained from Sigma-Aldrich, USA.
2.5. Characterization of CEA through Agglutination Assays
Colocasia lectin was allowed to agglutinate rabbit erythrocytes as described by Mondal et al. [9]. Rabbit erythrocytes were collected, washed extensively in a 0.9% saline solution, and finally made up to 20% (v/v) with 0.9% saline. Twenty microliters of erythrocyte suspension was dispensed in each well of microtiter plate. Eighty microliters of serially diluted lectin solutions (starting from 4 to 1 μg per well) were added to each well. Control well was supplemented with same amount of 0.9% saline water. Agglutination reaction was monitored visually, after incubation for 1 h at 37˚C.
2.6. Insect Bioassay in Artificial Diet
Bioassay has been set up on an artificial diet formulated with some modifications [6] from the original description by Dadd and Mittler [17]. Second instar nymphs of Aphis gossypii [Cowpea aphid], Dysdercus cingulatus [Red cotton bug] and Aphis craccivora [Cowpea aphid] were incubated for each set in liquid diet of 200 μl supplemented with different doses of CEA (0, 5, 10, 15, 20, 25 μg/ml each). The experimental set up was designed according to Mondal et al. [9]. Entire experiment was done in triplicate. Data on survival of the insects were collected at every 24 h. The LC50 value of each toxin corresponding to each insect had been determined by statistical probit analysis [18]. Statistical significance level used was 0.05 [α ≤ 0.05]. The statistical evaluation was done using the χ2 method as specified by the Probit analysis software [11,18].
2.7. Mass Spectrometric Analysis of Purified CEA
CEA protein band form SDS PAGE gel was excised and was reduced with TCEP [tris(2-carboxyethyl)phosphine], alkylated by using iodoacetamide and finally digested with trypsin. Generated peptides were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) on a Orbitrap Velos. MASCOT searching UNIPROT_ 2011_08 was used for protein identification with a minimum of 5 peptide match and over 95% probability cut off. The results were visualized with scaffold.
For getting better sequence coverage the identified proteins were selected and searched again with an error tolerant search allowing for amino acid mutations and modifications.
2.8. Isolation of Genomic DNA from C. esculenta Tubers
Genomic DNA was isolated from young tubers of C. esculenta using Qiagen DNeasy Plant Mini kit (GmbH, Hilden, Germany) following manufacturer’s instructions.
2.9. Isolation of RNA from C. esculenta Tubers
RNA was isolated from young tubers of C. esculenta following the protocol described by Kumar et al. [19]. Diethylpyrocarbonate (DEPC) treated water was used to prepare all the chemicals and reagents. Two grams of fresh tuber tissue of C. esculenta was ground to a fine powder in liquid nitrogen using a mortar pestle and transferred to a Falcon tube. RNA was extracted by the addition of 1.0 ml 5 M NaCl, 0.5 ml 10% (w/v) SDS, 1.65 ml 1.95% (w/v) Na2SO3, 1.75 ml borate-Tris buffer (0.2 M, pH 8.0 containing 10 mM EDTA) and 0.1 ml β-mercaptoethanol in a sequential manner. Following vortexing the extract was incubated at 65˚C for 5 min. Then the extract was centrifuged (1800 g, 5 min, room temperature), equal volume of Tris-saturated phenol (pH 7.9) was mixed with the supernatant and centrifuged further for phase separation. The upper phase was extracted with equal volume of chloroform-isoamyl alcohol (24:1) and centrifuged again for phase separation. To 500 µl of the upper phase 450 µl of isopropyl alcohol was added in a microfuge tube and incubated at 4˚C for 1 h. RNA was pelleted at 20,000 g for 15 min at 4˚C. The RNA pellet was washed with 70% ethanol; residual ethanol was evaporated in laminar air flow and the pellet was solubilised in DEPC treated water.
2.10. Preparation of cDNA from Total RNA Isolated from C. esculenta Tubers
cDNA was prepared from the total RNA isolated from C. esculenta tubers using SuperScriptTM III Reverse Transcriptase (Invitrogen, CA, USA) following manufacturer’s instructions.
2.11. Genome Walking to Determine the Complete Coding Sequence of CEA:
Genome walking was performed using BD Genome WalkerTM Universal Kit (BD Biosciences Clontech, California, USA) following manufacturer’s instruction. Primers were designed from the corresponding peptide data obtained from LC-MS/MS analysis from the protein which showed best coverage (Table 1; primers PF and PR were designed from the peptide information generated from the LC-MS/MS analysis of the protein, primers
Table 1. Sequence of primers used.

5’ GSP1 and 2 and 3’ GSP1 and 2 were used for 5’ and 3’ genome walk respectively, CDSF and CDSR were used to finally clone the complete coding sequence). PCR product after purification from agarose gel using Qiaquick Gel Extraction kit (Qiagen, Hilden, Germany) was cloned into pGEMT Easy Vector (Promega, Wisconsin, USA) and nucleotide sequencing was performed using Applied Biosystems 3130 xl Genetic Analyzer at the sequencing facility of Bose Institute, Kolkata, India.
3. Results
3.1. Purification, SDS-PAGE & Western Blot Analysis
Colocasia tuber lectin purified through affinity chromatography and subsequent ion-exchange chromatography was subjected to 15% SDS-PAGE. Two bands of ~25 kDa and ~12.5 kda resolved after affinity purification (Figure 1: Panel 1, lane 1) and a single band of purified CEA (~12.5 kDa) is visible in ion-exchange chromatography purified fraction (Figure 1: Panel 1, lanes 2 & 3). Western blot analysis authenticated the purified lectin fraction by showing its cross-reactivity to anti-CEA antibody (Figure 1: Panel 2, lane 4).
3.2. Agglutination Assay
The agglutination assay was performed with different concentrations of purified CEA (4 µg to 1 µg per well) on rabbit erythrocytes that demonstrated agglutination activity at all the given concentrations after the incubation time of 1 h confirming the functional activity of the purified protein (Figure 2).
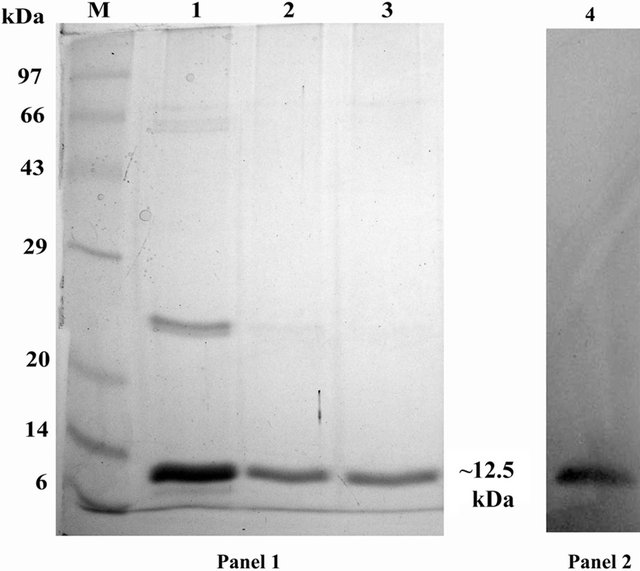
Figure 1. Purification of native CEA. SDS PAGE (15%) profile of purified fractions of native CEA. Panel 1: Lane M: Standard protein molecular weight marker; Lane 1: Affinity chromatography purified CEA fraction; Lanes 2 & 3: Ion exchange chromatography purified CEA fractions; Panel 2: CEA showing ~12.5 kDa band after western blot analysis with anti-CEA antibody.
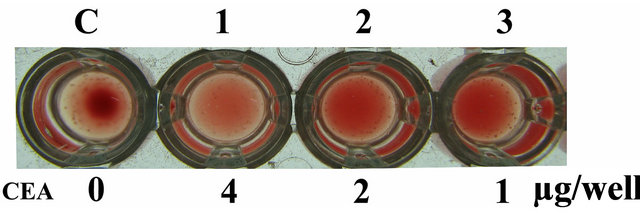
Figure 2. Agglutination assay. Wells represent agglutination pattern of rabbit erythrocytes with various doses of native CEA started with 4 µg/well to 1 µg/well.
3.3. Insect Bioassay of CEA on Hemipteran Insects in Artificial Diet
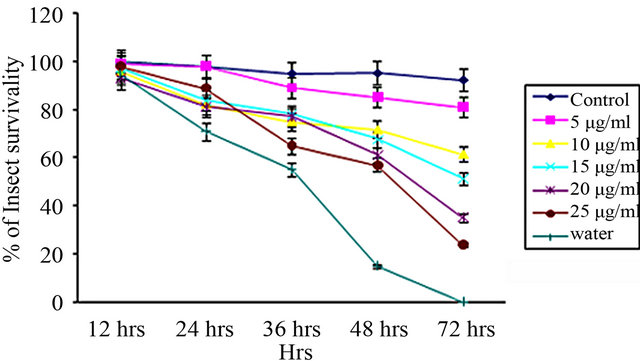
From insect bioassay experiments, it was evident that the effect of CEA was more with increasing dose over a period of 72 hours [Figures 3(a)-(c)].
The LC50 values of CEA calculated by Probit analysis against Aphis gossypii, Dysdercus cingulatus and Aphis craccivora were 9.98 ± 0.239 μg∙ml–1, 16.95 ± 0.279 μg∙ml–1 and 15.21 ± 0.274 μg∙ml–1, respectively (Table 2).
3.4. Mass Spectrometric Analysis
LC-MS/MS analysis of the lectin under study shows best 3 protein hits (Figure 4). Best coverage (81%) was observed with Colocasia esculenta lectin (A5HMM7_ COLES, GenBank Accession: EF541132.1) followed by 73% coverage in case of (B5LYJ9_9ARAE, GenBank Accession: EU924066.1, [20]) which is a mannose-binding lectin from Remusatia vivipara. The third best hit with 50% coverage was found with Q39487_COLES (GenBank Accession: D16173.1 [21]) corresponding to a partial cds of a 12 kD storage protein from tubers of C. esculenta.
3.5. Determination of the Complete Coding Sequence of CEA
Complete coding sequence was derived from the corresponding peptide mass fingerprinting data of purified CEA. Subsequently the full length CEA gene was cloned following genome walking method and the 795 bp sequence was submitted to GenBank under accession No. JX435122 (Figure 5).
Comparison of the sequence information obtained from the genomic DNA and cDNA showed that they are identical and the genomic sequence is devoid of any intron (data not shown).
The alignment of 5’ upstream region with tar1 gene from C. esculenta gave a possible assumption of the putative cis-acting elements like TATA box, major and minor transcription start sites reported in case of tar1 gene by Bezerra et al. [22] (Figure 6).
From the 3’ downstream sequence putative polyadenylation motifs [23] were also found (data not shown) similar to those of tar1 gene from C. esculenta [22].
CEA vs. Aphis gossypii
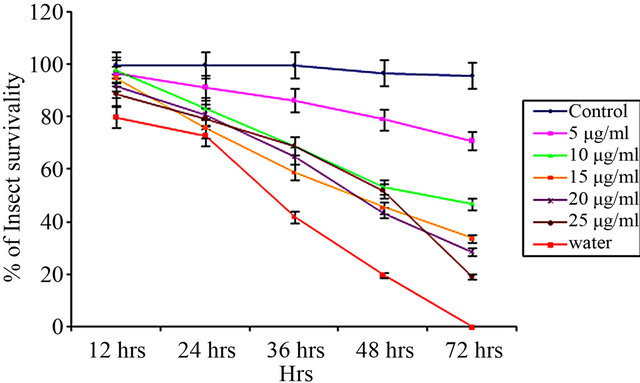
(a)
CEA vs. Dysdercus cingulatus
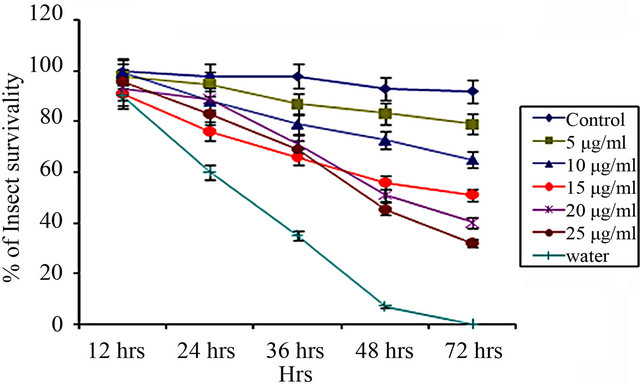
(b)
CEA vs. Aphis craccivora
 (c)
(c)
Figure 3. Insect bioassay in lectin supplemented artificial diet on second instar nymphs of various insects. Insect survivability graph at different concentrations of CEA (0, 5, 10, 15, 20, 25 μg/ml) recorded over 72 hours in (a) Aphis gossypii (Cotton aphid); (b) Dysdercus cingulatus (Red cotton Bug); and (c) Aphis craccivora (Cowpea aphid).
4. Discussion
Mannose binding plant lectins have been proven to be potent control agents against hemipteran insect pests in last couple of years. Galanthus nivalis agglutinin (GNA) is the most studied among them [24-27]. After GNA several other lectins viz., Allium sativum leaf agglutinin
Table 2. Insect bioassay of native CEA determined against Aphis gossypii, Dysdercus cingulatus and Aphis craccivora in artificial diet.
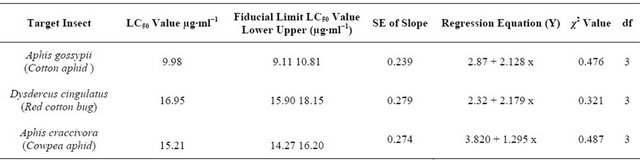
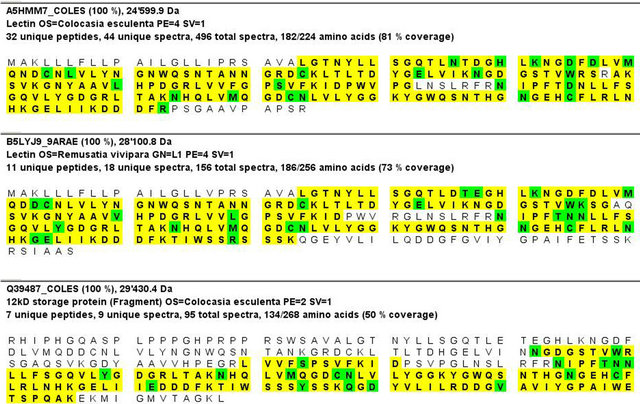
Figure 4. Mass spectrometric analysis of CEA. Best three protein hits obtained by LC MS/MS analysis after tryptic digestion of CEA protein band eluted from the SDS PAGE gel. Sequences shown in yellow were identified by MS/MS, marked in green are modification/mutation sites.
(ASAL), Amorphophallus paeonifolius Tuber Agglutinin (AMTL), Arum maculatum Tuber Lectin (ATL), PTA (Pinellia ternata agglutinin) from different sources have been enlisted in the group. Effectiveness of many of them have been established in transgenic situation also [2,27- 34]. It has also been established by now that lectins affect the insects by interacting with their receptors present in the insect midgut [9,12].
Previous studies using Colocasia esculenta tuber agglutinin (CEA) [11,12] has already established itself as a potent insecticidal agent. In the present study insect bioassay was conducted with CEA including another devastating hemipteran insect pest of cotton-Aphis gossypii. In the recent past the outbreak of A. gossypii has become a striking phenomenon throughout most of the cotton growing areas. The study was elaborated further to know the complete coding sequence of CEA.
The effectiveness of purified CEA on three important pests-Cotton aphid (Aphis gossypii), red cotton bug (Dysdercus cingulatus) and cowpea aphid (Aphis craccivora) was investigated through artificial diet based insect bioassay. Interestingly, among the three insects studied CEA was found to be the most effective against cotton aphid showing lowest LC50 value (Table 2). Effect of CEA on Aphis craccivora found in this study was comparable with that observed by Majumder et al. [12] and Chakraborti et al. [34]. Similarly, effect of CEA on A. craccivora was also comparable to that of Arum maculatum tuber lectin (ATL) [8]. CEA was found to be more effective on red cotton bug when compared with the effect of Allium sativum leaf agglutinin (ASAL) on red cotton bug observed by Bandyopadhyay et al. [6]. LC50 value of CEA is comparable with another mannose specific lectin Amorphophallus paeonifolius tuber agglutinin (AMTL) on D. cingulatus and A. gossypii [9].
Effectiveness of CEA on such an array of insects encouraged the present group to further clone its complete coding sequence for developing transgenic plants, resistant to target pests. Database search revealed that very few reported sequence from Colocasia sp. are available. Confusion further aroused while scrutinising the reported gene sequences corresponding to our protein of interest. To get rid of the confusion, CEA protein was analyzed in detail through LC-MS/MS and obtained peptide information was used to determine the complete coding sequence. The complete coding sequence was cloned for its further utilisation using primers designed on the basis of derived peptide information and applying PCR based genome walk method.

Figure 5. The complete coding sequence (cds) of CEA. The start codon is in green colour and stop codon is in red colour.
The LC-MS/MS data obtained using the ~12.5 kD SDS-PAGE band of purified CEA showed homology with two proteins from Colocasia esculenta-one is a lectin (A5HMM7_COLES) and another, a storage protein from tuber (Q39487_COLES). The second best hit in LC-MS/MS was also a lectin from Remusatia vivipara which belongs to the same family of araceae.
The presently derived sequence has an open reading freame of 795 bp (Figure 5). Interestingly, the presently dealt CEA genomic sequence does not contain any intron as it was in the case of tar1 gene from Colocasia esculenta [22] and another lectin gene from Remusatia vivipara [20].
The protein sequence deduced (using ExPaSy SIB Bioinformatics Resource Portal) from the presently reported gene sequence has 264 amino acids (Figure 7). The deduced amino acid sequence showed significant similarity with LC-MS/MS data (Figure 4).
The deduced amino acid sequence also showed 96% identity with the signal peptide region of tar1 gene (Figure 8) responsible for its vaculor transport [22]. This region also fulfils the criteria of signal peptide of storage protein [35].
Although BLASTp analysis (NCBI BLAST) with database showed that CEA has only 41% identity (query coverage 85%) with the most well studied mannose binding lectin Galanthus nivalis agglutinin (GNA) but it showed a significant identity (80% - 94%) with lectins reported from Colocasia esculenta, Remusatia vivipara, Alocasia macrorrhizos, Pinellia pedatisecta, Pinellia ternata belonging to family araceae (data not shown).
Multiple alignment (clastalW) of the deduced amino acid sequence with those of the three best hit proteins in the LC-MS/MS (Figure 9) showed that all of these proteins share common conserved motif (QXDXNXVXY) responsible for carbohydrate binding.
The present study establishes CEA as a potent insecticidal agent and provides itself as a new candidate for a better hemipteran pest management programme. The present study also reports a novel strategy to clone an important gene from the desired peptide information of the corresponding protein from LC MS/MS analysis.
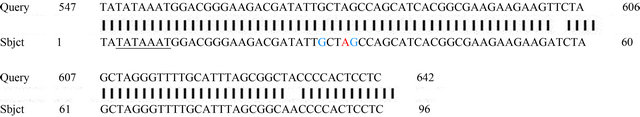
Figure 6. Alignment of 5’ upstream region of CEA with tar1 gene. Query: 5’ upstream region of CEA, Subject: 5’ upstream region of C. esculenta tar1 gene. TATA box in case of C. esculenta tar1 gene is underlined. Major transcription start site in case of C. esculenta tar1 gene is in red and minor transcription start sites are shown in green colour.

Figure 7. Deduced amino acid sequence of CEA.
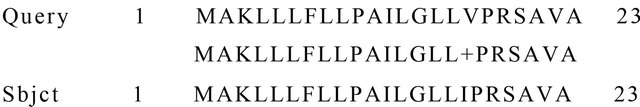
Figure 8. BLASTp alignment of the signal peptide region of tar1 gene with that of the deduced CEA protein sequence. Query: Portion of deduced amino acid sequence of CEA, Subject: Signal peptide portion of tar1 protein.
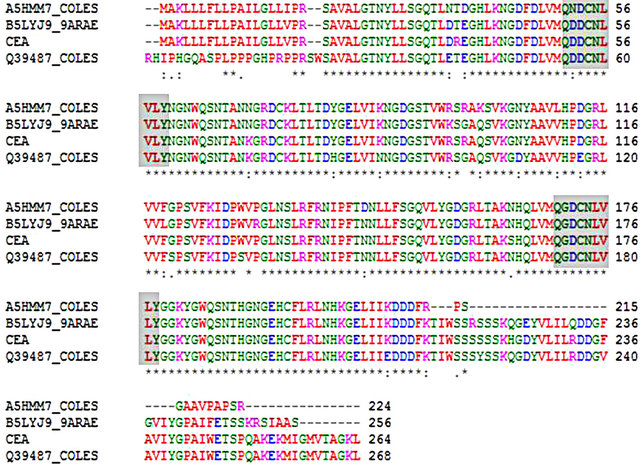
Figure 9. Multiple alignment (ClustalW) of the deduced amino acid sequence of CEA with deduced amino acid sequence of the three best hit proteins found in LC MS/MS. Conserved carbohydrate binding motif (QXDXNXVXY) of these proteins are shaded.
5. Acknowledgements
Authors are thankful to Bose Institute for providing necessary infrastructural facility. AD and AR are thankful to Council of Scientific and Industrial Research for providing fellowship. AD also acknowledges Dr. D. Basu, Bose Institute for his technical suggestions. Authors also acknowledge technical support of Mr. Arup Kumar Dey, Mr. Swarnava Das and Mr. Sudipta Basu.
REFERENCES
- N. P. Chougule and B. C. Bonning, “Toxins for Transgenic Resistance to Hemipteran Pests,” Toxins, Vol. 4, No. 6, 2012, pp. 405-429. doi:10.3390/toxins4060405
- K. V. Rao, K. S. Rathore, T. K. Hodges, X. Fu, E. Stoger, S. Sudhakar, P. Williams, P. Christou, M. Bharathi, D. P. Bown, K. S. Powell, J. Spence, A. Gatehouse and J. A. Gatehouse, “Expression of Snowdrop Lectin (GNA) in Transgenic Plants Confers Resistance to Rice Brown Plant Hopper,” Plant Journal, Vol. 15, No. 4, 1998, pp. 469-477. doi:10.1046/j.1365-313X.1998.00226.x
- C. Deraison, I. Darboux, L. Duportets, T. Gorojankina, Y. Rahbe and L. Jouanin, “Cloning and Characterization of a Gut-Specific Cathepsin L from the Aphid Aphis gossypii,” Insect Molecular Biology, Vol. 13, No. 2, 2004, pp. 165-177. doi:10.1111/j.0962-1075.2004.00474.x
- W. J. Peumans and E. J. M. Van Damme, “Lectins as Plant Defense Proteins,” Plant Physiology, Vol. 109, No. 2, 1995, pp. 347-352. doi:10.1104/pp.109.2.347
- N. Sauvion, Y. Rahbe, W. J. Peumans, E. J. M. V. Damme, J. A. Gatehouse and A. M. R. Gatehouse, “Effects of GNA and Other Mannose Binding Lectins on Development and Fecundity of the Peach-Potato Aphid Myzus persicae,” Entomologia Experimentalis Applicata, Vol. 79, No. 3, 1996, pp. 285-293. doi:10.1111/j.1570-7458.1996.tb00836.x
- S. Bandyopadhyay, A. Roy and S. Das, “Binding of Garlic (Allium sativum) Leaf Lectin to the Gut Receptors of Hemipteran Pests Is Correlated to Its Insecticidal Activity,” Plant Science, Vol. 161, No. 5, 2001, pp. 1025-1033. doi:10.1016/S0168-9452(01)00507-6
- S. Banerjee, D. Hess, P. Majumder, D. Roy and S. Das, “The Interactions of Allium sativum Leaf Agglutinin with a Chaperonin Group of Unique Receptor Protein Isolated from a Bacterial Endosymbiont of the Mustard Aphid,” Journal of Biological Chemistry, Vol. 279, No. 22, 2004, pp. 23782-23789. doi:10.1074/jbc.M401405200
- P. Majumder, H. A. Mondal and S. Das, “Insecticidal Activity of Arum Maculatum Tuber Lectin and Its Binding to the Glycosylated Insect Gut Receptors,” Journal of Agriculture and Food Chemistry, Vol. 53, No. 17, 2005, pp. 6725-6729. doi:10.1021/jf051155z
- H. A. Mondal, A. Roy, S. Gupta and S. Das, “ Exploring the Insecticidal Potentiality of Amorphophallus paeonifolius Tuber Agglutinin in Hemipteran Pest Management,” American Journal of Plant Sciences, Vol. 3, No. 6, 2012, pp. 780-790. doi:10.4236/ajps.2012.36094
- M. S. Chen, “Inducible Direct Plant Defence against Insect Herbivores: A Review,” Insect Science, Vol. 15, No. 2, 2008, pp. 101-114. doi:10.1111/j.1744-7917.2008.00190.x
- A. Roy, S. Banerjee, P. Majumder and S. Das, “Efficiency of Mannose-Binding Plant Lectins in Controlling a Hemipteran Insect, the Red Cotton Bug,” Journal of Agricultural Food Chemistry, Vol. 50, No. 23, 2002, pp. 6775-6779. doi:10.1021/jf025660x
- P. Majumder S. Banerjee and S. Das, “Identification of Receptors Responsible for Binding of the Mannose Specific Lectin to the Gut Epithelial Membrane of the Target Insects,” Glycoconjugate Journal, Vol. 20, No. 9, 2004, pp. 525-530.
- E. J. M. V. Damme, K. Goossens, K. Smeets, F. V. Leuven, P. Verheart and W. J. Peumans, “The Major Tuber Storage Protein of Araceae Species Is a Lectin,” Plant Physiology, Vol. 107, No. 4, 1995, pp. 1147-1158. doi:10.1104/pp.107.4.1147
- M. M. Bradford, “Rapid and Sensitive Method for Quantitation of Microgram Quantities of Protein Utilizing Principle of Protein Dye Binding,” Anual Review of Biochemistry, Vol. 72, No. 1-2, 1976, pp. 248-254. doi:10.1016/0003-2697(76)90527-3
- U. K. Laemmli, “Cleavage of Structural Proteins during Assembly of Head of Bacteriophage-T4,” Nature, Vol. 227, No. 5259, 1970, pp. 680-688. doi:10.1038/227680a0
- E. Harlow, “Antibodies: A Laboratory Manual,” Cold Spring Harbour Laboratory, New York, 1988, pp. 60-67.
- R. Dadd and T. Mittler, “Permanent Culture of an Aphid on a Totally Synthetic Diet,” Cellular and Molecular Life Sciences, Vol. 22, No. 12, 1976, pp. 832-833. doi:10.1007/BF01897447
- H. Chi, “Computer Program for the Probit Analysis,” National Chung Hsing University, Taichung, 1997.
- G. N. M. Kumar, S. Iyer and N. R. Knowles, “Extraction of RNA from Fresh, Frozen, and Lyophilized Tuber and Root Tissues,” Journal of Agriculture and Food Chemistry, Vol. 55, No. 5, 2007, pp. 1674-1678. doi:10.1021/jf062941m
- G. G. Bhat, K. N. Shetty, N. N. Nagre, V. V. Neekhra, S. Lingaraju, R. S. Bhat, S. R. Inamdar, K. Suguna and B. M. Swamy, “Purification, Characterization and Molecular Cloning of a Monocot Mannose-Binding Lectin from Remusatia vivipara with Nematicidal Activity,” Glycoconjugate Journal, Vol. 27, No. 3, 2010, pp. 309-320. doi:10.1007/s10719-010-9279-0
- M. Hirai, K. Nakamura, T. Imai and T. Sato, “cDNAs Encoding for Storage Proteins in the Tubers of Taro (Colocasia esculenta Schott),” Japanese Journal of Genetics, Vol. 68, No. 3, 1993, pp. 229-236. doi:10.1266/jjg.68.229
- I. C. Bezerra, L. A. Castro, G. Neshich, E. R. de Almeida, M. F. de Sa, L. V. Mello and D. C. Monte-Neshich, “A Corm-Specific Gene Encodes Tarin, a Major Globulin of Taro (Colocasia esculenta L. Schott),” Plant Molecular Biology, Vol. 28, No. 1, 1995, pp. 137-144. doi:10.1007/BF00042045
- C. P. Joshi, “Putative Polyadenylation Signals in Nuclear Genes of Higher Plants: A Compilation and Analysis,” Nucleic Acids Research, Vol. 15, No. 23, 1987, pp. 9627- 9640. doi:10.1093/nar/15.23.9627
- A. Gatehouse, R. Down, K. Powell, N. Sauvion, Y. Bahbe, C. A. Newell, A. Merryweather, W. D. O. Hamilton and J. A. Gatehouse, “Transgenic Potato Plants with Enhanced Resistance to Peach Potato Aphid Myzus persicae,” Entomologia Experimentalis et Applicata, Vol. 79, No. 3, 1996, pp. 295-307. doi:10.1111/j.1570-7458.1996.tb00837.x
- K. S. Powell, J. Spence, M. Bharathi, J. A. Gatehouse and A. M. R. Gatehouse, “Immunohistochemical and Developmental Studies to Elucidate the Mechanism of Action of the Snowdrop Lectin on the Rice Brown Planthopper, Nilaparvata lugens (Stal),” Journal of Insect Physiology, Vol. 44, No. 7-8, 1998, pp. 529-539. doi:10.1016/S0022-1910(98)00054-7
- N. T. Loc, P. Tinjuangjun, A. M. R. Gatehouse, P. Christou and J. A. Gatehouse, “Linear Transgene Constructs Lacking Vector Backbone Sequences Generate Transgenic Rice Plants Which Accumulate Higher Levels of Proteins Conferring Insect Resistance,” Molecular Breeding, Vol. 9, No. 4, 2002, pp. 231-244. doi:10.1023/A:1020333210563
- S. Ramesh, D. Nagadhara, V. D. Reddy and K. V. Rao, “Production of Transgenic Indica Rice Resistant to Yellow Stem Borer and Sap Sucking Insects, Using Super-Binary Vectors of Agrobacterium tumefaciens,” Plant Science, Vol. 166, No. 4, 2004, pp. 1077-1085. doi:10.1016/j.plantsci.2003.12.028
- V. A. Hilder, K. S. Powell, A. M. R. Gatehouse, J. Gatehouse, L. N. Gatehouse, Y. Shi, W. Hamilton, A. Merryweather, C. A. Newell and J. C. Timans, “Expression of Snowdrop Lectin in Transgenic Tobacco Plants Results in Added Protection against Aphids,” Transgenic Research, Vol. 4, No. 1, 1995, pp. 18-25. doi:10.1007/BF01976497
- I. Dutta, P. Saha, P. Majumder, A. Sarkar, D. Chakraborti, S. Banerjee and S. Das, “The Efficacy of a Novel Insecticidal Protein, Allium sativum Leaf Lectin (ASAL) against Homopteran Insect Monitored in Transgenic Tobacco,” Plant Biotechnology Journal, Vol. 3, No. 6, 2005, pp. 601- 611. doi:10.1111/j.1467-7652.2005.00151.x
- I. Dutta, P. Majumder, P. Saha, K. Ray and S. Das, “Constitutive and Phloem Specific Expression of Allium sativum Leaf Agglutinin (ASAL) to Engineer Aphid (Lipaphis erysimi) Resistance in Transgenic Indian Mustard (Brassica juncea),” Plant Science, Vol. 169, No. 6, 2005, pp. 996-1007doi:10.1016/j.plantsci.2005.05.016
- P. Saha, D. Chakraborti, A. Sarkar, I. Dutta, D. Basu and S. Das, “Characterization of Vascular Specific RSs1 and rolC Promoters for Their Utilization in Engineering Plants to Develop Resistance against Hemipteran Insect Pests,” Planta, Vol. 226, No. 2, 2007, pp. 429-442. doi:10.1007/s00425-007-0493-3
- A. Sadeghi, S. Broeders, H. D. Greve, J. P. Hernalsteens, W. J. Peumans, E. J. M. Van Damme and G. Smagghe, “Expression of Garlic Leaf Lectin under the Control of the Phloem-Specific Promoter Asus1 from Arabidopsis thaliana Protects Tobacco Plants against the Tobacco Aphid (Myzus nicotianae),” Pest Management Science, Vol. 63, No. 12, 2007, pp. 1215-1223. doi:10.1002/ps.1455
- A. Sadeghi, G. Smagghe, S. Broeders, J. P. Hernalsteens, H. De Greve, W. J. Peumans and E. J. M. Van Damme, “Ectopically Expressed Leaf and Bulb Lectins from Garlic (Allium sativum L.) Protect Transgenic Tobacco Plants against Cotton Leafworm (Spodoptera littoralis),” Transgenic Research, Vol. 17, No. 1, 2008, pp. 9-18. doi:10.1007/s11248-007-9069-z
- D. Chakraborti, A. Sarkar, H. A. Mondal and S. Das, “Tissue Specific Expression of Potent Insecticidal, Allium sativum Leaf Agglutinin (ASAL) in Important Pulse Crop, Chickpea (Cicer arietinum L.) to Resist the Phloem Feeding Aphis craccivora,” Transgenic Research, Vol. 18, No. 4, 2009, pp. 529-544. doi:10.1007/s11248-009-9242-7
- J. Messing, D. Geraghty, G. Heidecker, N.-T. Hu, J. Kridl and I. Ruenstein, “Plant Gene Structure,” In: C. P. Kosuge, A. Meredith and K. Hollaender, Eds., Genetic Engineering of Plants, Plenum Press, New York, 1983, pp. 211-227. doi:10.1007/978-1-4684-4544-2_16
NOTES
*Equal Contribution.
#Corresponding author.

