Natural Resources
Vol.5 No.11(2014), Article ID:49400,19 pages
DOI:10.4236/nr.2014.511056
An Online mtDNA Tool for Identification of Neotropical Psittacid Species and Taxonomic Issues: A Study Case of the Amazona ochrocephala Complex
Anderson Vieira Chaves1, Rodrigo Octavio de Paiva Queiroz-Filho1,2, Fabiano Augusto Assunção Silva1, Cristina Yumi Miyaki3, Fabrício Rodrigues dos Santos1*
1Departamento de Biologia Geral, Instituto de Ciências Biológicas (ICB), Universidade Federal de Minas Gerais, Belo Horizonte, Brazil
2Polícia Federal, Belo Horizonte, Brazil
3Departamento de Genética e Biologia Evolutiva, Instituto de Biociências, Universidade de São Paulo, São Paulo, Brazil
Email: avc.bio@gmail.com, paiva.ropqf@gmail.com, fabianoaas@gmail.com, cymiyaki@usp.br, *fsantos@icb.ufmg.br
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 19 June 2014; revised 21 July 2014; accepted 6 August 2014
ABSTRACT
Parrots are among the most popular pets in the world and they are also some of the most illegally traded, particularly in Brazil. Some computational tools were recently developed by researchers based on molecular databases for taxonomy support, forensic identification and conservation purposes. In this study, the DNA Surveillance platform was used to build an online database tool for molecular identification of Brazilian Psittacids using DNA sequences of six mitochondrial genes. To illustrate possible taxonomic issues of the online tool due to interspecific hybridization or unresolved taxonomy, we focused on Amazona aestiva that is considered as one of the most common parrots in Brazil, commonly bred as pets, and considered to be part of a species complex with Amazona ochrocephala from South America. We provide three curated sequence databases, which allow the species identification of individuals or tissue samples of birds of the Psittacidae family using mitochondrial DNA markers, and a comprehensive description of a taxonomic issue involving the A. ochrocephala complex. The results obtained corroborate previous studies suggesting that these species are not reciprocally monophyletic, due to either an ancient hybridization in central Brazil, or, they maybe just are morpho-varieties of the same species. Alternatively, if A. aestiva and A. ochrocephala were considered as sister species, the data could be interpreted either as a result of secondary contact or incipient speciation. Beyond the use of mtDNA for species identification, the high mtDNA haplotype diversity observed in A. aestiva indicates its potential use in discrimination of lineages that could be an important auxiliary tool to certify the captive origin of legally commercialized parrots.
Keywords:Amazona aestiva, Illegal Trade of Birds, Psittaciformes, Phylogeography, DNA-Surveillance

1. Introduction
Psittacidae is the avian family with the highest percentage of endangered species, with 27% of its species listed as vulnerable, endangered or critically endangered [1] . Psittacids of genus Amazona are popularly known as parrots and are among the most threatened of the order Psittaciformes [2] . This genus is comprised of 33 species; 16 of them are threatened and two (A. martinicana and A. violacea) are extinct, according to the IUCN Red List [3] . All species are listed in Appendix I or II of CITES [4] . The evolutionary history of genus Amazona was investigated in a few studies [5] -[7] . The study by Russelo and Amato [5] indicated that Amazona is not a monophyletic group. Amazona aestiva, which is considered as one of the most common parrots in Brazil, forms a species complex with A. ochrocephala from South America [5] [7] .
Parrots, by virtue of their high ability to imitate the human voice, besides their beauty, gentleness and intelligence, are among the most sought birds as pets in the world and they are common in the illegal traded [8] . It is estimated that only about 5% of psitacids traded in Brazil are captively bred, the remainder being withdrawn from nature [9] [10] . Seizures between 1999 and 2000 by the environment agency from Brazil (IBAMA), indicated that parrots were the 3rd most common animal group illegally traded [9] .
Some global efforts aim to expedite and streamline molecular approaches to help taxonomy, forensic identification and conservation. One initiative is the Barcoding of Life Data Systems, which links curated specimens and their corresponding molecular data (DNA barcodes), and is publicly accessible on the web [11] . Since 1990’s, genetic tracking techniques were introduced in the surveillance of protected animals, in the attempt to inhibit the illegal trade and trafficking. Important studies detected meat from the illegal hunting of whales in popular markets in Japan and Korea, triggering a revision of international agreements controlling whaling, and promoting new resolutions of the International Whaling Commission (IWC) [12] -[14] . Besides, genetic monitoring was adopted for these animal products’ control, and it was also used on the surveillance of countries under the jurisdiction of CITES [15] , which reported recently that illegal trade routes of whale meat between Japan, Korea and the United States remain active [14] .
Custom databases for molecular identification of particular taxa in the DNA Surveillance online tool [16] can be used by forensic genetics against biopiracy and illegal trade. This surveillance started with a tool for identification of cetaceans that includes molecular data organized in a database and identification protocols [13] . The applicability of this tool has been expanded to other taxa, such as for the identification of morphologically similar rat species (Rattus spp.) from Southeast Asia and Pacific region [17] , and the identification of rodent preys in the stomach content of Carnivores based on the technique of mini-barcodes obtained from DNA extracted from carnivore faeces [18] .
In this study, we added in the DNA Surveillance online tool three databases for molecular identification of Brazilian psitacids based on mitochondrial sequences. We also discuss possible limitations of this tool due to taxonomic issues, using as example the species complex Amazona aestiva/A. ochrocephala. We sequenced three regions of two mitochondrial genes of various captive specimens from these taxa and analyzed them in the online tool to identify their species. The results showed that phylogenetic and taxonomic issues are essential information to allow a correct species identification.
2. Material and Methods
2.1. Samples, Amplifications and Sequencing
The analyzed blood samples were taken from 46 specimens of Amazona aestiva (two A. aestiva xanthopterix, and the remaining, A. aestiva aestiva) kept in captivity in the Vale Verde Ecological Park, a legal commercial breeding facility that is located in the city of Betim, Minas Gerais state, Brazil. All birds are banded, which allows for maintenance of individual records of the birds. The majority of the animals used in this study are founders or matrices from nature, which were originally apprehended by environmental monitoring agents of the Brazilian Government.
Segments of two mitochondrial genes (ND2 and COI) were amplified and sequenced. Early studies with psitacids used the 3’ end of COI gene, but with the DNA-Barcode initiative, a growing number of sequences of the 5’ end of COI are being produced. Thus, we generated and analyzed these two sections of this gene. The nomenclature and abbreviations used for phylogenetic groups and clades found in our results, tables and figures follow the same ones used in previous studies [7] [19] [20] .
For DNA extraction, we used a modified phenol-chloroform-isoamyl alcohol protocol [21] . DNA samples were stored in the collection of the Universidade Federal de Minas Gerais (BD-LBEM), licensed by the Brazilian Ministry of Environment (MMA/CGEN).
Two mitochondrial genes were entirely amplified to avoid amplification of NuMts [22] . The ND2 gene (1041 bp) was amplified with specific primers: H6313 and L5216 [23] , and the COI gene (1540pb) was amplified with primers L6615 and H8121 [24] . PCR reactions of ND2 were performed under the following conditions: 94˚C for 2 min, 35 cycles of 94˚C for 30 s, 63˚C for 40 s, 72˚C for 2 min, 94˚C for 40 s and a final extension of 10 min at 72˚C. For COI, we used the same cycle but with annealing temperatures ranging from 58˚C to 61˚C for 40 seconds. The amplifications were carried out in 12.5 μL reactions containing 0.5 U of Taq polymerase (Phoneutria), 1 × buffer with 1.5 mM MgCl2 (Phoneutria), 200 μM dNTPs 0.5 μM each primer, and 2 μL of genomic DNA (~40 ng). The amplification products were purified by precipitation in PEG 8000 (20% polyethyleneglycol, 2.5 M NaCl) and finally dissolved in ultrapure water [21] . The sequencing reactions consisted of 35 cycles of 95˚C for 25 s, 50˚C for 15 s, 60˚C for 3 min in a total volume of 10 μL, which contained 4 μL of the sequencing Kit (ET DYE Terminator Kit for Mega BACE, Amersham Biosciences), 3 μL of ultrapure water, 2 μL of purified PCR amplicons, and 1 μL of each primer (0.5 μM final concentration). The following primers were used for sequencing reactions: H6313 and L5216 for ND2 gene, and socoiF1 and H6035COI_Tyr [21] , and LCO1490, HCO2198 [24] for COI gene. Sequencing products were purified using ammonium acetate and ethanol, then dissolved with formamide-EDTA buffer and run in the automatic sequencer MegaBACE 1000 (Amersham Biosciences).
2.2. Molecular Analyses
Contigs for each sample were obtained from a total of four forward and reverse sequences, derived from at least two different PCR products, using programs Phred v 0.20425 [25] , Phrap v0.990319 and Consed v19.0 [26] . High quality consensus ND2 and COI sequences presented at least a Phred 20 score (99% confidence) for every nucleotide position. Final consensus sequences for each individual are deposited in GenBank (accession numbers JX476306 to JX476426). ND2 and/or COI sequences from 86 individuals of A. aestiva and 53 of A. ochrocephala (30 of South American subspecies and 23 of Central American subspecies) were downloaded from GenBank [5] [7] [20] . Sequence alignments were obtained using MUSCLE [27] implemented in the program MEGA v5.01 [28] .
The programs Arlequin v3.15 [29] and DnaSP v5 [30] were used to estimate the hierarchical distribution of genetic diversity through the Analysis of Molecular Variance (AMOVA) [31] , and genetic differences between species (A. aestiva, A. ochrocephala), subspecies (A. a. aestiva, A. a. xanthopteryx, A o. ochrocephala, A. o. xantholaema, A. o. nattereri, A. o. panamensis, A. o. tresmariae, A. o. oratrix, A. o. auropalliata, A. o. belizensis and “A. ochocephala CO-VE”) and groups (South America [SA], Central America [CA] and Northen South America [NSA]). Arlequin also calculated the haplotype diversity (h), average number of nucleotide differences (k), nucleotide diversity (π), the number of sites with substitutions (S), the composition of nucleotides, frequency haplotypes, Tajima’s D and Fu’s Fs neutrality tests, which may reveal signs of population expansion (when negative, and P value is less than 0.05). In Arlequin, analyses were performed with the model of pairwise distance (p). We built a haplotype network with the median-joining algorithm (MJ) [32] available in the software NETWORK 4.6 for inferring phylogenetic relationships among haplotypes and their possible geographical correlation.
2.3. Molecular Identification Databases and Tools
Mitochondrial sequences from GenBank (Supplementary Material SM2) were used in the three databases. The “Brazilian Parrots” database holds all mitochondrial sequences from all Psittacidae species, and some Amazona aestiva and A. ochrocephala sequences generated by our approaches. The database “Amazona Identification” presents ND2 and COI sequences of individuals from known geographic origin of the Amazona aestiva/A. ochrocephala complex. The “All Amazona” database has ND2 and COI sequences from genus Amazona. These databases were added to the DNA Surveillance platform (http://www.dna-surveillance.auckland.ac.nz/). ClustalX 2.012 [33] was used to align and generate these datasets in PHYLIP format.
The query results were extracted in FASTA format and TinySeqXML flatfiles. FASTA files were concatenated with the “Amazona Identification” dataset, aligned with ClustalX 2.012, manually corrected with MEGA v5.01 [28] , generated the “All Amazona” database haplotypes on DnaSP v5 [30] and realigned at ClustalX to generate DNA Surveillance datasets. With MS Excel 2007, the TinySeqXML flatfiles were converted as spreadsheets and used as database for bulk annotation of DNA Surveillance datasets, exported as Tabbed Separated Values format (TSV).
The online identification protocol aligns the user input sequence by a simple profile alignment against the prealigned dataset of reference sequences, using the penalty values: transitions = 1, transversions = 2, gap creation = 3, gap extension = 1, with the F84 model of substitution to calculate genetic distances and a neighbor-joining (NJ) tree, which is built from the table of distances, graphically and in Newick text format [34] .
To test the species identification performance of the domain “Brazilian Psittacidae v1.0” database we used non-curated GenBank sequences obtained with the Entrez queries: “txid9224[Organism:exp] COI”, “txid9224 [Organism:exp] ND2”, “txid9224 [Organism:exp] CytB”, “txid9224 [Organism:exp] 16S”and “txid9224 [Organism:exp] Control Region”. Functionality and confidence tests were performed using the “fvv039” Amazona sp. individual as input.
3. Results
3.1. Brazilian Parrots Platform
The three databases and the identification tool are available in the DNA-Surveillance website in the following link: http://dna-surveillance.fos.auckland.ac.nz:23060/page/parrots/title.
The “All Amazona species” database had 40 COI sequences from 23 species and 16 ND2 sequences from seven species. The “Amazona Identification” database had 11 COI sequences and 11 ND2 haplotypes from the A. aestiva/A. ochrocephala species complex. The “Brazilian Psittacidae v1.0” database had 55 COI sequences from 19 species, 107 ND2 sequences from 25 species, 229 CytB sequences from 68 species, 62 16SrDNA sequences from 25 species and 149 Control Region sequences from 44 species.
Figure 1 shows the result of the identification test of the COI sequence from sample fvv039 using the “Amazona Identification” database. This input sequence (query) grouped with those from A. aestiva and A. ochrocephala, confirming its identification (Supplementary Material SM1). The COI sequence from this same sample (fvv039) was tested using the “Brazilian Psittacidae v1.0” database and it correctly grouped with sequences of other samples of Amazona (Supplementary Material SM2).
3.2. Genetic Diversity and Phylogenetic Relationships
Sequence alignment matrices of genus Amazona contained 502 characters of the 5’ end of ND2, and 401 characters of the 3’ end and 474 of 5’ end of COI. ND2 showed the greatest number of variable sites, as well as the highest values of haplotype and nucleotide diversities (0.842 +/−0.032 and 0.012 +/−0.006 respectively). The 5’ end COI segment (DNA Barcode) showed the lowest genetic variation, presenting only two parsimony informative sites among eight variable ones, but it was informative for phylogenetic reconstructions.
Diversity indices, numbers of transitions and transversions, and neutrality test results are shown in Table1
Based on 903 characters of the concatenated sequence of ND2 and the 3’ end of COI of genus Amazona, we found 34 haplotypes with 74 substitutions (69 synonymous and five non-synonymous) in 73 polymorphic sites, of which 46 were parsimony informative sites, and 27 corresponded to singleton sites. Genetic diversity was estimated to be 0.016 ± 0.006 and haplotype diversity, 0.884 ± 0.028 (Table 1). The neutrality tests showed no significant values.
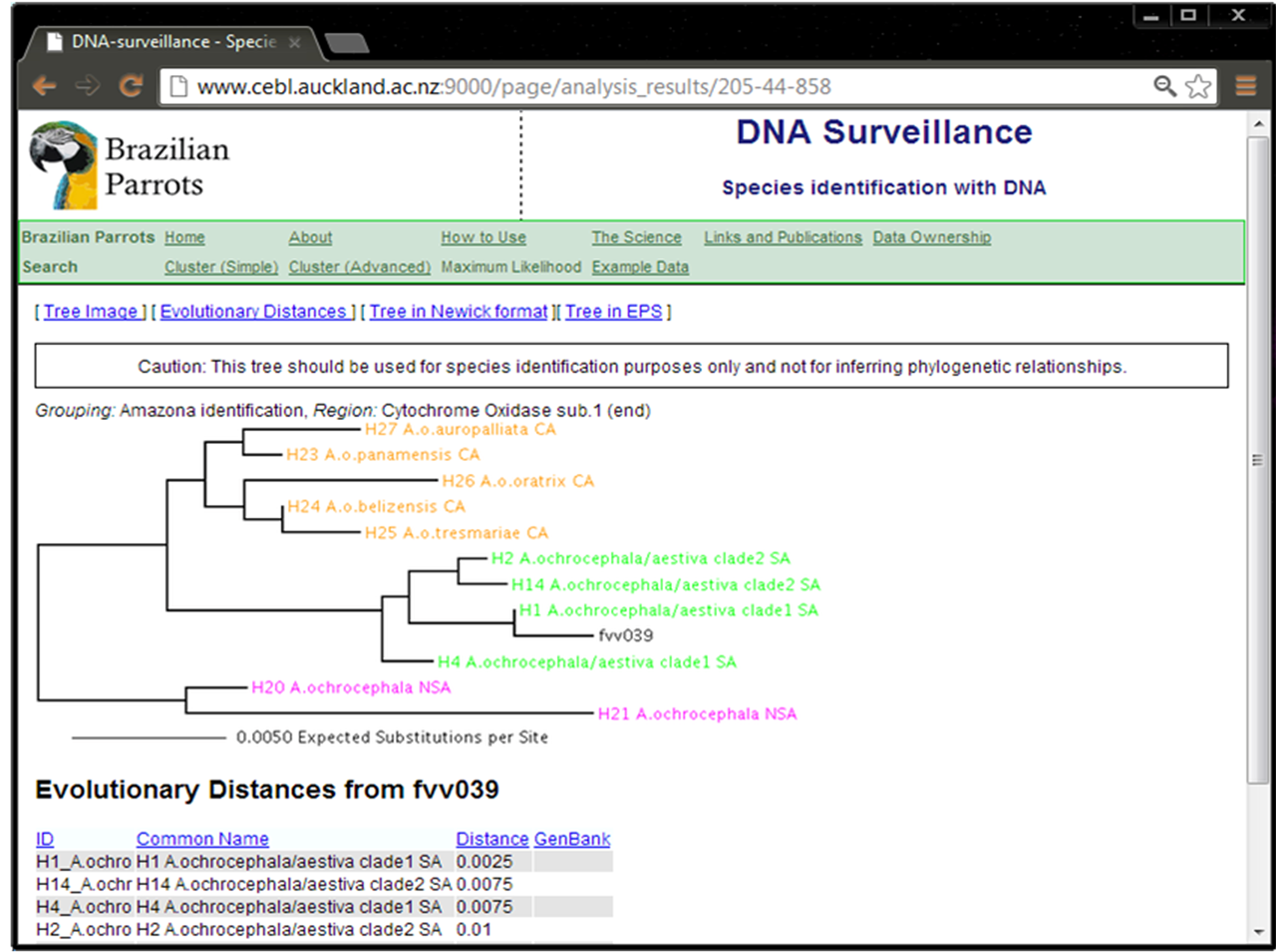
Figure 1. Identification result using sample fvv039 as input data and the “Amazona Identification” database.
Table 1. Indices of genetic diversity, numbers of transitions and transversions, and results of neutrality test indices. H: number of haplotypes, h: haplotype diversity, K: mean number of pairwise differences, = π: nucleotide diversity (average over loci), S: no. of sites with substitutions, ts: no. transitions, tv: no. transversions, D: Tajima’s D, and Fu’s FS test. Statistically significant values are highlighted in gray.
AMOVA based on the concatenated ND2 and 3’ end COI matrix showed moderate Fst and ΦST values (6.9% and 13.5% respectively) between A. aestiva and A. ochrocephala, which suggested that the greatest genetic diversity is found among individuals and populations within the complex, and not between these taxa (Table 2).
Among the 502 characters of ND2 from the 45 captive A. aestiva individuals, 13 haplotypes were found, 10 of which were not yet described for this species. The remaining three haplotypes are shared with A. ochrocephala from South America (haplotypes H1, H5 and H10). Genetic diversity was estimated to be 0.003 ± 0.001and haplotype diversity was 0.656 ± 0.093. For the COI DNA Barcode segment (474 characters), 10 haplotypes were found, seven of them were only found in the captive specimens, sequenced here and two haplotypes (H1 and H4) were shared with two of three specimens previously sequenced [19] .
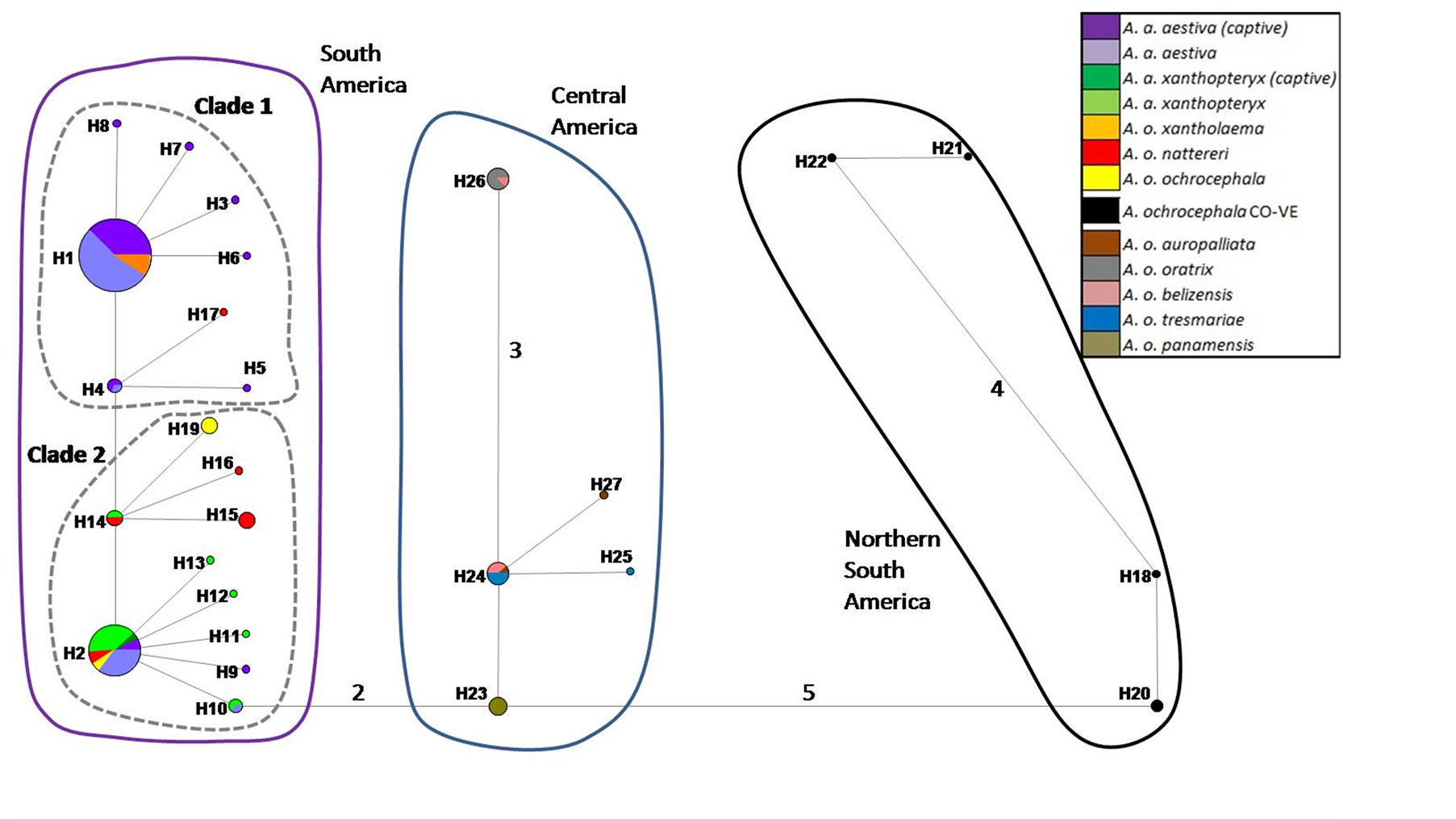
The haplotype network based on the concatenated sequence matrix of the A. aestiva/A. ochocephala complex showed three main divergent groups (South America [SA], Central America [CA] and northern South America [NSA]) (Figure 2). SA showed many highly divergent haplotypes, with two subgroups (Clade 1and Clade2). The star-shaped topology observed in Clade 1 suggests recent population expansion. The most common haplotypes were H1 (n = 27) and H5 (N = 11), which altogether account for the majority of the A. aestiva specimens.
NSA corresponds to a highly divergent A. ochrocephala individual from Colombia, which is frequently observed in close relationship with other specimens from Venezuela using other molecular markers in previous
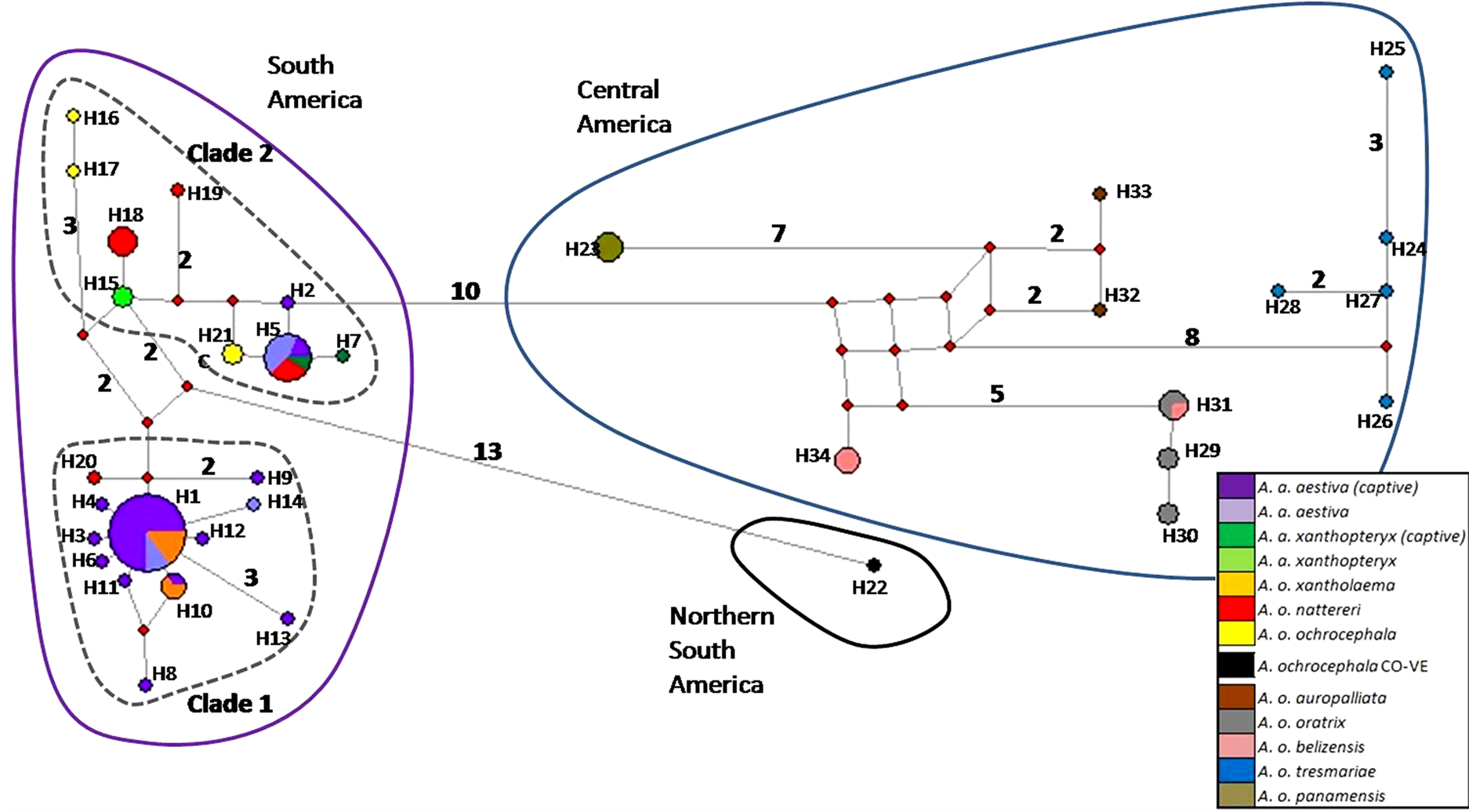
Figure 2. Haplotype network based on 902 bp of ND2 and COI of captive A. aestiva and other specimens of the A. aestiva/ A. ochocephala complex. The number of substitutions is proportional to the length of the line connecting haplotypes and is also shown. The size of the circles is proportional to the frequency of the haplotype obtained in this sample. Red diamonds represent median vectors.
studies (Supplementary Material SM3 and Supplementary Material SM4). CA group corresponds to A. ochrocephala auropalliata, A. o. oratrix, A. o. belizensis, A. o. tresmariae, and A. o. panamensis and it forms a clearly separated group from the A. aestiva/A. ochrocephala complex from South America. All haplotypes found in captive specimens sequenced here were grouped in Clades 1 and 2 from South America.
The haplotype network based on the 3’ end COI segment showed the same topology as previously obtained by Caparroz et al. [20] , but included more branches and new haplotypes (Supplementary Material SM3). The same general group arrangement was also found based only on ND2 (Supplementary Material SM4). The haplotype network based on COI-DNA Barcode segment showed no genetic structure (Supplementary Material SM5), but its star-like shape is congruent with the results of tests of neutrality tests, thus indicating recent population expansion (Table 1 and Figure 1).
Based on the sampling localities of previously sequenced samples of the A. aestiva/A. ochrocephala complex [6] [7] [20] we tried to assign the possible geographical origin of the captive parrots sequenced here (Table 3). However, as the majority of the haplotypes presents wide distribution, this analysis not conclusive.
4. Discussion
4.1. Phylogenetic Relationships and the Limits of Species in the Amazona ochrocephala Complex
Our data endorse previous studies [6] [7] [20] as members of the A.aestiva/A. ochrocephala complex are more closely related amongst themselves than to other A. ochrocephala subspecies from Central America.We confirmed previous results [6] [7] [20] [35] that indicated that within this species complex there are three lineages (South America—SA, Central America—CA and North South America—NSA). The current taxonomy of these (once) subspecies from Central America recognizes three species (A. tresmarie, A. oratrix, and A. auropalliata) [36] . In addition, Amazona barbadensis [35] (from several isolated populations in dry forest in the coastal areas of Southern Venezuela and Caribbean region) has a strong and basal affiliation with the NSA lineage of the A. aestiva/A. ochrocephala complex from South America.
Eberhard and Bermingham [6] recovered a polytomy involving these three lineages. In the study of Ribas et al. [9] the SA lineage is sister to the CA lineage, and NSA appears in a basal position, suggesting an ancestral distribution of the group in northern South America with subsequent diversification to the north and south. Urantówka et al. [35] tested combinations of genetic markers and exclusion of rapidly evolving sites, and provided support for the close relationship between NSA and SA.
Our results indicate that none of the subspecies of the A. aestiva/A. ochrocephala complex from South America (A. o. ochrocephala, A. o. natereri, A. o. xantholaema, A. a. aestiva, and A. a. xanthopteryx) formed reciprocally monophyletic groups. Instead, A. aestiva was paraphyletic in relation to A. ochrocephala, as observed in previous studies [6] [7] [20] [35] .
Sampling four places representing a lineage that is distributed across the whole Amazon Basin, Eberhard and
Brazilian States (TO: Tocantins, BA: Bahia, MG: Minas Gerais, DF: Distrito Federal, PA: Pará, MS: Mato Grosso do Sul, AC: Acre).
Bermingham [6] considered that there is a strong phylogenetic structure in the CA lineage, contrasting with a lack of structure in the SA lineage. Ribas et al. [7] showed in a larger sampling (three out of 12 localities represented A. aestiva) that there is genetic structure in the SA lineage, with two well-supported and closely related clades. Caparroz et al. [20] sampled over eight locations for A .aestiva, including six in central Brazil and two in northern Argentina, but the increase of sampling did not show a more structured phylogeographic pattern. However, Caparroz et al. [20] observed a major divergence between two groups: “north-eastern” (corresponding to central Brazil states of Bahia, Tocantins, Minas Gerais, Distrito Federal, and Goias), and “southwestern” (Mato Grosso do Sul in Brazil, and northern Argentina). These groups are respectively related to clades1 and 2 observed here and by Ribas et al. [7] .
Urantówka et al. [35] suggest that NSA represent the descendants of the most ancient lineage of A. aestiva/A. ochrocephala complex, that about 1 Mya colonized Venezuela. Thereafter, it colonized Central America originating the CA lineage and, southern regions of South America originating the SA clade. Within the SA lineage, Caparroz et al. [20] suggested that Mato Grosso do Sul could represent the ancestral distribution of the two A. aestiva clades identified in their study and the current distribution of lineages could be the result of a recent population expansion towards the northeast and the southwest. Considering that the divergence between these two clades occurred in the end of Middle Pleistocene (about 300,000 years ago [20] ), this expansion could be related to climatic changes and the cycles of expansion and contraction of the savannah areas at that time [20] .
As in [7] [20] [35] , we also consider the process of incomplete lineage sorting unlikely to explain the current distribution of the genetic variability of the A. aestiva/A. ochrocephala complex from South America, because if this was the case, A. aestiva groups should have maintained the same pattern of haplotype distribution found in A. ochrocephala during the population expansion events, which is not observed in our data. However, it is possible that hybridization followed by recurrent introgression after isolated diversification could be a better hypothesis. Gene flow due to migration and introgression may have prevented genetic differentiation within the SA clade [7] [35] . The presence of haplotypes shared between A. aestiva and South American A. ochrocephala (Figure 2) support the hypothesis of historical hybridization/introgression. Alternatively, incipient speciation accompanied by morphological differentiation during population expansion could be an alternative hypothesis. In any case, our results do not support that A. aestiva and South American A. ochrocephala are two distinct species.
4.2. Species Identification and Traceability of the mtDNA Lineages in Captivity
The illegal removal of animals from their natural environments is one of the main problems to be solved by wildlife protection agencies [37] . Most of the animals that are seized or voluntarily delivered in Brazil are sent to screening centers of wild animals (CETAS) [37] -[39] . Recently, a study using the mitochondrial marker Cyt-b, reported the success of identification of species of 17 specimens (seven carcasses, and ten eggs) of birds seized in an operation of the Brazilian Federal Police [40] . Some specimens were identified as psitacids: Alipiopsitta xanthops, Ara chloropterus, Aratinga aurea, and also A. aestiva/A. ochrocephala complex. In a study of all animals sent to CETAS/BH (IBAMA) in the state of Minas Gerais (MG) during the period of 1992-2012 it was shown that several endangered and endemic bird species seized in MG came from other regions of the country [36] .
The main objective of the “Brazilian Parrots” tool is to allow species identification of individuals or tissue samples of birds of the Psittacidae family. Unfortunately, the identification of the origin of the sample is a more complex issue as the taxa under investigation should have a well characterized population genetic structure.
The high mtDNA haplotype diversity (H) observed in the captive A. aestiva specimens (Table 1) could be used to identify specific maternal lineages. In the case of a legal commercial breeding facility this data could help to certify the captive origin of the commercialized progeny. According to the Brazilian law, only the F1 or subsequent generations of captive bred A. aestiva and A. ochrocephala can be legally traded. Therefore, the mtDNA could be used as a marker of the maternal lineage of the breeding females used as matrices. This matrilineal genetic marker can be used as a lineage certificate for the progeny that is born in captivity and is legally sold. This molecular certificate can be used to identify, for example, illicit trafficking due to commercialization of wild animals that were illegally implanted with chips or rings, to legalize animals as captive-born. The combination of mitochondrial DNA analyses with other tools and devices such as microchip (properly controlled by IBAMA-Brazil) could help to reduce illegal trafficking, or at least could facilitate the detection of illegal specimens. In this scenario, a mtDNA database of captive-bred females and their descendants could be created for this purpose, which could be publicly consulted by breeders, animal trade organizations, importers, and institutions worldwide.
Acknowledgements
We thank Paulo Machado and Fazenda Vale Verde, who provided blood samples of captive animals used in this study. Thanks to Camila Clozato Lara and Ricardo França who helped in the execution of some laboratory experiments. This study was supported by FAPEMIG (PRONEX), FAPEMIG-Vale SA (RDP-00043-10/2010-1), CNPq (473428/2004-0; 303558/2009-1; 473809/2008-7 and Pro-Reitoria de Pesquisa da UFMG.). A.V.C. received studentship from FAPEMIG and CNPq; F.A.A.S. was supported by FAPEMIG and CNPq; C.Y.M. was supported by FAPESP, CNPq, and CAPES. F.R.S was supported by CNPq.
References
- Snyder, N., McGowan, P., Gilardi, J. and Grajal, A. (2000) Parrots: Status Survey and Conservation Action Plan 2000-2004. IUCN, Gland, Switzerland and Cambridge.
- Lopes, I.F., Del Lama, M.A. and Del Lama, S.N. (2007) Genetic Variability in three Amazon Parrot Species. Brazilian Journal of Biology, 67, 883-887. http://dx.doi.org/10.1590/S1519-69842007000500011.
- IUCN (2007) The IUCN Red List of Threatened Species. http://www.iucnredlist.org
- CITES (2013) Convention on International Trade of Endangered Species of Wild Flora and Fauna. http://www.cites.org
- Russello, M.A. and Amato, G. (2004) A Molecular Phylogeny of Amazona: Implications for Neotropical Parrot Biogeography, Taxonomy, and Conservation. Molecular Phylogenetics and Evolution, 30, 421-437. http://dx.doi.org/10.1016/S1055-7903(03)00192-1
- Eberhard, J.R. and Bermingham, E. (2004) Phylogeny and Biogeography of the Amazona ochrocephala (Aves: Psittacidae) Complex. The Auk, 121, 318-332. http://dx.doi.org/10.1642/0004-8038(2004)121[0318:PABOTA]2.0.CO;2
- Ribas, C.C., Tavares, E.S., Yoshihara, C. and Miyaki, C.Y. (2007) Phylogeny and Biogeography of Yellow-Headed and Blue-Fronted Parrots (Amazona ochrocephala and Amazona aestiva) with Special Reference to the South American Taxa. Ibis, 149, 564-574. http://dx.doi.org/10.1111/j.1474-919X.2007.00681.x
- Schunck, F., Somenzari, M., Lugarini, C. and Soares, E.S. (2001) Plano de Acao Nacional Para a Conservacao dos Papagaios da Mata Atlantica, Série Espécies Ameacadas No. 20, ICMBio, Brasília.
- RENCTAS (2001) Rede Nacional de Combate ao Tráfico de Animais Silvestres, 1’ Relatório Nacional Sobre o Tráfico de Fauna Silvestre, Brasília. http://www.RENCTAS.org.br
- IBAMA (2005) Relatório de Apreensoes em 2005. http://www.ibama.gov.br
- Ratnasingham, S. and Hebert, P.D.N. (2007) BOLD: The Barcode of Life Data System (http://dx.doi.org/10.1111/j.1471-8286.2007.01678.x
- Baker, S.C. and Palumbi, S.R. (1994) Which Whales Are Hunted? A Molecular Genetic Approach to Monitoring Whaling. Science, 265, 1538-1539. http://dx.doi.org/10.1126/science.265.5178.1538
- Baker, S.C., Cipriano, F. and Palumbi, S.R. (1996) Molecular Genetics Identification of Whale and Dolphin Products from Commercial Markets in Korea and Japan. Molecular Ecology, 5, 671-685. http://dx.doi.org/10.1111/j.1365-294X.1996.tb00362.x
- Baker, S.C., Steel, D., Choi, Y., Lee, H., Kim, K.S., Choi, S.K., Ma, Y.U., Hamleton, C., Psihoyos, L., Brownell, R.L. and Funahashi, N. (2010) Genetic Evidence of Illegal Trade in Protected Whales Links Japan with the US and South Korea. Biology Letters, 6, 647-650. http://dx.doi.org/10.1098/rsbl.2010.0239
- Cipriano, F. and Palumbi, S.R. (1999) Genetic Tracking of a Protected Whale. Nature, 397, 307-308. http://dx.doi.org/10.1038/16823
- Baker, S.C., Dalebout, M.L., Lavery, S. and Ross, H.A. (2003) www.DNA-surveillance: Applied Molecular Taxonomy for Species Conservation and Discovery. Trends in Ecology & Evolution, 18, 271-272. http://dx.doi.org/10.1016/S0169-5347(03)00101-0
- Robins, J.H., Hingston, M., Matisoo-Smith, E. and Ross, H.A. (2007) Identifying Rattus Species Using Mitochondrial DNA. Molecular Ecology Notes, 7, 717-729. http://dx.doi.org/10.1111/j.1471-8286.2007.01752.x
- Chaves, P.B., Graeff, V., Lion, M.B., Oliveira, L.R. and Eizirik, E. (2012) DNA Barcoding Meets Molecular Scatology: Short mtDNA Sequences for Standardized Species Assignment of Carnivore Noninvasive Samples. Molecular Ecology Resources, 12, 18-35. http://dx.doi.org/10.1111/j.1755-0998.2011.03056.x.
- Kerr, K.C.R., Lijtmaer, D.A., Barreria, A.S., Herbert, P.D.N. and Tubaro, P.L. (2009) Probing Evolutionary Patterns in Neotropical Birds through DNA Barcodes. PLoS ONE, 4, Article ID: e4379. http://dx.doi.org/10.1371/journal.pone.0004379
- Caparroz, R., Seixas, G.H.F., Berkunsky, I. and Colevatti, R.G. (2009) The Role of Demography and Climatic Events in Shaping the Phylogeography of Amazona aestiva (Psitaciformes, Aves) and a Definition of Management Units for Conservation. Diversity and Distribution, 15, 459-468. http://dx.doi.org/10.1111/j.1472-4642.2009.00558.x
- Chaves, A.V., Clozato, C.L., Lacerda, D.R., Sari, H.E.R. and Santos, F.R. (2008) Molecular Taxonomy of Brazilian Tyrant-Flycatchers (Passeriformes, Tyrannidae). Molecular Ecology Resources, 8, 1169-1177. http://dx.doi.org/10.1111/j.1755-0998.2008.02218.x
- Sorenson, M.D. and Quinn, T.W. (1998) Numts: A Challenge for Avian Systematics and Population Biology. The Auk, 115, 214-221. http://dx.doi.org/10.2307/4089130
- Sorenson, M.D. (2003) Avian mtDNA Primers. http://people.bu.edu/msoren/Bird.mt.Primers.pdf
- Folmer, O., Black, M., Hoeh, W., Lutz, R. and Vrijenhoek, R. (1994) DNA Primers for Amplification of Mitochondrial Cytochrome C Oxidase Subunit I from Diverse Metazoan Invertebrates. Molecular Marine Biology and Biotechnology, 3, 294-299.
- Ewing, B., Hillier, L., Wendi, M. and Green, P. (1998) Base-Calling of Automated Sequencer Tracer Using Phred. I. Accuracy Assessment. Genome Research, 8, 175-185. http://dx.doi.org/10.1101/gr.8.3.175
- Gordon, D., Abajian, C. and Green, P. (1998) Consed: A Graphical Tool for Sequence Finishing. Genome Research, 8, 195-202. http://dx.doi.org/10.1101/gr.8.3.195
- Edgar, R.C. (2004) MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Research, 32, 1792-1717. http://dx.doi.org/10.1093/nar/gkh340
- Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M. and Kumar, S. (2011) MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution, 28, 2731-2739. http://dx.doi.org/10.1093/molbev/msr121
- Excoffier, L., Laval, G. and Schneider, S. (2005) Arlequin Ver. 3.0: An Integrated Software Package for Population Genetics Data Analysis. Evolutionary Bioinformatics Online, 1, 47-50.
- Rozas, J., Sánches-Delbarrio, J.C., Messeguer, X. and Rozas, R. (2003) DnaSP, DNA Polymorphism Analysis by the Coalescent and Other Methods. Bioinformatics, 19, 2496-2497. http://dx.doi.org/10.1093/bioinformatics/btg359
- Excoffier, L., Smouse, P.E. and Quattro, J.M. (1992) Analysis of Molecular Variance Inferred from Metric Distances among DNA Haplotypes: Application to Human Mitochondrial DNA 15 Restriction Data. Genetics, 131, 479-491.
- Bandelt, H.J., Forster, P. and Rohl, A. (1999) Median-Joining Networks for Inferring Intraspecific Phylogenies. Molecular Biology and Evolution, 16, 37-48. http://dx.doi.org/10.1093/oxfordjournals.molbev.a026036
- Larkin, M.A., Blackshields, G., Brown, N.P., Chenna, R., McGettigan, P.A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J.D., Gibson, T.J. and Higgins, D.G. (2007) Clustal W and Clustal X Version 2.0. Bioinformatics, 23, 2947-2948. http://dx.doi.org/10.1093/bioinformatics/btm404
- Ross, H.A., Lento, G.M., Dalebout, M.L., Goode, M., Ewing, G., McLaren, P., Rodrigo, A.G., Lavery, S. and Baker, C.S. (2003) DNA Surveillance: Web-Based Molecular Identification of Whales, Dolphins, and Porpoises. Journal of Heredity, 94, 111-114. http://dx.doi.org/10.1093/jhered/esg027
- Urantówka, A.D., Mackiewicz, P. and Strzala, T. (2014) Phylogeny of Amazona barbadensis and the Yellow-Headed Amazon Complex (Aves: Psittacidae): A New Look at South American Parrot Evolution. PLoS ONE, 9, Article ID: e97228. http://dx.doi.org/doi:10.1371/journal.pone.0097228
- Clements, J.F., Schulenberg, T.S., Iliff, M.J., Sullivan, B.L., Wood, C.L. and Roberson, D. (2013) The eBird/Clements Checklist of Birds of the World: Version 6.8. http://www.birds.cornell.edu/clementschecklist/download
- Souza, T.S. and Vilela, D.A.R. (2013) Espécies Ameacadas de Extincao víTimas do Tráfico e Criacao Ilegal de Animais Silvestres. Atualidades Ornitológicas, 176, 64-68.
- Gogliath, M., Bisaggio, E.L., Ribeiro, L.B., Resgalla, A.E. and Borges, R.C. (2010) Avifauna Apreendida e Entregue Voluntariamente ao Centro de Triagem de Animais Silvestres (Cetas) do Ibama de Juiz de Fora, Minas Gerais. Atualidades Ornitológicas, 154, 55-59.
- Pagano, I.S.A., Sousa, A.E.B.A., Wagner, P.G.C., and Ramos, R.T.C. (2009) Aves Depositadas no Centro de Triagem de Animais Silvestres do IBAMA na Paraíba: Uma Amostra do tráfico de aves Silvestres no Estado. Ornithologia, 3, 132-144.
- Carvalho, C.B.V. (2012) Identificacao Genética de Aves Vítimas do Tráfico de Animais Silvestres. Atualidades Ornitológicas, 165, 40-44.
Supplementary Material
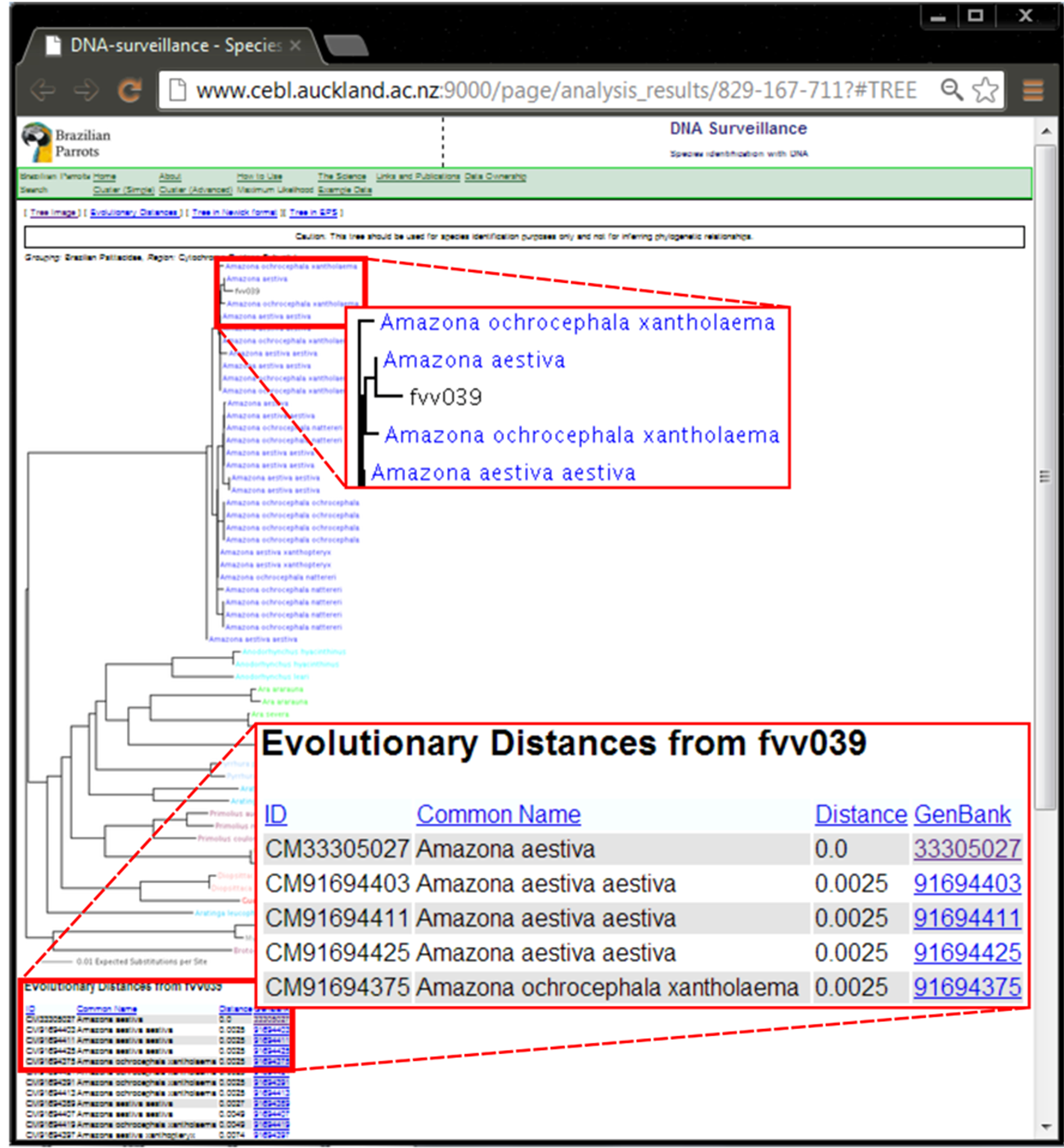
SM 1. Result of the analysis of the ND2 sequence from sample fvv039 in the DNA Surveillance Brazilian parrots database.

SM 2. Samples, origin and Genbank accession numbers of Amazona aestiva used in this study.
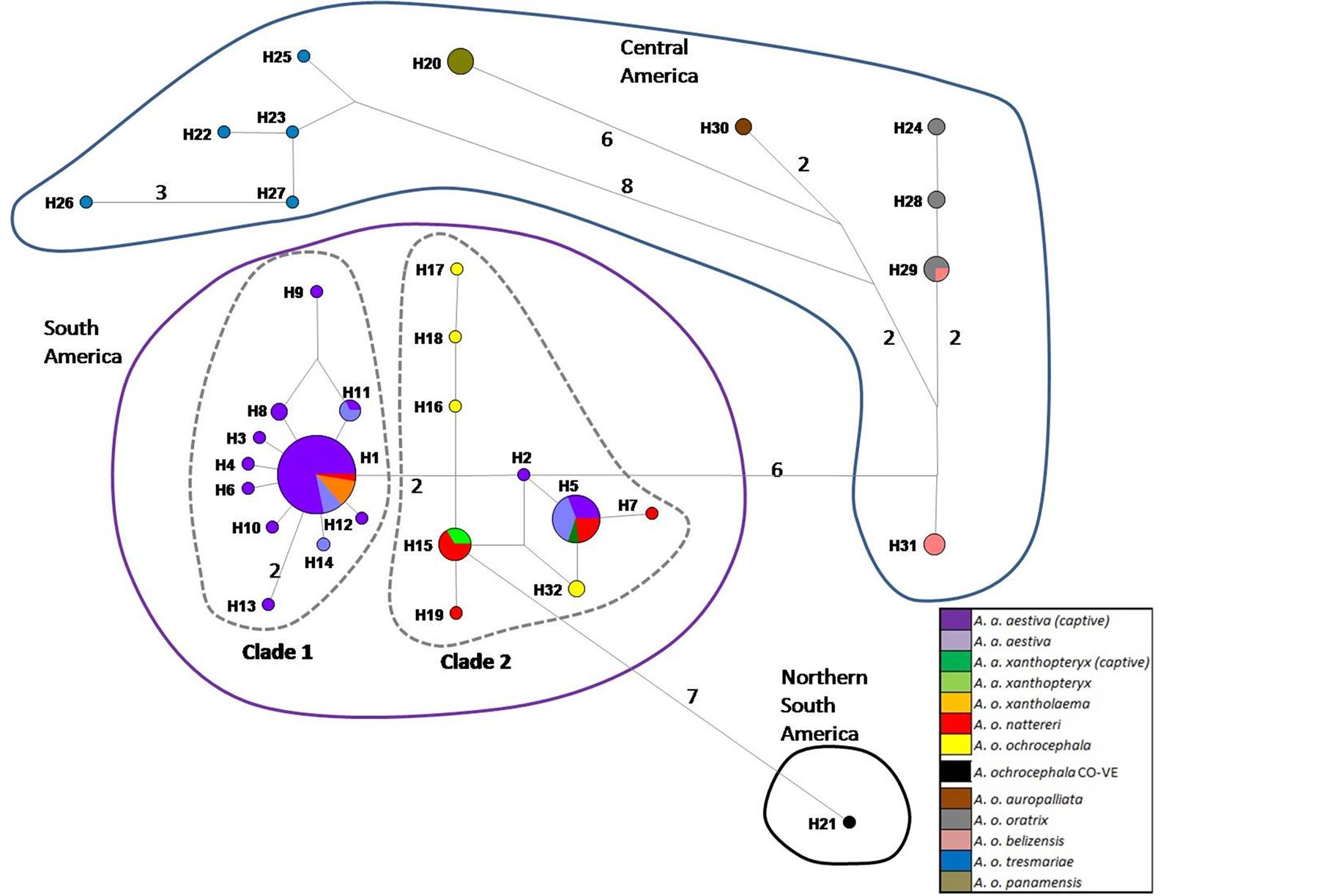
SM 3. Haplotype network based on 502 bp of ND2 of captive A. aestiva and other specimens of the A. aestiva/A. ochocephala complex. The number of substitutions is proportional to the length of the line connecting haplotypes and is also shown. The size of the circles is proportional to the frequency of the haplotype obtained in this sample. Red diamonds represent median vectors.
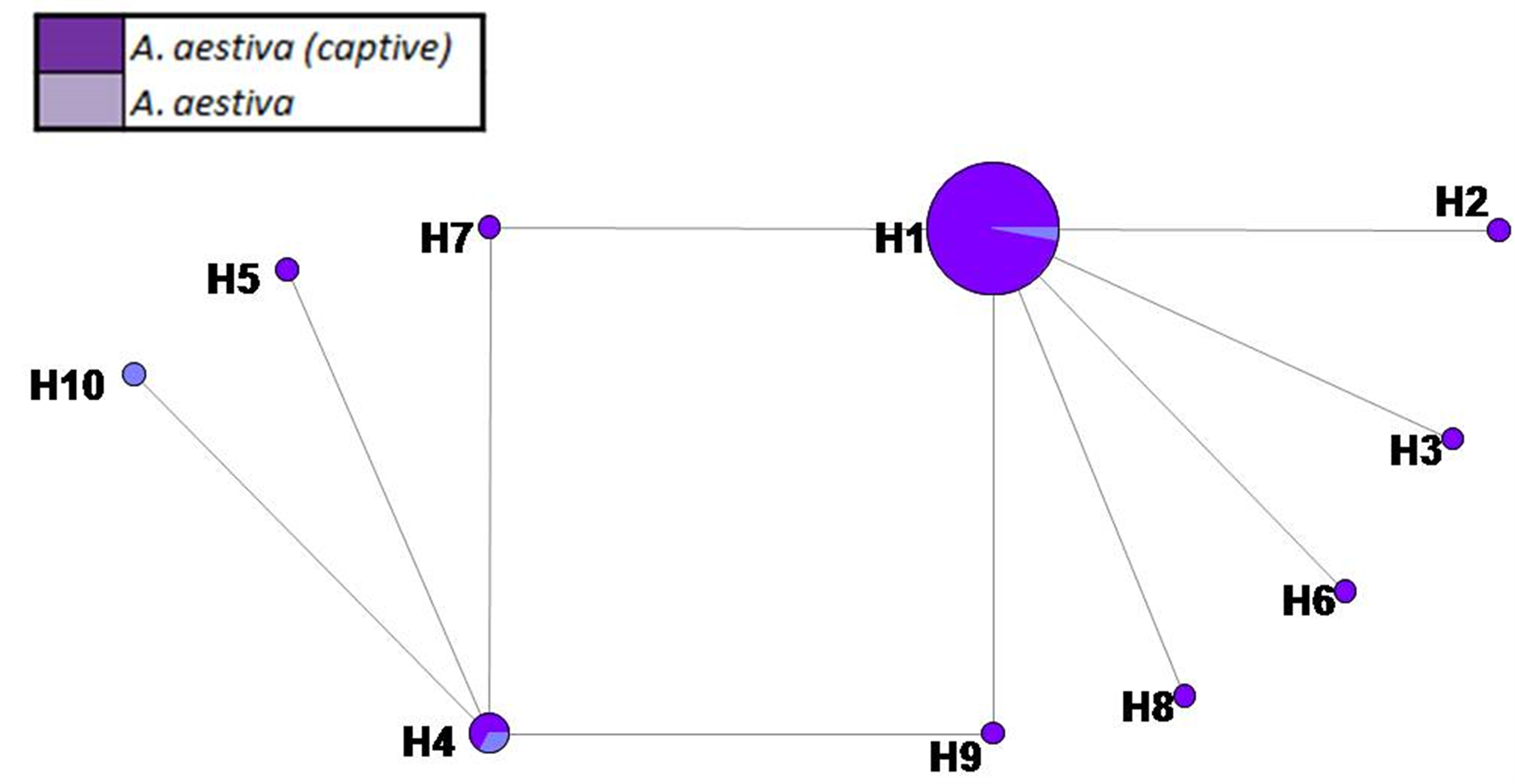
SM 4. Haplotype network based on 474 bp of COI of captive A. aestiva and other specimens of the A. aestiva/A. ochocephala complex. The number of substitutions is proportional to the length of the line connecting haplotypes and is also shown. The size of the circles is proportional to the frequency of the haplotype obtained in this sample.
SM 5. Haplotype network based on 474 bp of COI-DNA Barcode of captive A. aestiva and other specimens of the A. aestiva/ A. ochocephala complex. The size of the circles is proportional to the frequency of the haplotype obtained in this sample.
NOTES

*Corresponding author.


