Advances in Microbiology
Vol.4 No.2(2014), Article ID:42304,10 pages DOI:10.4236/aim.2014.42016
Molecular and Pathogenic Study of Guignardia spp. Isolates Associated to Different Hosts
1Estação Experimental de Itajaí, EPAGRI, Itajaí, Estado de Santa Catarina, Brazil
2Departamento de Tecnologia, FCAV/UNESP, Jaboticabal, Estado de São Paulo, Brazil
3UFGD, Faculdade de Ciências Biológicas e Ambientais, Dourados, Estado de Mato Grosso do Sul, Brazil
4Departamento de Genética e Evolução, UFSCAR, São Carlos, Estado de São Paulo, Brazil
5Departamento de Fitossanidade, FCAV/UNESP, Jaboticabal, Estado de São Paulo, Brazil
Email: *esterwickert@epagri.sc.gov.br
Copyright © 2014 Ester Wickert et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accordance of the Creative Commons Attribution License all Copyrights © 2014 are reserved for SCIRP and the owner of the intellectual property Ester Wickert et al. All Copyright © 2014 are guarded by law and by SCIRP as a guardian.
Received November 6, 2013; revised December 6, 2013; accepted December 13, 2013
KEYWORDS
Fungi; Endophytes; Molecular Markers; Plant Disease
ABSTRACT
Fungi of Guignardia genus are commonly isolated from different plant species and most of the time, they are characterized as endophytes. However, some species of this genus, like G. citricarpa and G. psidii are known as causal agents of serious diseases that affect important crops such as Citrus Black Spot and guava fruit rot, respectively. They are also responsible for diseases that cause foliar spots in different fruit species and also in other crops, but cause minor damages. Despite evidences that G. mangiferae colonizes different plant species, there are few studies about its genetic diversity associated with different hosts. This work has the objective to characterize Guignardia isolates obtained from different hosts and tissues by RAPD, fAFLP and DNA sequence of ITS1-5.8S-ITS2 region, as well as to develop pathogenicity tests through cross inoculation in citrus and guava fruits. It was observed that molecular markers were able to discriminate isolates of different Guignardia species. Pathogenicity tests showed that G. citricarpa caused CBS symptoms on citrus fruits, but it did not produce any symptoms in guava fruits. G. mangiferae isolates were able to cause rot symptoms on guava fruits, but they have not produced any symptoms on citrus fruits. Guignardia isolates obtained from mango leaves that have not been classified in species have not presented any symptoms in citrus and guava fruits. Although G. mangiferae is commonly isolated asymptomatically in different plants, this work supports the evidence that this species has a latent pathogen behavior, at least for guava plants.
1. Introduction
The Guignardia Genus (Kingdom Fungi, Phylum Ascomycota, Class Dothideomycetes, Order Botryosphaeriales, Family Botryosphaeriaceae) encompasses around 330 known species, many of them with unknown anamorphic phase [1]. Many species considered plant endophytic fungi are classified in this family and genus, among them, the G. mangiferae species, the causal agents of Citrus Black Spot (CBS) G. citricarpa [2], and G. psidii species causing fruit rot in guava. The G. psidii species is considered responsible for fruit rot disease in different plants, mainly in postharvest conditions. This fungus species is responsible for great losses in guava fruits in Brazil [3]. However, studies using molecular techniques suggested that fungi isolates identified as G. psidii could be in fact G. mangiferae, or also could be conspecific to this cosmopolitan species [4]. It is very common that organisms belonging to the same species, when obtained from different hosts tend to be identified according the host, resulting in taxonomy mistakes and redundancies in databases.
Despite causing foliar spots in mango (Mangifera indica), G. mangiferae was isolated in a wide range of different hosts, and it was considered endophytic because of the symptomless tissues from where it was isolated. These hosts include Brazilian tropical plants like Apidosperma polineuron, Anacardium giganteum, Myracrodreun urundeuva, Spondias mombin, Bowdichia nítida and Cassia occidentalis [5]. Citrus plants are also known as hosts of G. mangiferae [4,6,7], and it is considered endophytic to this plant species because none symptom is related to this fungus in this host. Isolates obtained by these authors were identified by DNA sequence of ITS rDNA (ITS1- 5.8S-ITS2). Other G. mangiferae plant hosts, like Surinam cherry (Eugenia uniflora) and Brazilian grape tree (Myrciaria cauliflora) [4] were also identified in Brazil. Other known G. mangiferae hosts are mango (Mangifera indica L.), banano (Musa sp.) [8], eucaliptus (Eucalytus sp.) [4], and different Ericaeae plants [9].
The use of molecular tools to identify and to characterize fungal isolates arouses interest, mainly because of its quickness when compared to conventional techniques. RAPD (Random Amplified Fragment Lenght Polymorphisms) are the mostly used classes of markers [10] because they are a good method to identify genetic diversity among different organisms in a short period of time and low costs. The fAFLP (fluorescent Amplified Fragment Lenght Polymorphism) markers possess advantages over other techniques as a tool to identify high levels of genetic diversity and also because it allows reproducibility, fastness and reliability [11].
Studies about phylogeny and molecular system of fungi have been done using ITS rDNA, because of the higher number of random copies of this sequence dispersed in the genome and the uniformity of them, which is generally maintained by harmonic evolution [12]. The use of this region proved to be efficient for classifying fungi of Guignardia genus in species and also to infer the genetic diversity among isolates [4].
This work has the objective to characterize Guignardia isolates from different hosts by rDNA ITS1-5.8S-ITS2, RAPD and fAFLP molecular markers, as well as to carry out pathogenic characterization tests by crossed inoculation in citrus and guava fruits with the same isolates.
2. Material and Methods
2.1. Isolates Sampling
Guignardia isolates for this study were obtained from different hosts and tissues shown in Table 1. This isolates were searched in order to represent the entire collection of Guignardia sp. of the Laboratório de Fitopatologia from the Faculdade de Ciências Agrárias e Veterinárias/UNESP, Campus de Jaboticabal/São Paulo State, Brazil.
2.2. Molecular Studies
DNA of the isolates was obtained according [13] protocol. Molecular markers were obtained according the conditions described below.
• RAPD markers. RAPD-PCR reaction was carried out using 30 ng of genomic DNA, PCR Buffer 1X (50 mM KCl, 200 mM Tris-HCl, pH 8,4) (Invitrogen, CA, USA), 2,5 mM MgCl2 (Invitrogen, CA, USA), 200 mM dNTP (Invitrogen, CA, USA), 1,5 U Taq DNA polimerase (Invitrogen, CA, USA), 10 pmol of each primer and sterile water to complete the final volume of 20 mL. Amplification was done on PTC 100 Programable Thermal Controler (MJ Research, Inc.,) termocycler with a initial denaturation step of 92˚C during 3 min., 47 cycles of (92˚C during 1 min., 36˚C during 1 min. e 45 s e 72˚C during 2 min.) and a final cycle of 72˚C during 7 min. Primer kit of Operon Technologies Inc. was tested to search for primers with good amplifications and polymorphism. Electrophoresis was done on 1.2 % agarose gel using TEB 1X buffer (89 mM Tris; 89 mM Boric acid; 2.5 mM EDTA, pH 8.3), with ethidium bromide (0.5 mg/ml) during 1 h 30 min. under 90 Volts. As ladders, 1 kb DNA ladder plus (Invitrogen, CA, USA) and 100 pb DNA ladder (Invitrogen, CA, USA) were used and gel images were visualized with Gel-Doc 1000 (BioRad, CA, USA) equipment. Data was scored in a binary matrix 0 representing absence of bands and 1 the presence of a band for each position. Binary matrix was analized by PAUP (Phylogenetic Analysis Using Parcimony-versão 3.01) [14] software to convert it in a distance matrix that was used to build the similarity phylogram by MEGA (version 4.0) [15] software using the Distance Method with Neighbour Joining [16] groupment algorithm.
• fAFLP markers. These markers were obtained using the “AFLP Microbial Fingerprinting Kit” (Applied Biosystems do Brasil Ltda.), according manufacturer instructions. fAFLP-PCR products were added with formamide-loading dye (1.5 ml final volume) and loaded onto an ABI Prism 377 DNA Sequencer (Applied Biosystems) along with an internal lane standard, GS-500 Rox on ABI Prism 3700 DNA Sequencer (Applied Biossytems, Foster City, USA). Fragments were detected and compiled by the ABI Prism Data CollectionTM (Applied Biosystems) software. Gel image files were generated and all the lanes were extracted for making individual electropherograms using GeneScan (ABI Prism version 1.0) and Geno-
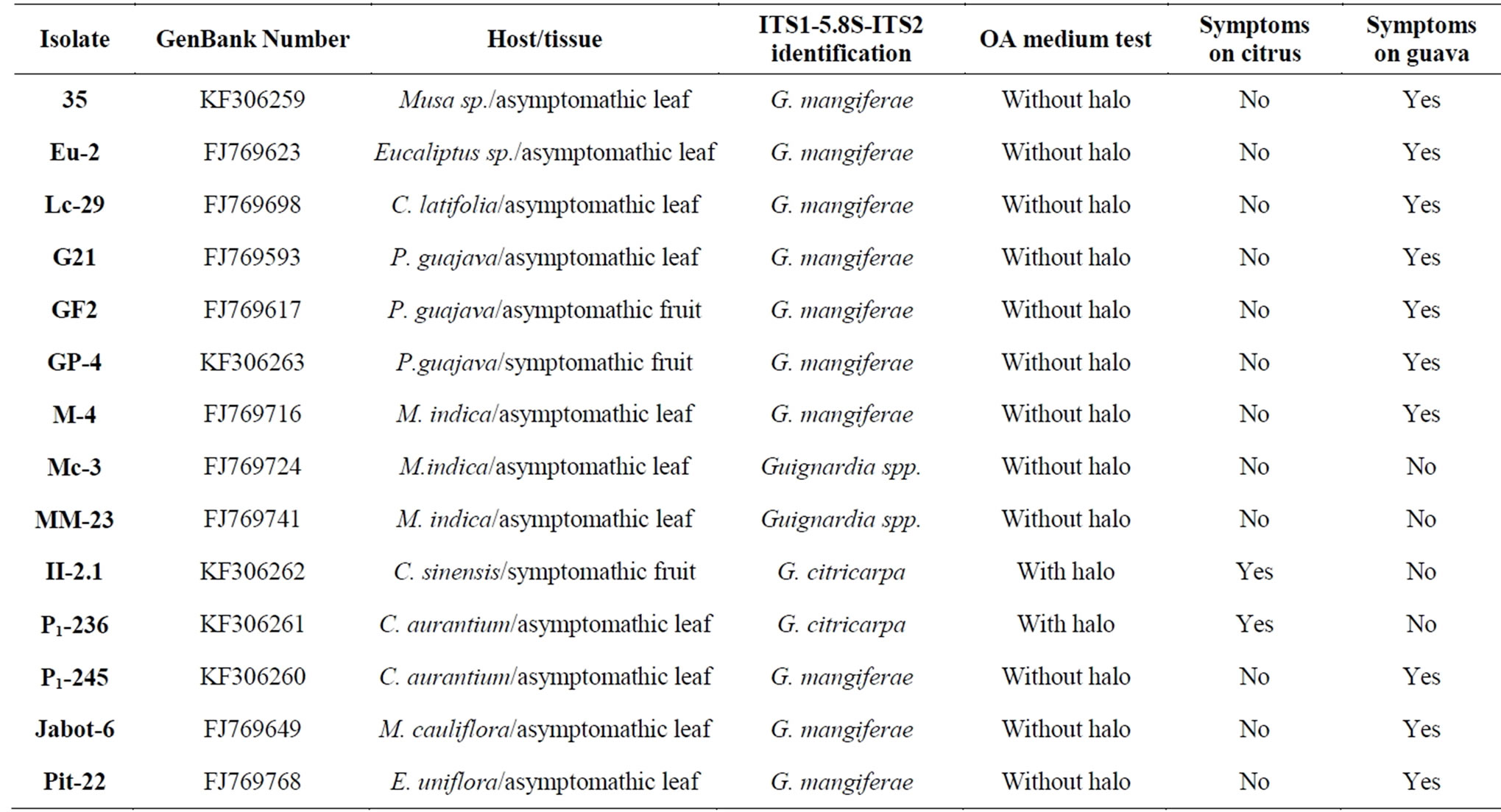
Table 1. Isolates characterization by molecular rDNA sequence, OA medium test and crossed innoculation pathogenicity test conducted in Field with citrus and guava fruits.
typer (ABI Prism version 1.03) softwares. Fragments between 50 and 500 base pair were selected and analyzed on PAUP (Phylogenetic Analysis Using Parcimony-version 3.01) [14] and phylogram was obtained according the same conditions of RAPD markers.
• Analysis of ITS1-5.8S-ITS2 regions. The ITS1-5, 8SITS2 regions were amplified by ITS1/ITS4 primers [12] and DNA sequence for each isolate was obtained using DYEnamic ET Dye Terminator Kit (GE Healthcare) according manufacturer instructions on ABI Prism 3700 DNA Sequencer (Applied Biossytems, Foster City, USA). Electropherograms were collected by ABI Analysis Data Collection software and converted on nucleotide sequences by DNA Sequencing Analysis Software (Version 3.3) software. The DNA sequences were submitted to Phred/Phrap/Consed [17] package and SequencherTM (version 4.05 (Gene Codes Corp, Ann Arbor, USA)) software for checking base quality, alignment and edition. By BLAST [18], the DNA sequences were compared for similarity search on GenBank (www.ncbi.nlm.nih.gov). The sequences from different Guignardia species that presented some similarity with isolates of this work were selected and used to build a phylogram. These sequences were aligned and edited by BioEdit 7.5.0.3 [19]. The phylogram was obtained by MEGA (versão 4.0) [15] software with Distance Method and Neighbour Joining [16] groupment algorithm and 1000 bootstrap. DNA sequences of the isolates were deposited on GenBank and ID numbers are disposable on Table 1.
3. Results
3.1. Molecular Studies
For the developing of RAPD markers, 220 primers were tested, and 14 were selected (OPA1, OPA4, OPA5, OPA8, OPA10, OPA12, OPA17, OPA18, OPA19, OPA20, OPB17, OPC9, OPC10, OPG5) according number of polymorphic bands and capacity to amplify all isolates. A total of 157 polymorphic markers were obtained (average of 11.6 markers/primer). RAPD markers separated the isolates in two main groups, one of them with the isolates P1-245, Lc-29, Eu-2, Jabot-4, GF2, GP1, G21, 35, M4 e Pit-22 (Figure 1). The other group was composed by isolates P1-236 and II2.1. The isolates from mango Mc-3 and MM-23 formed separated branches.
For the fAFLP markers, 36 selective combinations were tested and 15 of them were selected (Fam Eco-RIMseI combinations ACT/CAC; ACA/CAC; ACA/CTC and ACA/CTA; Ned EcoRI-MseI combinations AAC/ CAT; ACC/CTT; AAC/CAC; ACG/CAA and AGC/CTC; Joe EcoRI-MseI combinations (AGG/CAC; AAG/CAC; ACG/CTG; ACG/CAC; AGG/CTT and AAG/CTA). A total of 268 polymorphic bands were detected (average of 17.9 markers/primer combination). This markers also separated isolates in two main groups, one of them formed by isolates P1-245, Lc-29, GF2, Eu-2, G21, Lc-29, G21, Jabot-6, GP-4, GF2, Pit-22, M4 e 35 (Figure 2). Other group was formed by isolates P1-236 and II-2.1. As in RAPD markers, isolates Mc-3 and MM-23 also formed separated branches.
Analysis of ITS1-5.8S-ITS2 rDNA regions allowed to classify most of the Guignardia isolates in Genera and species. All isolates presents similarity higher than 90% with sequences deposited in GenBank, and this is a strong evidence that all of them belong to the Guignardia Genera. Only isolates that presented sequences with similarity higher than 96% were classified in species. Assuming this condition, isolates obtained from asymptomatic leaves of Sour Orange and from CBS symptomatic fruit of sweet orange Valencia were classified as G. citricarpa (with similarities of 99% and 100%, respectively).
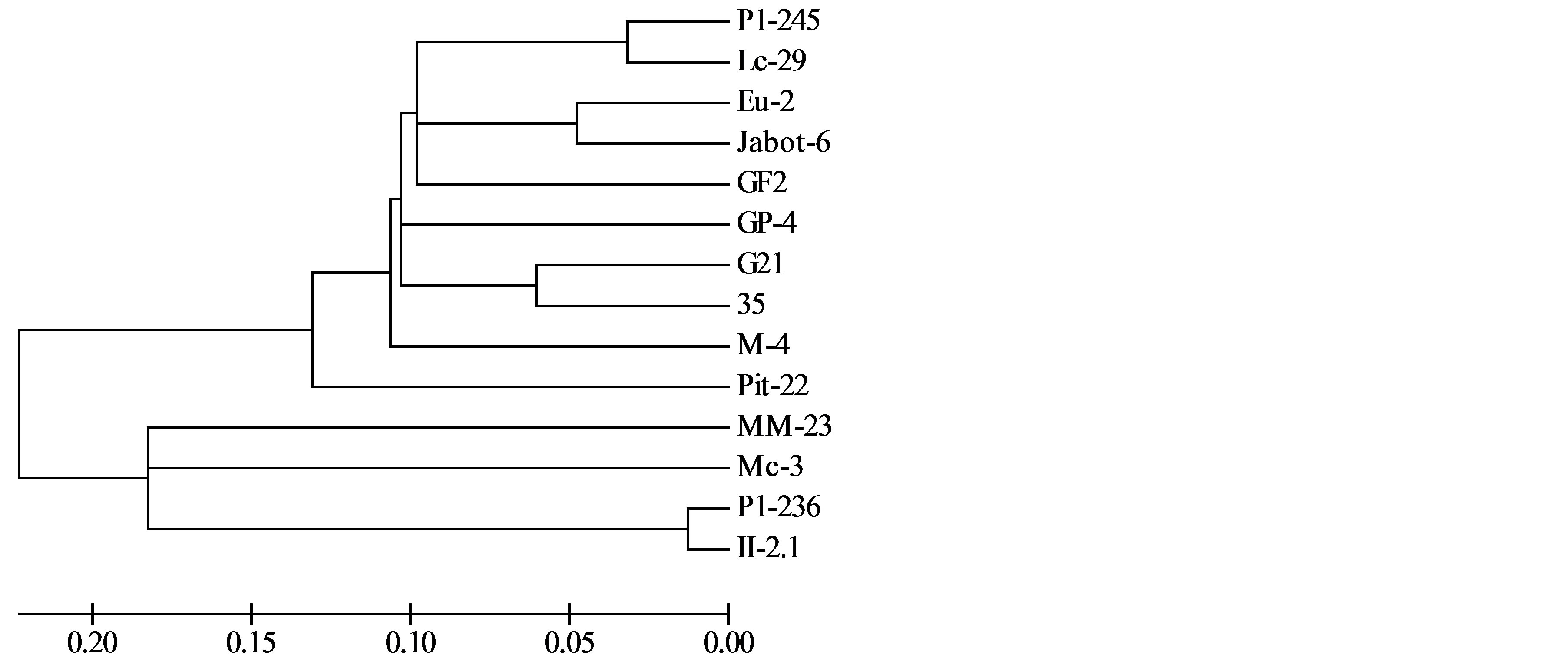
Figure 1. Genetic relationships among Guignardia isolates from different hosts and tissues, evaluated by RAPD markers. These markers were able to separate G. mangiferae and G. citricarpa isolates.
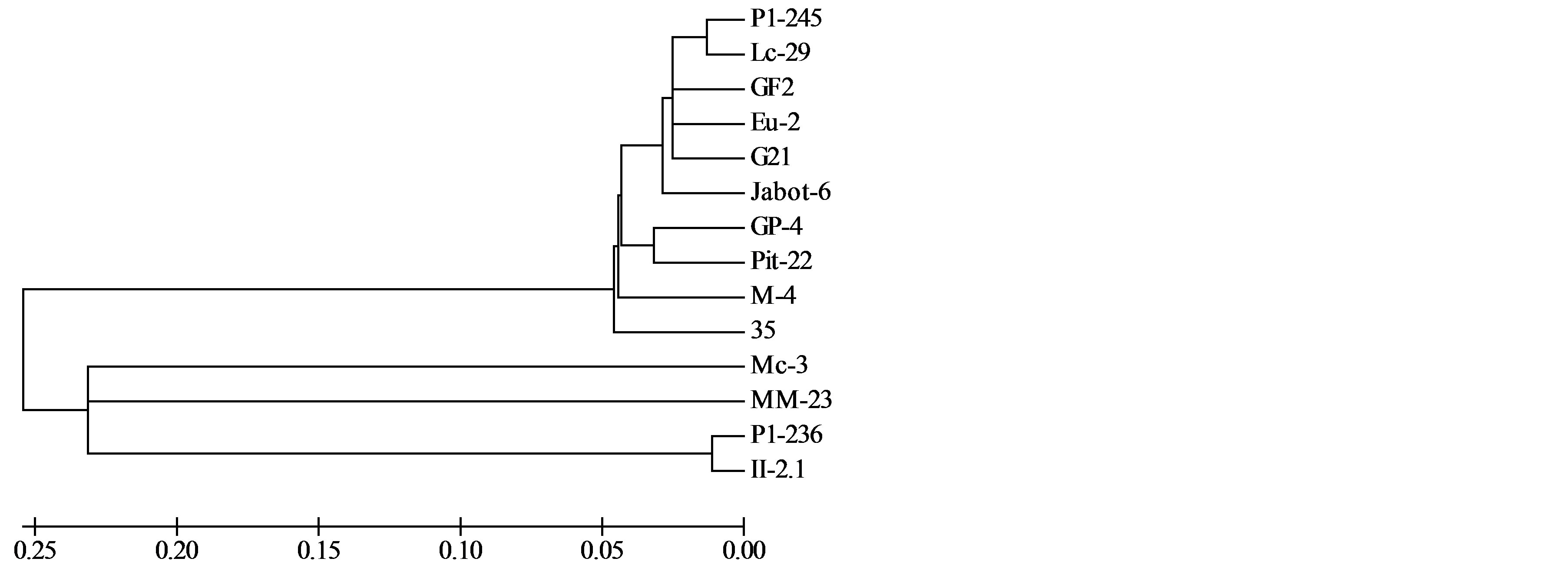
Figure 2. Genetic relationships among Guignardia isolates from different hosts and tissues, evaluated by fAFLP markers. Isolates grouped according its similarity. These markers were also able to separate G. mangiferae and G. citricarpa isolates.
Isolates obtained from asymptomatic leaves of Tahiti acid lime, Sour orange, guava, mango, banana, eucalyptus, Surinam cherry and Brazilian grape tree showed high similarities with rDNA deposited sequences representing G. mangiferae, G. psidii, G. alliacea and G. camelliae species. The same occurred with the isolate obtained from an asymptomatic guava fruit (GF2) and the isolate obtained from a symptomatic guava fruit rot (GP1).
One of the isolates obtained from mango, Mc-3 showed high similarity with Phyllosticta brazilianiae and the other, MM-23, despite placed near G. citricarpa (MM-23 presented similarity of 92% with this species) and P. citriasiana (MM-23 presented similarity of 93% with this species) on phylogram presented no sufficient similarity to be classified as species (Figure 3).
3.2. Pathogenic Studies
When the isolates of this study were submitted to OA medium test, only the isolates classified as G. citricarpa showed presence of the characteristic halo around the fungal colonies (Table 1). This method has demonstrated the usefulness of this medium for discriminate citrus isolates of G. citricarpa species from other Guignardia species.
When mango isolates were inoculated in citrus and guava fruits, none of them were able to cause CBS or fruit rot symptoms (Figure 4). But isolates classified as G. citricarpa by rDNA produced CBS symptoms in citrus fruits, but did not it in guava fruits (Figure 5 and Table 1). Isolates that showed high similarity in rDNA regions with G. mangiferae/G. psidii/G. alliaceae/G. camelliae did not caused symptoms on citrus fruits (Figure 7 and Table 1), but despite its different original hosts and tissues, all of them caused identical fruit rot symptoms on guava (Figure 6 and Table 1).
4. Discussion
DNA markers are powerful tools to discriminate different Guignardia species and evaluate its genetic diversity. Because of its capacity on searching variations over the whole genome, RAPD and fAFLP markers proved to be efficient on evaluating the genetic similarity among the isolates of this study. These two molecular markers showed similar results, but fAFLP markers showed more clearly the grouping of the fungi isolates than RAPD markers. It could be probably a result of the fAFLP higher number of polymorphic markers.
The use of DNA sequences as ITS1-5.8S-ITS2 regions has revitalized the fungal systematic, mainly because of the recent studies of global genetic diversity [20]. Despite that one of the limiting factors to the increase of molecular phylogenetics of fungi is the few number of
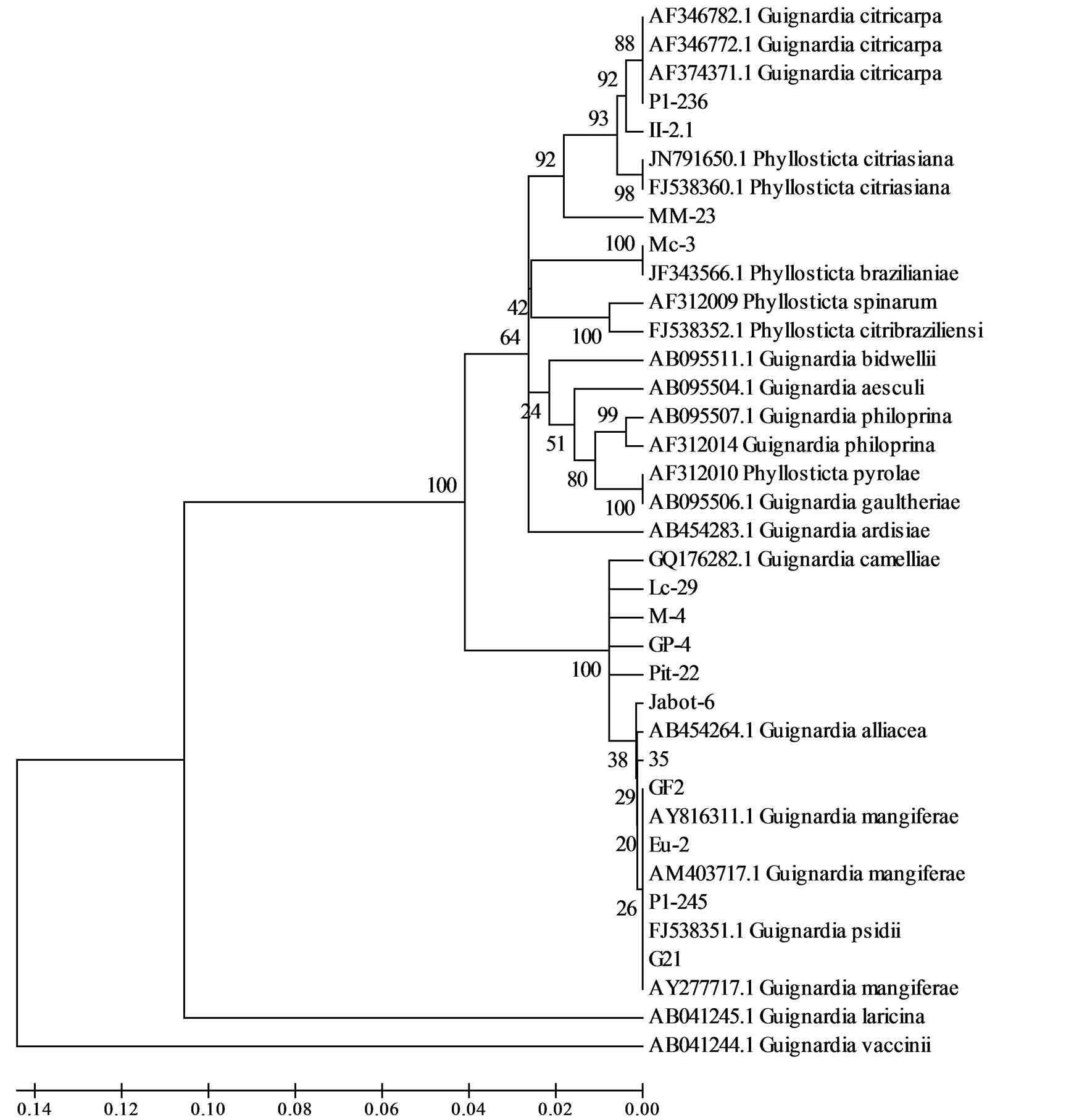
Figure 3. Genetic relationships among Guignardia isolates from different hosts and tissues according ITS1-5,8S-ITS2 DNA sequence. SNP markers were able to discriminate and identify almost all isolates of this study.


Figure 4. Pathogenicity tests conducted with crossed innoculation using isolate Mc-3 from mango on guava (left) and citrus (right) fruits. None of inoculated fruits showed symptoms.


Figure 5. Pathogenicity tests conducted with crossed innoculation using isolate P1-236 from “Tahiti” acid lime on guava (left) and citrus fruits. All citrus fruits inoculated with this G. citricarpa isolate showed CBS symptoms.




Figure 6. Pathogenicity tests conducted with crossed innoculation using G. mangiferae isolates from different hosts and tissues on guava fruits. All guava fruits inoculated with these isolates isolate showed rot symptoms. The only fruit without symptoms corresponds to the control (identified as “Testemunha”).




Figure 7. Pathogenicity tests conducted with G. mangiferae isolates from different hosts innoculated on citrus fruits. None of the innoculated fruits showed CBS symptoms.
easily available genes—mainly the ribosomal genes and its internal spacers—it is undeniable that the use of these regions enabled substantial progress in this field [20]. In our study, only one isolate from mango leaves could not be classified in species using DNA sequence of ITS1- 5.8S-ITS2 regions, because of the low similarity of this isolate with the sequences deposited on GenBank. All the other isolates showed high similarity with sequence of GenBank and could be classified in species using this rDNA sequence, showing that the use of these region is very helpful.
Although restricted to one genome regions, variation on DNA sequence of rDNA ITS1-5.8S-ITS2 were efficient in classify the isolates of this study in species and also to infer genetic diversity among the isolates. These variations (also called SNP’s or Snips-Single Nucleo-tide Polymorphism) proved also to be more accurate to demonstrate the genetic similarity of the two mango isolates with other Guignardia isolates than RAPD and fAFLP markers. This was done because the high number of rDNA sequences available on GenBank. This kind of information is not disposable when DNA fragment polymorphism markers were used. While RAPD and fAFLP markers placed isolates Mc-3 and MM-23 in separated branches, SNPs could demonstrate that these isolates were more similar to species present in databases, but not sampled in this study, as is was the case of P. braziliensis and P. citriasiana. SNP markers clearly showed that isolate MM-23 was more similar to G. citricarpa and P. citriasiana species than to the other species represented on this study. This was not so evident using RAPD and fAFLP markers. Despite this greater similarity to G. citricarpa species, it is probably that MM-23 isolate do not belong to none of this species because it does not show yellow halo in OA medium and its colony morphology is different (data not showed) if compared to the other Guignardia isolates of the pathogenicity tests. A similar result was obtained with isolate Mc-3, which was placed in separated branches by RAPD and fAFLP markers, but was classified as P. brazilianiae by rDNA sequence. Previous studies showed that some Guignardia isolates from Mangiferae indica on Brazil belongs to P. brazilianeae [21], also stating that this species is common of this plant species.
The pathogenic characterization that was carried out using OA medium showed that the isolates of this study that presented a yellow halo around the colonies also caused CBS symptoms when citrus fruits were inoculated at the field. This corroborated with the importance of this simple and fast test to differentiate G. citricarpa isolates [4,6,8] from other Guignardia species.
The G. mangiferae species is considered cosmopolitan because it was found colonizing assymptomatically a great number of hosts [9]. This was also observed in this work, as all of the hosts had been colonized by G. mangiferae. But the DNA sequence of ITS1-5.8S-ITS2 of these isolates was not able to differentiate them from G. mangiferae, G. psidii, G. alliaceae and G. camelliae. This occurs probably because different authors, working in different places had isolated the same fungi and named it according the original host. It is conceivable that all these species are in fact the same species, G. mangiferae. In this work, we adopted the criteria of the older and well documented references that classify this fungus as G. mangiferae [6]. Therefore, all isolates of this study that showed high similarity with these sequences, were classified as G. mangiferae.
All isolates obtained from asymptomatic tissues on different hosts that were classified as G. mangiferae caused symptoms in guava fruits, even though some references report that G. psidii species is responsible for the guava fruit rot [3]. Assuming that the patogenicity tests and classification in species by ITS1-5.8S-ITS2 DNA sequence are reliable methods, there are strong evidences that G. psidii and G. mangiferae are either the same species or they are conspecific. Banana and eucalyptus plants present the same conditions. Literature review shows these two species plant as hosts of G. musae and G. eucaliptorum, where they supposedly cause spots on leaves [22, 23]. Recently, the identity of the casual agent of freckle disease of banana was investigated and based on morphological and molecular data from a global set of banana specimens it was found that several species were associated with the disease [24]. These authors introduced P. maculata as the new name of G. musae, and named P. musarum to represent a distinct species from India and Taiwan, as G. stevensii is confirmed as distinct species from Hawaii and G. musicola from northern Thailand was shown to contain different taxa. The same authors described P. cavendishii as a new, widely distributed species, appearing primarily on Cavendish, but also on non-Cavendish banana cultivars. It is also true that the increase in the number of new species introduced is largely a result of the widespread use of DNA sequence data, but is also due to the exploration of new geographic regions and habitats [25]. But in our work, the isolates obtained in banana and eucalyptus were classified as G. mangiferae and caused fruit rot symptoms when inoculated in guava, but none symptom was induced in citrus fruits.
As all of G. mangiferae isolates obtained in citrus, mango, banana, Surinam cherry and Brazilian grape tree has caused the same symptoms of fruit rot in guava, this could supports the idea that probably all isolates belongs to the same species that is spread on different hosts. Considering that it is frequent the same organism to receive different name according the host, it is probable that this also occurs with G. mangiferae. Despite the fact that molecular markers evidenced the presence of genetic diversity among G. mangiferae isolates from this study, this can be credited to intraspecific variations, possibly as a response to host selection pressure, mainly because the original hosts belong to different botanical Families and Genera, and not because the isolates are from different species.
We believe that this could be supported by the pathogenic tests, at least for citrus and guava plants.
G. mangiferae is considered a fungal endophyte. The concept of endophytic organisms is widely discussed in different reviews [26-29]. Most authors describe endophytes as organisms that colonizes intercellular spaces of different plant tissues in some period of their lives, without causing apparent damages to the host.
Many genera belonging to the Botryosphaeriaceae family include species that have been described as endophytes [1]. There are examples in the genus Guignardia, Botryosphaeria (anamorph Fusicoccum), Dothidotthia (anamorph Dothiorella), Neofusicoccum, Pseudofusicoccum, Lasiodiplodia and Diplodia. Some of these endophytes are very common and sometimes they overcome endophytic communities in Eucalyptus and Pinus plants. It seems that endophytism is common to most species of the family [28] in some environments. Although detected fungi in plant symptomless tissue are arbitrarily classified as endophytes, they could have in fact different survival strategies. Some of them could be pathogens in a non-host, latent or quiescent pathogens, saprophytes “hiding”, e.g. in a stomatal cavity and “awaiting” host senescence as spores, or virulent pathogens in a latent phase [29].
Endophytes were isolated from almost all studied plants until now [26] and most of the time they take advantages to the host, establishing mutual relationships, increasing plant resistance to diseases and drought and enhancing plant development [27,30]. Some endophytes are known as latent pathogens, or in some cases, endophytes that became pathogens under specific conditions [31], as is the example of Phomopsis citri, Fusicoccum aesculi and Lasiodiplodia theobromae in citrus and Phomopsis viticola in grape [26]. This behavior change can be produced by host physiology alterations caused by the fungi activity, by the environmental changes that stress the host, or by the developmental stage of the plant or the fungi, or by both of them [22].
Latent pathogens (endophytic pathogens) have coevolved with their hosts and became less virulent, using senescent leaves, some host stress period or eventually the ripe fruit rot for sporulating [30]. This was demonstrated for G. mangiferae species, which was able to produce the same degradating enzymes as its saprobic counterpart [32]. This is probably the case of the G. mangiferae isolates of this study that were obtained from symptomless tissues of different hosts, as well as the isolates obtained in guava ripe fruit rot symptoms. However, the same isolates present an apparently endophyte behavior in citrus plants, but symptoms associated to the presence of G. mangiferae in this plant species are unknown until now.
The endophyte/host relationship status can be transient and the stability and variability of this interaction depends of factors as the environment and host conditions [29]. So, in spite the fungi isolates were isolated from symptomless tissues does not exclude the possibility of this fungi become a pathogen when the host is stressed or in senescence [33].
The fungi isolates of this study that were obtained from symptomless tissues of mango did not produce CBS symptoms on citrus fruits, nor fruit rot symptoms on guava. These isolates were detected only in mango tissues and are different between each other. Additional pathogenicity tests are necessary with this isolates in mango fruits.
Additional pathogenicity tests are also necessary to elucidate if the G. mangiferae isolates from the different Brazilian hosts and the G. citricarpa isolates from citrus of this study also have the ability to cause diseases on mango fruits and leaves, as is it reported in literature (available on http://www.apsnet.org/publications/commonnames/Pages/Mango.aspx).
5. Conclusion
In this study, the RAPD and fAFLP markers, and DNA sequences showed similar results, and we believe that they are complementary methods for evaluating the interaction pathogen-host-disease, as the first two allow accessing genetic variation within the genome as a whole and the other focuses on small changes in a few diagnostic sequences. As the trees generated by these complementary techniques yield similar groupings and branching patterns, their congruence could be interpreted as independent confirmation of the proposed evolutionary phylogenies. It is feasible that the used molecular markers added to rDNA sequences were efficient tools to evaluate genetic diversity and to classify unidentified fungi isolates into species. As DNA sequence databases are frequently and continuously fed with more information about different organisms, mainly with rDNA sequences, the future works to classify organisms will be easier and more accurate.
Acknowledgements
We are grateful to FAPESP (Fundo de Amparo à Pesquisa do Estado de São Paulo) for financial support to projects 04/10560-4 and 01/10993-0, as well as postdoctoral fellowship of first author. We also thanks to CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) and CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), for grant fellowship to the other authors.
REFERENCES
- P. W. Crous, B. Slippers, M. J. Wingfield, J. Rheeder, W. F. O. Marasas, A. J. L. Philips, A. Alves, T. Burgess, P. Barber and J. Z. Groenewald, “Phylogenetic Lineages in the Botryosphaeriaceae,” Studies in Mycology, Vol. 55, No. 1, 2006, pp. 235-253. http://dx.doi.org/10.3114/sim.55.1.235
- B. C. Sutton and J. M. Waterston, “Guignardia citricarpa,” In: C. M. I. Commonwealth Mycological Institute (Descriptions of Pathogenic Fungi and Bacteria, 85) Surrey, England, Commonwealth Mycological Institute, Kew, 1966, No. 85, pp. 1-2.
- L. J. Tozello and W. R. C. Ribeiro, “Tratamento Póscolheita de Goiaba (Psidium guajava L.) Contra Podridão de Guignardia Psidii,” Revista Brasileira de Fruticultura, Vol. 20, No. 2, 1998, pp. 229-234.
- E. Wickert, A. Goes, E. G. M. Lemos, A. Souza, E. L. Silveira, F. D. Pereira and D. Rinaldo, “Relações Filogené- ticas e Diversidade de Isolados de Guignardia spp. Oriundos de Diferentes Hospedeiros nas Regiões ITS1-5, 8SITS2,” Revista Brasileira de Fruticultura, Vol. 31, No. 2, 2009, pp. 360-380. http://dx.doi.org/10.1590/S0100-29452009000200010
- K. F. Rodrigues, T. N. Sieber, C. Grünig and O. Holdenrider, “Characterization of Guignardia mangiferae Isolated from Tropical Plants Based in Morphology, ISSRPCR Amplifications and ITS1-5.8S-ITS2 Sequences,” Mycological Research, Vol. 108, No. 1, 2004, pp. 45-52. http://dx.doi.org/10.1017/S0953756203008840
- R. P. Baayen, P. J. M Bonants, G. Verkley, G. C. Carroll, H. A. van Der Aa, M. de Weerdt, I. R. van Brouwershaven, G. C. Schutte, W. J. R. Maccheroni, C. G. de Blanco and J. L. Azevedo, “Nonpathogenic Isolates of the Citrus Black Spot Fungus, Guignardia citricarpa, Identified as a Cosmopolitan Endophyte of Woody Plants, G. mangiferae (Phyllosticta capitalensis),” Phytopathology, Vol. 92, No. 5, 2002, pp. 464-477. http://dx.doi.org/10.1094/PHYTO.2002.92.5.464
- P. J. M. Bonants, G. C. Carroll, M. de Weerdt, I. R. van Brouwershaven and R. P. Baayen, “Development and Validation of a Fast PCR-Based Detection Method for Pathogenic Isolates of the Citrus Black Spot Fungus, Guignardia citricarpa,” European Journal of Plant Pathology, Vol. 109, No. 5, 2003, pp. 503-513. http://dx.doi.org/10.1023/A:1024219629669
- R. B. Baldassari, E. Wickert and A. de Goes, “Pathogenicity, Colony Morphology and Diversity of Isolates of Guignardia citricarpa and G. mangiferae Isolated from Citrus spp.,” European Journal of Plant Pathology, Vol. 120, No. 2, 2008, pp. 103-110. http://dx.doi.org/10.1007/s10658-007-9182-0
- I. Okane, A. Nakagiri and T. Ito, “Identity of Guignardia sp. Inhabiting Ericaceous Plants,” Canadian Journal of Botany, Vol. 79, No. 1, 2001, pp. 101-109.
- M. T. Sadder and A. F. Ateyyeh, “Molecular Assessment of Polymorphism among Local Jordanian Access of Common Fig (Ficus carica L.),” Scientiae Horticulturae, Vol. 107, No. 4, 2006, pp. 347-351. http://dx.doi.org/10.1016/j.scienta.2005.11.006
- C. Yang, J. Zhang, Q. Xu, C. Xiong and M. Bao, “Establishment of AFLP Technique and Assessment of Primer Combinations for Mei Flower,” Plant Molecular Biology Reporter, Vol. 23, No. 1, 2005, pp. 79-80. http://dx.doi.org/10.1007/BF02772651
- T. J. White, T. Bruns, S. Lee and J. Taylor, “Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics,” In: M. A. Innis, D. H. Gelfand, J. J. Sninsky and T. J. White, Eds., PCR Protocols: A Guide to Methods and Applications, Academic Press, New York, 1990, pp. 315-322.
- E. E. Kuramae-Izioka, “A Rapid, Easy and High Yield Protocol for Total Genomic DNA Isolation of Colletotrichum gloesporioides and Fusarium oxysporum,” Revista Unimar, Vol. 19, No. 3, 1997, pp. 683-689.
- D. L. Swofford, “PAUP: Phylogenetic Analysis Using Parcimony (*and Other Methods),” Version 4.0b10, Sinauer Associates, Sunderland, 2002.
- K. Tamura, J. Dudley, M. Nei and S. Kumar, “MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0,” Molecular Biology and Evolution, Vol. 24, No. 8, 2007, pp. 1596-1599. http://dx.doi.org/10.1093/molbev/msm092
- N. Saitou and M. Nei, “The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees,” Molecular Biology and Evolution, Vol. 4, No. 4, 1987, pp. 406-425.
- D. Gordon, C. Abajian and P. Green, “Consed: A Graphical Tool for Sequence Finishing,” Genome Research, Vol. 8, No. 3, 1998, pp. 195-202. http://dx.doi.org/10.1101/gr.8.3.195
- S. F. Altschul, T. L. Madden, A. A. Schaffer et al., “Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs,” Nucleic Acids Research, Vol. 25, No. 17, 1997, pp. 3389-3402. http://dx.doi.org/10.1093/nar/25.17.3389
- T. A. Hall, “BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/ 98/NT,” Nucleic Acids Symposium Series, Vol. 41, 1999, pp. 95-98.
- J. Xu, “Fundamentals of Fungal Molecular Population Genetic Analyses,” Current Issues in Molecular Biology, Vol. 8, No. 2, 2006, pp. 75-90. http://www.citeulike.org/user/jszekely/article/80546
- C. Glienke, O. L. Pereira, D. Stringari, J. Fabris, V. KavaCordeiro, L. Galli-Terasawa, J. Cunnington, R. G. Shivas, J. Z. Groenewald and P. W. Crous, “Endophytic and Pathogenic Phyllosticta Species, with Reference to Those Associated with Citrus Black Spot,” Persoonia, No. 26, 2011, pp. 47-56. http://dx.doi.org/10.3767/003158511X569169
- W. Photita, S. Lumyong, P. Lumyong, E. H. C. McKenzie and K. D. Hyde, “Are Some Endophytes of Musa acuminate Latent Pathogens?” Fungal Diversity, Vol. 16, 2004, pp. 131-140. http://www.fungaldiversity.org/fdp/sfdp/16-10.pdf
- P. W. Crous, M. J. Wingfield, F. A. Ferreira and A. Alfenas, “Mycosphaerella parkii and Phyllosticta eucaliptorum, Two New Species from Eucalyptus Leaves in Brazil,” Mycological Research, Vol. 97, No. 5, 1993, pp. 582-584. http://dx.doi.org/10.1016/S0953-7562(09)81181-7
- M.-H. Wong, P. W. Crous, J. Henderson, J. Z. Groenewald and A. Drenth, “Phyllosticta Species Associated with Freckle Disease of Banana,” Fungal Diversity, Vol. 56, 2012, pp. 173-187. http://dx.doi.org/10.1007/s13225-012-0182-9
- M. W. Marques, N. B. Lima, M. A. Morais Jr., S. J. Michereff, A. J. L. Phillips and M. P. S. Câmara, “Botryosphaeria, Neofusicoccum, Neoscytalidium and Pseudofusicoccum Species Associated with Mango in Brazil,” Fungal Diversity, Vol. 61, 2013, pp. 195-208. http://dx.doi.org/10.1007/s13225-013-0258-1
- K. D. Hyde and K. Soytong, “The Fungal Endophyte Dilemma,” Fungal Diversity, Vol. 33, 2008, pp. 163-173. http://www.fungaldiversity.org/fdp/sfdp/33-9.pdf
- A. E. Arnold, “Understanding the Diversity of Foliar Endophytic Fungi: Progress, Challenges, and Frontiers,” Fungal Biology Reviews, Vol. 21, No. 2-3, 2007, pp. 51-66. http://dx.doi.org/10.1016/j.fbr.2007.06.002
- B. Slippers and M. J. Wingfield, “Botryosphaeriaceae as Endophytes and Latent Pathogens of Woody Plants: Diversity, Ecology and Impact,” Fungal Biology Reviews, Vol. 21, No. 2-3, 2007, pp. 90-106. http://dx.doi.org/10.1016/j.fbr.2007.06.002
- B. Schulz and C. Boyle, “The Endophytic Continuum,” Mycological Research, Vol. 109, No. 6, 2005, pp. 661- 686. http://dx.doi.org/10.1017/S095375620500273X
- T. SIEBER, “Endophytic Fungi in Forest Trees: Are They Mutulists?” Fungal Biology Reviews, Vol. 21, No. 2-3, 2007, pp. 75-89. http://dx.doi.org/10.1016/j.fbr.2007.05.004
- K. H. Kogel, P. Franken and R. Hückelhoven, “Endophyte or Parasite—What Decides?” Current Opinion in Plant Biology, Vol. 9, No. 4, 2006, pp. 358-363. http://dx.doi.org/10.1016/j.pbi.2006.05.001
- I. Promputtha, K. D. Hyde, E. H. C. McKenzie, J. F. Peberdy and S Lumyong, “Can Leaf Degrading Enzymes Provide Evidence That Endophytic Fungi Becoming Saprobes?” Fungal Diversity, Vol. 41, 2010, pp. 89-99. http://dx.doi.org/10.1007/s13225-010-0024-6
- G. A. Kuldau and I. E. Yates, “Evidence for Fusarium Endophytes in Cultivated and Wild Plants,” In: C. W. Bacon and J. F. White, Eds., Microbial Endophytes, Marcel Dekker, New York and Basel, 2010, pp. 85-120.
NOTES
*Corresponding author.

