Open Journal of Medical Microbiology
Vol.3 No.1(2013), Article ID:28574,9 pages DOI:10.4236/ojmm.2013.31006
Molecular Epidemiology of Nontuberculous Mycobacteria in a German CF Center and Clinical Course of NTM Positive Patients
1Paediatric Pneumology, Dr. von Hauner Children’s Hospital, Ludwig-Maximilians-University, Munich, Germany
2Institute for Clinical Chemistry, Molecular Diagnostics, Medical Microbiology and Hygiene Department, Ludwigshafen General Hospital, Ludwigshafen, Germany
3Institute for Medical Microbiology and Infection Control, University Hospital, Johann-Wolfgang-Goethe-University, Frankfurt/Main, Germany
4Max von Pettenkofer Institute of Hygiene and Medical Microbiology, Ludwig-Maximilians-University, Munich, Germany
Email: #matthias.griese@med.uni-muenchen.de
Received December 3, 2012; revised January 6, 2013; accepted January 22, 2013
Keywords: Mycobacteria; nontuberculous; Mycobacterium abscessus; Cystic Fibrosis; Molecular Epidemiology; Multilocus Sequence Analysis (MLSA); Interferon Gamma
ABSTRACT
Goal of this study was to analyse the clinical course of cystic fibrosis (CF) patients with nontuberculous mycobacteria (NTM) in their respiratory secretions and to investigate the molecular epidemiology of the most prevalent NTM species by multi-locus sequence analysis (MLSA). The respiratory specimen and the clinical parameters forced expiratory volume in one second (FEV1), body-mass-index (BMI), erythrocyte sedimentation rate (ESR) 1 h and immunoglobulin G (IgG) of 357 CF patients, 0 - 52.4 years, mean FEV1 2009 81.5% pred were analysed between 1998 and 2010. In 13 patients NTM were detected. 12 of 13 patients carried M. abscessus, for one patient the NTM species was not characterized. 4 patients carried a second NTM species (M. avium, M. chelonae (2x), M. intracellulare). 6 patients exhibited a significant decline in FEV1, however changes in BMI, IgG and ESR were discordant. Molecular genotyping of M. abscessus isolates revealed a unique MLSA pattern in 6 patients. 2 patients harboured identical strains, and one patient a closely related strain. Whether the presence of identical strains is attributed to the acquisition of NTM clones from common environmental sources or to patient-to-patient transmission cannot be definitely clarified. Although cross-infection of the three patients with identical/closely related strains in the present cohort is highly unlikely, we recommend strict hygiene measures for all CF patients harbouring NTM.
1. Introduction
Nontuberculous mycobacteria (NTM) are acid-fast bacilli (AFB) which are ubiquitous in environment, found in soils, water, animals, milk, and food products. Over 140 different species have been described. NTM cause 4 different disease entities: pulmonary disease in people with or without underlying lung disease, especially cystic fibrosis (CF) patients [1,2], lymphadenitis especially in children possibly on the basis of specific immune deficiencies [3-5], disseminated mycobacterial disease in immune-compromised patients, especially HIV patients, and localized skin and soft tissue infections. Transmission is believed to occur via inhalation or direct inoculation. In contrast to Mycobacterium (M.) tuberculosis human-tohuman transmission appears unlikely for NTM, however, this view was challenged recently [6].
The improved survival of CF patients in the past decades has led to an increasing relevance of resistant and virulent pathogens including NTM. Several studies have reported prevalence rates of NTM in CF patients between 2% and 28% [1,7-10] . The results regarding the effect of the NTM infection on the clinical course are controversial [11,12].
In this study, the detection rate of NTM was determined in a cohort of CF patients for which strict preventive anti-cross infection policies were in place [13] and the individual clinical course of NTM positive patients was analysed in detail. Multi-locus sequence analysis (MLSA) was applied to investigate the molecular epidemiology of the NTM clones in the cohort.
2. Materials and Methods
All respiratory samples of on average 357 CF patients who attended the CF Centre at the Children’s University Hospital in Munich (159 male, 210 patients <18 years, mean age 2009 18.03 years, range 0 - 52.4 years, mean FEV1 81.5% pred) were studied between January 1st, 1998 and January 1st, 2010 for the presence of AFB. Patient visits took place on average every 3 months, if indicated more often. At each visit sputum samples or throat swabs were taken, the patients were investigated clinically, routine laboratory examinations including inflammatory parameters and lung-function test were performed. Informed consent was obtained from all participants and the study was approved by the institutional review board of the University of Munich. The clinical course of NTM positive patients was analysed retrospectively based on the parameters forced expiratory volume in one second (FEV1, % pred), body mass index (BMI, kg/m2) and the inflammatory markers immunoglobulin G (IgG, g/l) and erythrocyte sedimentation rate (ESR 1h, mm). Decline in FEV1 was defined as a decline of >1% pred/year (based on an average yearly drop of FEV1 values of 1% pred/year in large CF registries) [14]. Decline in BMI was defined based on a change in the BMI percentile, increase in IgG as an increase by >3 g/l and increase in ESR as an increase by >5 mm/h throughout the observation period.
2.1. Specimen Preparation
Processing of throat swabs and sputum samples was performed on sheep blood agar and on selective agars. The selective agars included McConkey-agar, mannitol salt agar, BD Cepacia Medium, Haemophilus selective agar (chocolate agar containing bacitracin) and Sabouraud dextrose agar. For the detection of NTM sputum samples of patients who were suspected to colonize NTM (e.g. due to prior NTM detection or due to unexplained clinical deterioration) were routinely decontaminated by the N-acetyl-L-cysteine + NaOH procedure. In addition, some M. abscessus isolates of patients without clinical suspicion of NTM were accidentally recovered from B. cepacia-selective agar due to its effective suppression of CF typic microflora and prolonged incubation. Plates were incubated for up to 72 hours, except the Cepacia Medium and the Sabouraud agar, which were incubated up to 5 days. The sheep blood agar was incubated in 5 to 7% CO2, the Haemophilus-selective agar under anaerobic conditions and the other plates under ambient air conditions. The throat swabs were streaked on the surfaces of the culture plates for semi-quantitative assessment. The sputum specimens were also processed semi-quantitatively after homogenisation with dithiothreitol-solution. The plates were examined daily for the presence of growth. All NTM strains were recovered after a minimum of 3 days incubation on the BD Cepacia Medium, where they grew as greyish white smooth to rough colonies.
2.2. Isolation and Identification of M. abscessus
Direct microscopy for mycobacteria and culture were done according to routine procedures at the laboratory [15,16]. Mycobacteria were identified by the ZiehlNeelsen staining method. M. abscessus was isolated on Lowenstein-Jensen (LJ) solid medium as well as in liquid medium using mycobacterial growth indicator tubes (MGIT) (Becton Dickinson, Heidelberg, Germany). Positive MGIT cultures were subcultured with 0.1 ml of a 1:10 dilution on Lowenstein-Jensen selective medium containing 5% NaCl (LJS, Oxoid GmbH, Wesel, Germany). Species were identified by using a biochemical test panel (tolerance to 5% NaCl, iron uptake, Tween hydrolysis, urease, nitrate reduction) and by sequencing of the 16S rRNA gene [17]. These methods were complemented by using the GenoType Mycobacterium CM test (HAIN Lifescience, Nehren, Germany) [18].
2.3. Molecular Genotyping of M. abscessus Isolates
Internal fragments from 5 housekeeping genes together with 16S-23S-rRNA gene internal transcribed spacer sequence (ITS) were selected for MLSA. A total of 2972 nucleotides, corresponding to 5 housekeeping genes and the ITS sequence, were determined for 11 clinical M. abscessus isolates from CF patients (two isolates were not conserved) and compared to those of the M. abscesssus reference strain (GeneBank accession number CU45 8896).
Fragments from 5 housekeeping genes together with 16S-23S-rRNA gene internal transcribed spacer sequence (ITS) [19] were amplified using the sets: argH (argininosuccinate lyase) [20], cya (adenylate cyclase) [20] , pgm (phosphoglucomutase) [20] , rpoB (beta-subunit of RNA polymerase) [21], and hsp65 (65-kDa heat shock protein) [20,22] (Table 1). The gene products for argH, cya, and pgm have been used previously in the multilocus enzyme electrophoresis (MLEE) analysis of mycobacteria [23,24]. Amplification was performed using 5 µl purified DNA plus 44 µl of PCR master mix (BIORON, Ludwigshafen, Germany) and 0.5 µl of each primer (20 pmol). The dideoxy sequencing of the amplified gene fragments was carried out on both strands with the Big
Table 1. Primers used for PCR and sequencing.

Dye Terminator cycle sequencing kit (Applied Biosystems), using the same primers as those for amplification. Sequencing products were purified and analyzed with an ABI 3700 DNA analyzer. The sequences were aligned and trimmed to defined start and end positions using the Lasergene software package, version 8 (DNASTAR Inc., Madison, WI, USA) according to previous studies [20]. Reference M. abscessus sequences of the 5 housekeeping genes as well as the ITS sequence were obtained from the whole genome sequence of M. abscessus CU458896 (accession number CU458896) [25]. Gene sequences were aligned with the closest relatives retrieved from the BLAST nucleotide sequence database. The alignment was done by a hierarchical method for multiple alignments implemented in the CLUSTAL V computer program [26] and nucleotide sequence relationships were presented as a rooted tree. Automatically aligned sequences were checked manually. Between the isolates the sequence similarities for the respective DNA fragments varied from 98.0% to 100% (ITS), 98.5% to 99.8% (pgm), 98.8% to 100% (hsp65), 85.05% to 100% (argH), 82.93% to 100% (rpoB), and 86.18% to 100% (cya).
3. Results
13 patients (8 male), i.e. 3.6% of the 357 patients attending the CF Center at the Children’s University Hospital Munich between January 1st, 1998 and January 1st, 2010 had at least one NTM positive respiratory specimen (Table 2). The mean age at first detection was 17.4 years (range 13.8 - 28 years). 9 patients were <18 years old at first detection. The mean follow-up period (years from first detection of NTM to the end of the observation period) was for 11 patients 5.2 years (range: 2 - 11.7 years), one patient (9) died shortly after first detection of M. abscessus, one patient (7) left the service shortly after the first detection of NTM and thus fell out of observation. M. tuberculosis was not detected during the observation period.
3.1. Clinical Assessment of NTM Positive Patients
3 patients (8, 12, 13) had only one NTM positive respiratory sample (Table 2), they all had a stable or a slightly increased BMI and a stable ESR. IgG increased in 2 of these patients but remained within the reference range. FEV1 remained stable with the exception of the FEV1 of patient 8, which declined. As the respiratory specimen 4 years after detection of NTM in patient 8 were NTM negative, a causal relationship between clinical condition and M. abscessus infection was unlikely.
9 patients had repeatedly positive NTM samples with a mean detection period of 3.7 years (range 0.1 - 11.4 years). All of these patients but patient 6 fulfilled the 2007 American Thoracic Society (ATS) clinical and microbiologic diagnostic criteria for NTM lung disease [27]. All patients received anti-mycobacterial therapy consisting of a combination of oral, nebulized and i.v. antibiotic and antimycobacterial treatment. Treatment duration was at least one year. 5 of these patients exhibited a prolonged NTM detection (>2 years) despite intensive treatment: 2 (2,4) had a stable FEV1, one (1) a decline in FEV1 and 2 (3,11) an especially strong decline. 3 patients (5,6,10) with repeated NTM detection were suc-
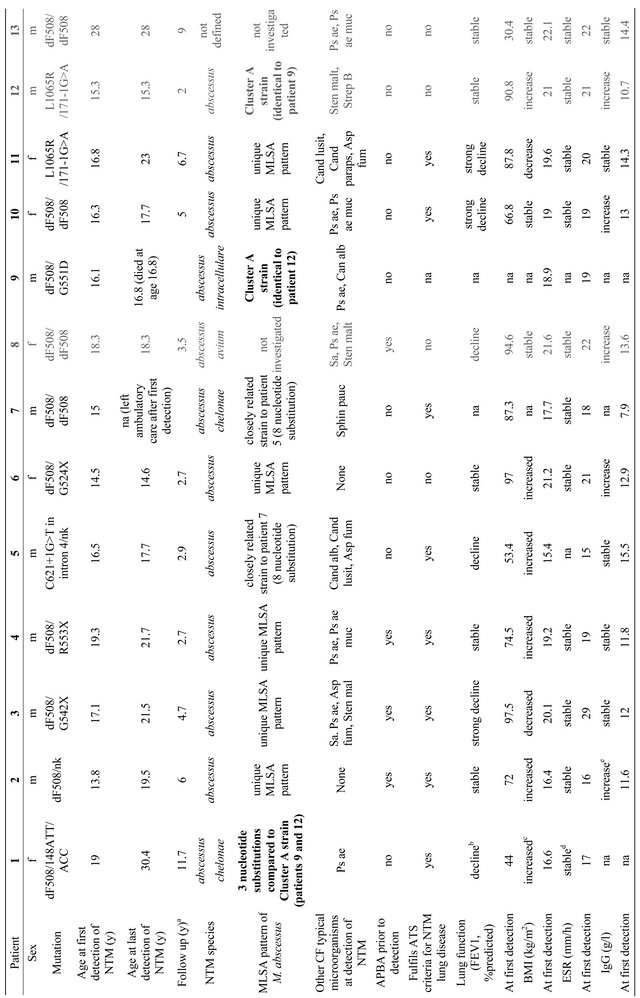
cessfully eradicated after 1.2, 0.1 and 1.4 years respectively: patient 5 exhibited a decline in FEV1, patient 6 had a stable FEV1 and patient 10 an especially strong decline. No clear trend could be observed for IgG, ESR and BMI.
One patient (9) died 0.7 years after first detection of NTM. The clinical course of this patient prior to NTM infection was stable with normal FEV1 values and minor respiratory infections (Table 2, Figures 1 and 2).
No patient had been treated with long-term macrolides prior to first NTM detection, no patient suffered from diabetes mellitus at first detection.
One patient (3) with a detection period of 4.4 years and a severe clinical deterioration was treated with interferon (IFN) gamma 1b 0.1 mg 3 times per week subcutaneously in addition to intensive i.v., oral and nebulized anti-mycobacterial treatment. This led to a clinical stabilization at very low FEV1 levels (mean 36.6% pred) compared to normal FEV1 levels (mean 102.9% pred) prior to infection.
3.2. Molecular Characterization of M. abscessus
The most prevalent NTM in the study population was M. abscessus: 12 of 13 NTM positive patients had at least one positive sample for M. abscessus, for one patient the NTM species was not characterized. 4 patients with M. abscessus carried a second NTM species (Table 2).
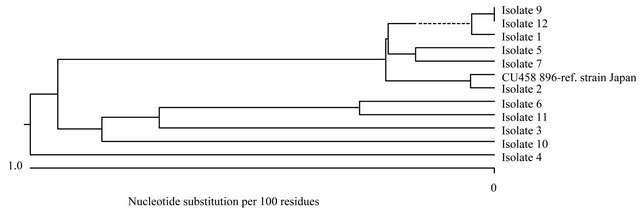
MLSA showed no dominant M. abscessus clone (Figure 3(a)). 6 patients had isolates with unique MLSA pattern. Patients 9 and 12 harboured strains that revealed identical nucleotide sequences in all DNA fragments analysed. We designated these strains cluster A. Patientto-patient transmission was highly unlikely (Figure 3(b)):
patient 9 was admitted from an external hospital, where he had been treated previously and where NTM were first detected, directly to our intensive care unit, where he died 2 months after admission. In patient 12 M. abscessus was detected once one year after patient 9 had died. Patient 1 carried a strain with almost identical sequences, revealing only 3 nucleotide substitutions within the 2773 bp concatenated sequence. However, patient 1 was seen in a different, associated clinic by a different physician and first detection had occurred 8 years prior to first detection in patient 9, also in an external hospital. Personal interaction between the 3 patients did not occur and places of residence were several 100 km apart.
The strains from patients 5 and 7 were closely related to each other (8 nucleotide substitutions), but cannot be assigned to a single clonal lineage. The sequences of the M. abscessus strain from patient 2 were identical to those of the M. abscessus reference strain previously isolated in Japan revealing 3 nucleotide substitutions and an 11 bp gap in the concatenated sequence 2 (Figure 3(a)).
4. Discussion
The detection rate of NTM-predominantly M. abscessus in this study was 3.6%. Reported prevalence rates for CF patients range between 2% and 28% [9,10,28,29]. The low detection rate in this cohort may have several reasons including the specimen preparation described above, the higher proportion of throat swabs compared to sputum in a largely paediatric cohort and the lack of routine use of long-term macrolides in this cohort [30].
The effect of chronic NTM infection on the clinical course of CF patients is variable [11,12]. Several studies
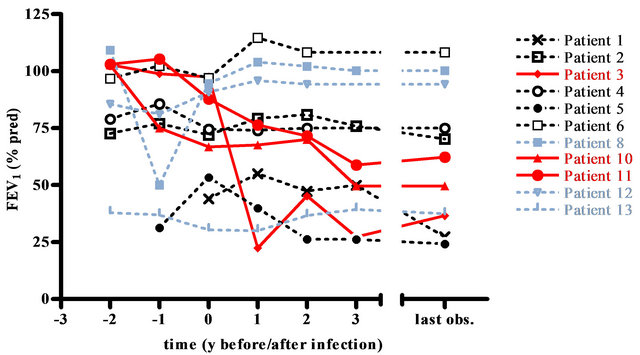
Figure 1. FEV1 (% pred) of NTM positive CF patients. Mean FEV1 (% predicted) values of 11 NTM positive CF patients (patients 7 and 9 are not shown due to insufficient data) during 7 different time intervals: 2 years and 1 year before first detection, at first detection, 1, 2 and 3 years after first detection and at the end of the observation period. Each symbol represents one patient; the red color marks the three patients with the most dramatic clinical deterioration, the grey lines indicate the patients with only a single NTM detection.
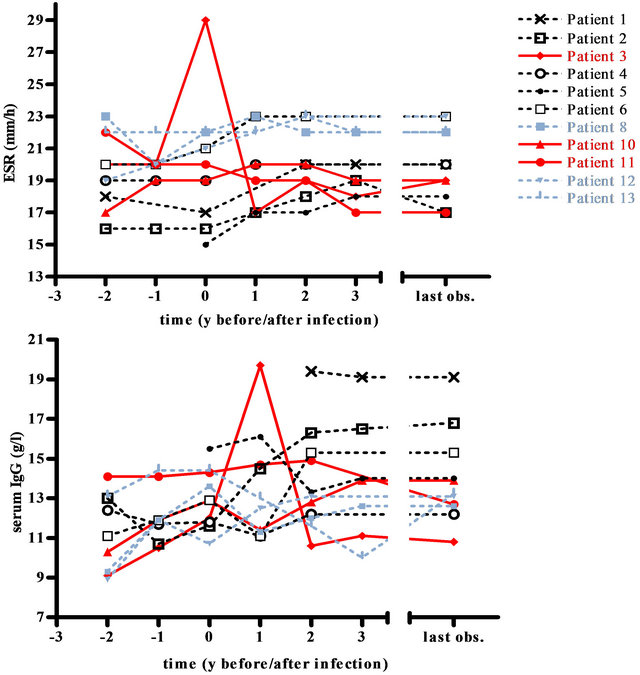
Figure 2. Inflammatory markers ESR (mm/h) and IgG (g/l) of NTM positive CF patients. Mean ESR (mm/h) and mean serum IgG (g/l) of 11 NTM positive CF patients (patients 7 and 9 are not shown due to insufficient data) during 7 different time intervals: 2 years and 1 year before first detection, at first detection, 1, 2 and 3 years after first detection and at the end of the observation period. Each symbol represents one patient; the red color marks the three patients with the most dramatic clinical deterioration, the grey lines indicate the patients with only a single NTM detection.
have shown that an infection with NTM, in particular chronic infection with M. abscessus, may be associated with a decline in lung function [31,32], while other large studies have failed to demonstrate this relationship [2,29, 33]. In the present study, patients with only a single NTM detection exhibited a stable clinical course. The clinical course of patients with longer NTM detection was variable. The inter-individual variance in the response to NTM infection was striking, the reasons remain elusive. Factors, such as genetic background, a more severe underlying CF disease, bacterial load, host defence and chronic exposure have to be considered. One NTM positive CF patient with a severe clinical deterioration was treated with IFN-gamma 1b 3 times per week subcutaneously in addition to the standard NTM-treatment. IFN-gamma therapy stabilized his clinical condition at however a very low level.
Systemic or inhaled IFN-gamma treatment for CF patients [34,35] can have a beneficial effect if administered in addition to antimycobacterial treatment. A few studies have reported that IFN-gamma, TNF-alpha and Il-12 are likely to protect against M. avium infection in non-CF patients, while Il-10 was considered immunosuppressive [3]. Another study demonstrated a decreased IFN-gamma response to mitogen stimulation [4].
Several previous studies have demonstrated a lack of patient-to-patient transmission for NTM [9,36]. The molecular analysis of 11 M. abscessus strains in this study showed that the majority of the strains were unique. The sequences of the M. abscessus strain from CF patient 2 were almost identical to those of the M. abscessus reference strain isolated in Japan. This may indicate that some clonal groups of M. abscessus are spread around the world representing natural reservoirs for M. abscessus colonisation or clinical infections of CF patients. 2 patients (9, 12) harboured identical strains, without evidence that patient-to-patient transmission can have occurred. Despite carrying identical strains patient 12 showed no clinical effect of the NTM infection, while patient 9 exhibited a fatal clinical course. Patient 1 carried a strain with almost identical sequences to patients 9 and 12 revealing only 3 nucleotide substitutions within
 (a)
(a)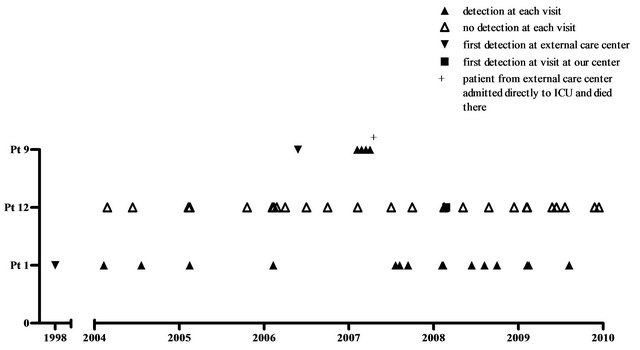 (b)
(b)
Figure 3. (a) Phylogenetic tree of M. abscessus-Mycobacterium abscessus MLST hsp65/ITS/rpoB/pgm/argH/cya (Clustal V). Phylogenetic tree of the concatenated DNA sequences for the 5 housekeeping genes and the ITS sequence of the different M. abscessus strains isolated from 11 CF patients. The sequences of reference M. abscessus strain CU458896 are given as comparator. The tree was constructed with ClustalV and the neighbor-joining method to distinguish the different isolates. The reliability of the tree topology was gauged by performing bootstrap analysis using 1000 replicates. Bootstrap values of 100% at nodes, where the configuration to the right was found, are shown. The bar shown in the graph provides scale to the branch lengths and represents the number of substitutions per 100 residues; (b) Overview of the clinical visits of the 3 patients with identical (pt 9 and 12)/closely related (pt 1) M. abscessus strains.
the 2773 bp concatenated sequence, however, patientto-patient transmission was also highly unlikely. A previous study using different molecular typing methods such as pulsed-field gel electrophoresis (PFGE) and enterobacterial repetitive intergenic consensus (ERIC) PCR identified closely related M. abscessus isolates in patients with no common epidemiological history. These isolates came from different sources and geographic locations and were isolated in different years [37]. A recent study also confirmed that several different CF patients in the same care center can share M. abscessus complex strains which are indistinguishable by variable-number tandem repeats (VNTR) and by targeting PCR primers to noncoding repetitive sequences (rep-PCR) [38]; the potential of patient-to-patient transmission was however not investigated.
The results of the current study speak in favour of the presence of distinct M. abscessus clones found in clinical samples and environmental sources as patient-to-patient transmission was unlikely. However, patient-to-patient transmission cannot generally be excluded if identical M. abscessus stains are present in a single cohort. Of interest in this regard is the report of a recent outbreak of a M. abscessus ss. massiliense in 5 patients with CF: transmission seems to have occurred by patient-to-patient respiratory spread or by contamination of the clinic or equipment [6] . Both repetitive unit-sequence of based PCR and pulsefield gel electrophoresis analysis of isolates from the 5 patients were identical.
We recommend that the standard transmission-based precautions applicable to all CF patients including NTM positive patients [39] should be extended. In addition to standard hand washing and repeated, rigorous surface and equipment cleaning after the consultation, we recommend separation of NTM positive patients and the use of face masks during their stay in the ambulatory care center or the hospital.
REFERENCES
- K. N. Olivier, J. R. Yankaskas and M. R. Knowles, “Nontuberculous Mycobacterial Pulmonary Disease in Cystic Fibrosis,” Seminars in Respiratory Infections, Vol. 11, No. 4, 1996, pp. 272-284.
- K. N. Olivier, D. J. Weber, R. J. Wallace Jr., A. R. Faiz, J. H. Lee, Y. Zhang, B. A. Brown-Elliot, A. Handler, R. W. Wilson, M. S. Schechter, L. J. Edwards, S. Chakraborti and M. R. Knowles, “Nontuberculous Mycobacteria. I: Multicenter Prevalence Study in Cystic Fibrosis,” American Journal of Respiratory and Critical Care Medicine, Vol. 167, No. 6, 2003, pp. 828-834. doi:10.1164/rccm.200207-678OC
- R. Vankayalapati, B. Wizel, B. Samten, D. E. Griffith, H. Shams, M. R. Galland, C. F. Von Reyn, W. M. Girard, R. J. Wallace Jr. and P. F. Barnes, “Cytokine Profiles in Immunocompetent Persons Infected with Mycobacterium avium Complex,” The Journal of Infectious Diseases, Vol. 183, No. 3, 2001, pp. 478-484. doi:10.1086/318087
- A. Safdar, D. A. White, D. Stover, D. Armstrong and H. W. Murray, “Profound Interferon Gamma Deficiency in Patients with Chronic Pulmonary Nontuberculous Mycobacteriosis,” American Journal of Medicine, Vol. 113, No. 9, 2002, pp. 756-759. doi:10.1016/S0002-9343(02)01313-X
- R. E. Colombo, S. C. Hill, R. J. Claypool, S. M. Holland and K. N. Olivier, “Familial Clustering of Pulmonary Nontuberculous Mycobacterial Disease,” Chest, Vol. 137, No. 3, 2010, pp. 629-634. doi:10.1378/chest.09-1173
- M. L. Aitken, A. Limaye, P. Pottinger, E. Whimbey, C. H. Goss, M. R. Tonelli, G. A. Cangelosi, M. A. Dirac, K. N. Olivier, B. A. Brown-Elliott, S. McNulty and R. J. Wallace Jr., “Respiratory Outbreak of Mycobacterium abscessus Subspecies Massiliense in a Lung Transplant and Cystic Fibrosis Center,” American Journal of Respiratory and Critical Care Medicine, Vol. 185, No. 2, 2012, pp. 231-232.
- J. M. Kilby, P. H. Gilligan, J. R. Yankaskas, W. E. Highsmith Jr., L. J. Edwards and M. R. Knowles, “Nontuberculous Mycobacteria in Adult Patients with Cystic Fibrosis,” Chest, Vol. 102, No. 1, 1992, pp. 70-75. doi:10.1378/chest.102.1.70
- M. L. Aitken, W. Burke, G. McDonald, C. Wallis, B. Ramsey and C. Nolan, “Nontuberculous Mycobacterial Disease in Adult Cystic Fibrosis Patients,” Chest, Vol. 103, No. 4, 1993, pp. 1096-1099. doi:10.1378/chest.103.4.1096
- K. N. Olivier, D. J. Weber, J. H. Lee, A. Handler, G. Tudor, P. L. Molina, J. Tomashefski and M. R. Knowles, “Nontuberculous Mycobacteria. II: Nested-Cohort Study of Impact on Cystic Fibrosis Lung Disease,” American Journal of Respiratory and Critical Care Medicine, Vol. 167, No. 6, 2003, pp. 835-840. doi:10.1164/rccm.200207-679OC
- I. Levy, G. Grisaru-Soen, L. Lerner-Geva, E. Kerem, H. Blau, L. Bentur, M. Aviram, J. Rivlin, E. Picard, A. Lavy, Y. Yahav and G. Rahav, “Multicenter Cross-Sectional Study of Nontuberculous Mycobacterial Infections among Cystic Fibrosis Patients, Israel,” Emerging Infectious Diseases, Vol. 14, No. 3, 2008, pp. 378-384. doi:10.3201/eid1403.061405
- S. Razvi and L. Saiman, “Nontuberculous Mycobacteria in Cystic Fibrosis,” The Pediatric Infectious Disease Journal, Vol. 26, No. 3, 2007, pp. 263-264. doi:10.1097/01.inf.0000256964.30181.df
- K. N. Olivier, “The Natural History of Nontuberculous Mycobacteria in Patients with Cystic Fibrosis,” Paediatric Respiratory Reviews, Vol. 5, No. A, 2004, pp. S213- S216.
- D. Schewe, M. Kappler and M. Griese, “Instructions for Infection Control in Outpatient Care of Patients with Cystic Fibrosis,” European Journal of Medical Research, Vol. 10, No. 8, 2005, pp. 345-351.
- K. De Boeck, F. Vermeulen, S. Wanyama and M. Thomas, “Inhaled Corticosteroids and Lower Lung Function Decline in Young Children with Cystic Fibrosis,” European Respiratory Journal, Vol. 37, No. 5, 2011, pp. 1091-1095. doi:10.1183/09031936.00077210
- P. K. Kent, “Public Health Mycobacteriology: A Guide for the Level III Laboratory,” US Department of Health and Human Services, Centers for Disease Control, Atlanta, 2011, in Print.
- E. Richter, J. Beer, R. Diel, D. Hillemann, H. Hoffmann, M. Klotz, H. Mauch and S. Rüsch-Gerdes, “MIQ 05: Qualitätsstandards in der Mikrobiologisch-Infektiologischen Diagnostik-Tuberkulose und Mykobakteriose,” Elsevier, Urban & Fischer, 2011, pp. 11-45.
- P. Kirschner, B. Springer, U. Vogel, A. Meier, A. Wrede, M. Kiekenbeck, F. C. Bange and E. C. Bottger, “Genotypic Identification of Mycobacteria by Nucleic Acid Sequence Determination: Report of a 2-Year Experience in a Clinical Laboratory,” Journal of Clinical Microbiology, Vol. 31, No. 11, 1993, pp. 2882-2889.
- J. Makinen, M. Marjamaki, H. Marttila and H. Soini, “Evaluation of a Novel Strip Test, GenoType Mycobacterium CM/AS, for Species Identification of Mycobacterial Cultures,” Clinical Microbiology and Infection, Vol. 12, No. 5, 2006, pp. 481-483. doi:10.1111/j.1469-0691.2006.01380.x
- A. Roth, U. Reischl, A. Streubel, L. Naumann, R. M. Kroppenstedt, M. Habicht, M. Fischer and H. Mauch, “Novel Diagnostic Algorithm for Identification of Mycobacteria Using Genus-Specific Amplification of the 16S- 23S rRNA Gene Spacer and Restriction Endonucleases," Journal of Clinical Microbiology, Vol. 38, No. 3, 2000, pp. 1094-1104.
- E. Macheras, A. L. Roux, S. Bastian, S. C. Leao, M. Palaci, V. Sivadon-Tardy, C. Gutierrez, E. Richter, S. Rusch-Gerdes, G. Pfyffer, T. Bodmer, E. Cambau, J. L. Gaillard and B. Heym, “Multilocus Sequence Analysis and rpoB Sequencing of Mycobacterium abscessus (Sensu Lato) Strains,” Journal of Clinical Microbiology, Vol. 49, No. 2, 2011, pp. 491-499. doi:10.1128/JCM.01274-10
- T. Adekambi, P. Colson and M. Drancourt, “rpoB-Based Identification of Nonpigmented and Late-Pigmenting Rapidly Growing Mycobacteria,” Journal of Clinical Microbiology, Vol. 41, No. 12, 2003, pp. 5699-5708. doi:10.1128/JCM.41.12.5699-5708.2003
- V. A. Steingrube, J. L. Gibson, B. A. Brown, Y. Zhang, R. W. Wilson, M. Rajagopalan and R. J. Wallace Jr., “PCR Amplification and Restriction Endonuclease Analysis of a 65-Kilodalton Heat Shock Protein Gene Sequence for Taxonomic Separation of Rapidly Growing Mycobacteria,” Journal of Clinical Microbiology, Vol. 33, No. 1, 1995, pp. 149-153.
- M. A. Yakrus, S. M. Hernandez, M. M. Floyd, D. Sikes, W. R. Butler and B. Metchock, “Comparison of Methods for Identification of Mycobacterium abscessus and M. chelonae Isolates,” Journal of Clinical Microbiology, Vol. 39, No. 11, 2001, pp. 4103-4110. doi:10.1128/JCM.39.11.4103-4110.2001
- Y. Zhang, M. A. Yakrus, E. A. Graviss, N. Williams- Bouyer, C. Turenne, A. Kabani and R. J. Wallace Jr., “Pulsed-Field Gel Electrophoresis Study of Mycobacterium abscessus Isolates Previously Affected by DNA Degradation,” Journal of Clinical Microbiology, Vol. 42, No. 12, 2004, pp. 5582-5587. doi:10.1128/JCM.42.12.5582-5587.2004
- F. Ripoll, S. Pasek, C. Schenowitz, C. Dossat, V. Barbe, M. Rottman, E. Macheras, B. Heym, J. L. Herrmann, M. Daffe, R. Brosch, J. L. Risler and J. L. Gaillard, “Non Mycobacterial Virulence Genes in the Genome of the Emerging Pathogen Mycobacterium abscessus,” PLoS One, Vol. 4, No. 6, 2009, e5660. doi:10.1371/journal.pone.0005660
- D. G. Higgins, A. J. Bleasby and R. Fuchs, “CLUSTAL V: Improved Software for Multiple Sequence Alignment,” Computer Applications in the Biosciences, Vol. 8, No. 2, 1992, pp. 189-191.
- D. E. Griffith, T. Aksamit, B. A. Brown-Elliott, A. Catanzaro, C. Daley, F. Gordin, S. M. Holland, R. Horsburgh, G. Huitt, M. F. Iademarco, M. Iseman, K. Olivier, S. Ruoss, C. F. von Reyn, R. J. Wallace Jr. and K. Winthrop, “An Official ATS/IDSA Statement: Diagnosis, Treatment, and Prevention of Nontuberculous Mycobacterial Diseases,” American Journal of Respiratory and Critical Care Medicine, Vol. 175, No. 4, 2007, pp. 367-416. doi:10.1164/rccm.200604-571ST
- L. Leitritz, M. Griese, A. Roggenkamp, A. M. Geiger, V. Fingerle and J. Heesemann, “Prospective Study on Nontuberculous Mycobacteria in Patients with and without Cystic Fibrosis,” Medical Microbiology and Immunology, Vol. 193, No. 4, 2004, pp. 209-217. doi:10.1007/s00430-003-0195-9
- C. R. Esther Jr., M. M. Henry, P. L. Molina and M. W. Leigh, “Nontuberculous Mycobacterial Infection in Young Children with Cystic Fibrosis,” Pediatric Pulmonology, Vol. 40, No. 1, 2005, pp. 39-44. doi:10.1002/ppul.20222
- M. Renna, C. Schaffner, K. Brown, S. Shang, M. H. Tamayo, K. Hegyi, N. J. Grimsey, D. Cusens, S. Coulter, J. Cooper, A. R. Bowden, S. M. Newton, B. Kampmann, J. Helm, A. Jones, C. S. Haworth, R. J. Basaraba, M. A. DeGroote, D. J. Ordway, D. C. Rubinsztein and R. A. Floto, “Azithromycin Blocks Autophagy and May Predispose Cystic Fibrosis Patients to Mycobacterial Infection,” Journal of Clinical Investigation, Vol. 121, No. 9, 2011, pp. 3554-3563. doi:10.1172/JCI46095
- B. Petrini, “Mycobacterium abscessus: An Emerging Rapid-Growing Potential Pathogen,” APMIS, Vol. 114, No. 5, 2006, pp. 319-328. doi:10.1111/j.1600-0463.2006.apm_390.x
- D. E. Griffith, “Emergence of Nontuberculous Mycobacteria as Pathogens in Cystic Fibrosis” American Journal of Respiratory and Critical Care Medicine, Vol. 167, No. 6, 2003, pp. 810-812. doi:10.1164/rccm.2301001
- C. Pierre-Audigier, A. Ferroni, I. Sermet-Gaudelus, M. Le Bourgeois, C. Offredo, H. Vu-Thien, B. Fauroux, P. Mariani, A. Munck, E. Bingen, D. Guillemot, G. Quesne, V. Vincent, P. Berche and J. L. Gaillard, “Age-Related Prevalence and Distribution of Nontuberculous Mycobacterial Species among Patients with Cystic Fibrosis,” Journal of Clinical Microbiology, Vol. 43, No. 7, 2005, pp. 3467-3470. doi:10.1128/JCM.43.7.3467-3470.2005
- S. M. Holland, E. M. Eisenstein, D. B. Kuhns, M. L. Turner, T. A. Fleisher, W. Strober and J. I. Gallin, “Treatment of Refractory Disseminated Nontuberculous Mycobacterial Infection with Interferon Gamma. A Preliminary Report,” The New England Journal of Medicine, Vol. 330, No. 19, 1994, pp. 1348-1355. doi:10.1056/NEJM199405123301904
- H. Mussaffi, J. Rivlin, I. Shalit, M. Ephros and H. Blau, “Nontuberculous Mycobacteria in Cystic Fibrosis Associated with Allergic Bronchopulmonary Aspergillosis and Steroid Therapy,” European Respiratory Journal, Vol. 25, No. 2, 2005, pp. 324-328. doi:10.1183/09031936.05.00058604
- F. C. Bange, B. A. Brown, C. Smaczny, R. J. Wallace Jr. and E. C. Bottger, “Lack of Transmission of Mycobacterium abscessus among Patients with Cystic Fibrosis Attending a Single Clinic,” Clinical Infectious Diseases, Vol. 32, No. 11, 2001, pp. 1648-1650. doi:10.1086/320525
- J. L. Sampaio, C. Viana-Niero, D. de Freitas, A. L. Hofling-Lima and S. C. Leao, “Enterobacterial Repetitive Intergenic Consensus PCR Is a Useful Tool for Typing Mycobacterium chelonae and Mycobacterium abscessus Isolates,” Diagnostic Microbiology and Infectious Disease, Vol. 55, No. 2, 2006, pp. 107-118. doi:10.1016/j.diagmicrobio.2006.01.006
- K. A. Harris, D. T. Kenna, C. Blauwendraat, J. C. Hartley, J. F. Turton, P. Aurora and G. L. Dixon, “Molecular Fingerprinting of Mycobacterium abscessus Strains in a Cohort of Pediatric Cystic Fibrosis Patients,” Journal of Clinical Microbiology, Vol. 50, No. 5, 2012, pp. 1758- 1761. doi:10.1128/JCM.00155-12
- L. Saiman and J. Siegel, “Infection Control in Cystic Fibrosis,” Clinical Microbiology Reviews, Vol. 17, No. 1, 2004, pp. 57-71. doi:10.1128/CMR.17.1.57-71.2004
NOTES
*Shared last authorship.
#Corresponding authors.

