Open Journal of Genetics
Vol.3 No.2(2013), Article ID:33619,12 pages DOI:10.4236/ojgen.2013.32015
Transferability and application of microsatellites (SSRs) from Juniperus communis L. to Juniperus procera Hochst. Ex endl.
![]()
1Ethiopian Institute of Agricultural Research, Holetta Center, Forestry Research Division, Addis Ababa, Ethiopia
2Department of Tree Breeding and Forest Genetics, Georg-August University of Goettingen, Göttingen, Germany
3School of Forest Resource and Environmental Science, Michigan Technological University, Houghton, USA
Email: *dmsertse@yahoo.com
Copyright © 2013 Demissew Sertse et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 6 March 2013; revised 6 April 2013; accepted 10 May 2013
Keywords: Ethiopia; Juniperus procera; Microsatellites; Transferability
ABSTRACT
Transferability of five nuclear microsatellite markers (Jc-16, Jc-31, Jc-32, Jc-35 and Jc-37) that were originally developed for J. communis was tested to J. procera. Jc-31 & Jc-37 showed successful amplifications and polymorphism in J. procera. Jc-35 which had been reported as polymorphic in J. communis was monomorphic in J. procera while the primer pair for Jc-32 failed to record any amplification. The remaining one primer pair (Jc-16) showed double loci amplification in both J. procera and the control J. communis suggesting further examination of the primer pair and its binding sites. Genetic variation of six Ethiopian J. procera populations: Chilimo, Goba, Menagesha-Suba, Wef-Washa, Yabelo and Ziquala was assessed based on the two polymorphic loci (Jc-31 & Jc-37) in 20 - 24 individuals of each population. From these two loci, a total of 41 alleles could be retrieved. Two populations that are located south east of the Great Rift Valley together harboured 75% of private alleles signifying their deviant geo-ecological zones and suggesting special consideration for conservation. Chilimo, which is at the western margin of Juniper habitat in Ethiopian central highlands scored the highest fixation (FIS = 0.584) entailing lower immigrant genes and hence higher inbreeding. The AMOVA revealed that 97% of the variation resided within the populations while still among population variation was significant (p < 0.05).
1. INTRODUCTION
Due to their high polymorphism and co-dominant mode of inheritance, microsatellite (SSR) markers provide high resolution and precise information in genetic analysis such as gene flow, mating system and paternity [1,2]. Hence, they have become one of the important genetic markers that are being widely used for genetic studies of many important plants and animals. However, microsatellites developed for a species usually are transferable to only closely related other species [3]. As a result, a specific species may require development of specific microsatellite markers. A number of microsatellites have been developed and employed in genetic studies of tree species [4-8].
Juniperus procera is one of the biggest trees in its genus reaching to a height of over 40 m and a diameter of above 3 m [9,10]. The tree naturally grows between the Arabian Peninsula in Asia to Zimbabwe in Africa [11]. It is believed to have been evolved from J. excelsa [12] or a pre-existing common ancestral species for both J. excelsa and J. procera [13]. J. procera is the only species that succeeded south of equator in the genus Juniperus which comprises 67 taxa [11]. The separate success of J. procera to the south may entail its divergent evolution to its unique geographic regions which has likely led it having deviant genomic structure. The possible distinct pattern of variation with different genome structure adapted and specialized to the unique geographical region may limit the transferability rate of microsatellite markers that have been developed for other species in the genus. However, some microsatellites primer developed for species in other genus: Chamaecyparis; Chamaecyparis nootkatensis [14] and Chamaecyparis obtusa [15]
were reported to score strong amplification in J. procera [16]. This on the other hand, may suggest a more successful transferability of microsatellites within the genus.
Recently, microsatellite markers were developed for three species in the genus Juniperus, namely, J. communis [17], J. przewalskii [18] and J. tibetica [19]. This has promised detail genetic studies of species in the genus for high resolution data generation which will have significant role in setting up of their conservation strategies. J. procera is a high use value tree chosen for a variety of constructions, furniture and outdoor uses [10,20]. Consequently, the species has been subjected to extensive logging which has led its current status to be threatened in many of its habitats [21-23]. This justifies the need for a systematic study of the genetic structure of the populations of J. procera in order to design appropriate conservation strategies. The objectives of the present study were therefore, to assess the transferability rate of microsatellites developed for J. communis to J. procera and to use transferable markers in genetic variation analysis of J. procera populations in Ethiopia and accordingly, to recommend possible conservation measures.
2. MATERIALS AND METHODS
2.1. Sampling Technique
Leaves (needles) of 20 - 24 trees from each six representtative J. procera populations: Chilimo, Goba, Menagesha-Suba, Wef-Washa, Yabelo and Ziquala (Figure 1) in Ethiopian highlands were collected in separate plastic bags containing silica gel. In order to avoid the collection of relatives, a minimum 50-meter distance between any sampled trees in a population was maintained using GPS. Further leaves of J. communis from Forestry Botanical Garden of the Georg-August University of Goettingen were collected as experimental control.
2.2. DNA Extraction
Total genomic DNA was extracted from silica gel dried leaf samples of both J. procera and J. communis using DNeasy 96 plant kit (Qiagen, Hilden, Germany). In order to check the quality and quantity of the DNA, 5 μl from each probe mixed with 2 μl of 6× orange loading dye solution (Fermentas) was electrophoresized for 25 minute

Figure 1. Map of the populations studied.
on 1% (w/v) agarose, in 1× Tris-Acetate-Ethylenediaminetetraacetic acid (TAE) buffer and about 0.003% (v/v) ethidium bromide as dye. After confirming the quality and quantity, the remaining DNA was stored at 20˚C for further investigation.
2.3. Primer Testing
Five primer pairs (Jc-16, Jc-31, Jc-32, Jc-35 and Jc-37) that were designed for microsatellite loci in Juniperus communis [17] were tested for amplification in a sample from each J. procera population and J. communis as positive control. PCR was performed in a final volume 15 μl containing 2 μl (10 ng) genomic DNA template, 1.5 μl 10× buffer (promega), 0.07 mM of each dNTP, 2 μl each primer (5 piko mol/μl), 2.5 mM MgCl2 for two primer pairs (Jc-16 and Jc-31) and 1.5 mM MgCl2 for the rest three primer pairs and 1 U Hot star Taq polymerase (Qiagen). PCR was performed following the profile reported in [17] using a Peltier Thermal Gradient Cycler (PTC-200 version 4.0, MJ Research).
In order to observe amplifications in the expected regions of the respective primer pairs, PCR products were subjected to gel electrophoresis. Fragments of PRC products that showed amplification in the expected size ranges were separated on ABI Genetic analyzer 3100 with internal size standard fluorescent dye ROX (Gene Scan 500 ROX) from Applied Biosystems. The above PCR and fragment separation procedures were performed for all samples for the primer pairs that showed clear amplification and polymorphism thereby variation analysis of the J. procera populations. The allele sizes of the polymorphic loci were scored using Gene scan 3.7® and Genotyper 3.7® software (Applied Biosystems) and the scores were appended to excel for further analysis.
2.4. Data Analysis for Polymorphic Loci
Population variation analysis was made based on polymorphic microsatellite loci. Genetic variation parameters such as, allele frequencies, private alleles, fixation indices (FIS) and genetic differentiation based on FST via AMOVA were computed using GenAlEx version 6.4. In order to compute population pair wise Nei’s unbiased genetic distance [24], the program POPGEN version 1.32 [25] was applied. Unweighted pair group method using arithmetic averages (UPGMA) dendrogram was generated following the method of SAHN clustering [26] using NTSYSpc 2.0 program based on the unbiased genetic distance.
3. RESULTS
3.1. Test of Primer Pairs
Two microsatellites (Jc-31, Jc-37) produced clear and polymorphic amplifications in J. procera. Another loci, Jc-35, which was polymorphic in J. communis [17] appeared to be monomorphic in J. procera (Figure 2). Jc-16 primer pair amplified double loci in both J. procera and the control J. communis which the additional amplification was stronger than the amplification in the expected size ranges (Figure 3). On the other hand, the rest prime pair, Jc-32 did not record any amplification in J. procera.
3.2. Allelic Frequencies
A total of 41 alleles were revealed from the two polymorphic loci Jc-31 and Jc-37 (See Appendix). Jc-37 was found to be a highly variable locus with a total of 35 alleles revealed within the expected size range of 164 to 232 bps. The population, Menagesha-Suba, with its origin from the central part of Ethiopia, stood the first in terms of the total number of alleles revealed, which was 24 in this case (Table 1). The other locus, Jc-31, which was found to be less polymorphic in this study, contributed six alleles in a confined sizes ranged between 155 to 161 bps. At microsatellite locus Jc-37, allele sizes 188, 196 and 200 bps were observed in all the populations, with the most frequent observation of alleles being at the size of 196 bps. Two alleles with sizes of 155 and 156 bps accounted over 90% of at locus Jc-31 (see Appendix).
3.3. Private Alleles
A total of 12 private alleles were revealed for the overall six populations at the two loci. All private alleles were observed at locus Jc-37 except one at locus Jc-31 in Yabelo, a population which is from the extreme southern block of Ethiopia (Table 2). A private allele with a size of 164 bps at Jc-37 contributed for more than 10% of allelic frequency in this same population (Table 2). Two populations which are south east of The Great Rift valley, namely Goba and Yabelo, har bored about 75% of the private alleles observed in this study.
3.4. Heterozygosity and Fixation Index
Among the populations tested in this study, Ziquala
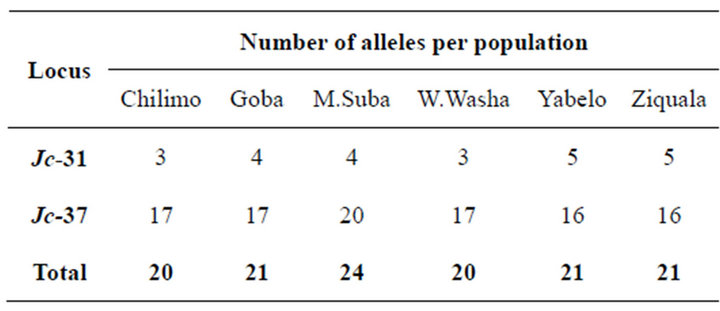
Table 1. Number of alleles in each population.

Figure 2. Monomorphic and homozygous locus Jc-35.
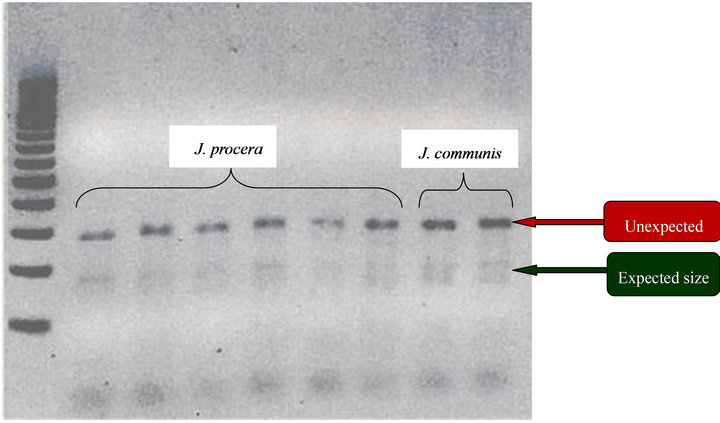
Figure 3. Loci amplified by single primer pair Jc-16.
scored the highest heterozygosity (Ho = 0.604) followed by Goba (Ho = 0.595) and congruently, these two populations showed lowest inbreeding, i.e. FIS = 0.283 & 0.296 in that order. The lowest heterozygosity (Ho = 0.354) and hence the highest fixation index (FIS = 0.584) was computed for Chilimo population (Table 3).
3.5. Among Populations Variation
3.5.1. AMOVA
AMOVA based on the two polymorphic loci combine puts 97% of the variation within the populations (Table 4). Nevertheless, the result indicated that there is highly
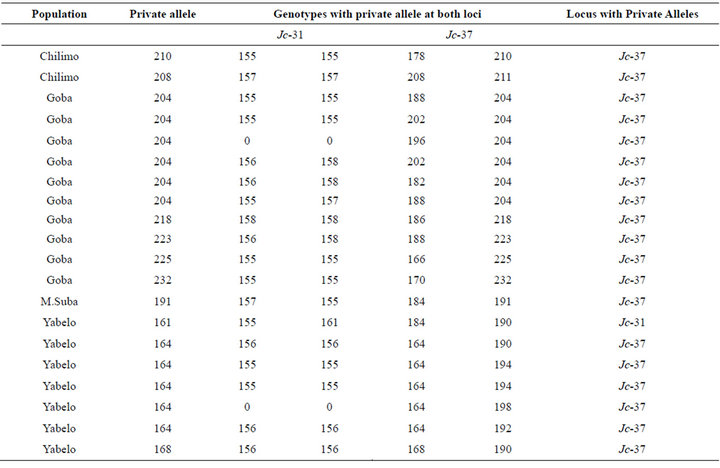
Table 2. Private alleles and their respective genotypes.
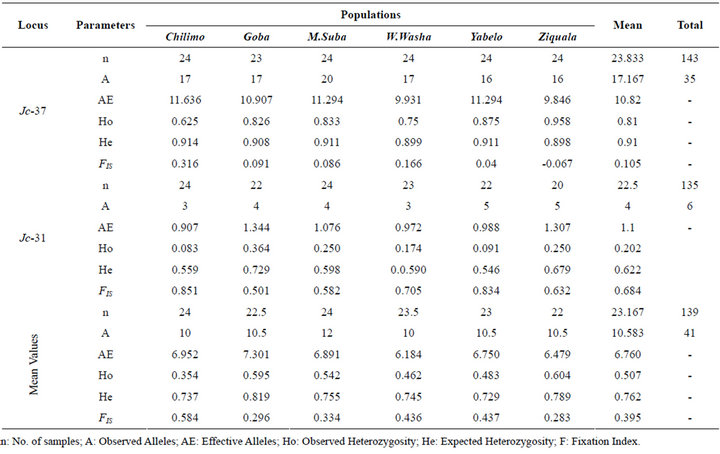
Table 3. Within population genetic variation based on the two microsatellite loci.
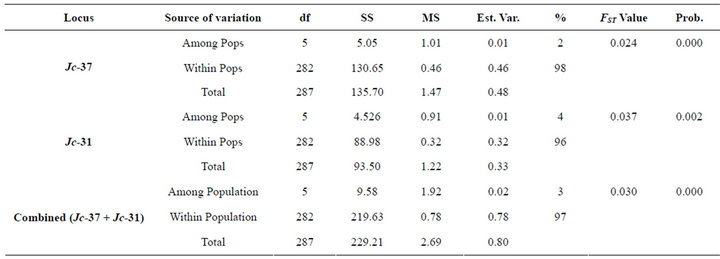
Table 4. AMOVA results at microsatellite loci.
significant (P < 0.01) variation among the populations.
3.5.2. Pair Wise Analysis
The pairwise FST analysis based on the polymorphic loci showed significant (P < 0.05) differentiations between all pairs except Chilimo with Yabelo, Goba with Ziquala and Menagesha-Suba with Wef-Washa (Table 5). Menagesha-Suba and Wef-Washa were the least differentiated (FST = 0.005).
3.5.3. Population Clustering
The populations from the different eco-geographical sources were grouped into two distinct clusters based on Nei’s unbiased genetic distance UPGMA (Figure 4). Two of the populations from the central part of Ethiopia, namely Menagesha-Suba and Wef-Washa, were grouped into one cluster, whereas the remaining four populations, i.e., Chilimo, Goba, Yabelo and Ziquala, were grouped into the second cluster.
4. DISCUSSION
4.1. Transferability of Microsatellites from J. communis to J. procera
The low transferability of the microsatellite markers from J. communis to J. procera may indicate the deviance in the genomic structure of the two species, which enabled the species to have success in their current respective eco-geographic zones. The success of J. procera in the south unlike the rest of the species in the genus [11,27] was already postulated to be due to evolutionary changes in the species to adapt to its unique ecogeographical region [11-13]. The polymorphism in J. communis [17] and the complete monomerphism in J. procera at locus Jc-35 may also indicate the degree of genetic divergence exist between the two species, justifying the taxonomic classification of the two species into different groups [11]. Reference [18] who checked poly-

Table 5. Pairwise population differentiation (FST) based on the two microsatellite loci.
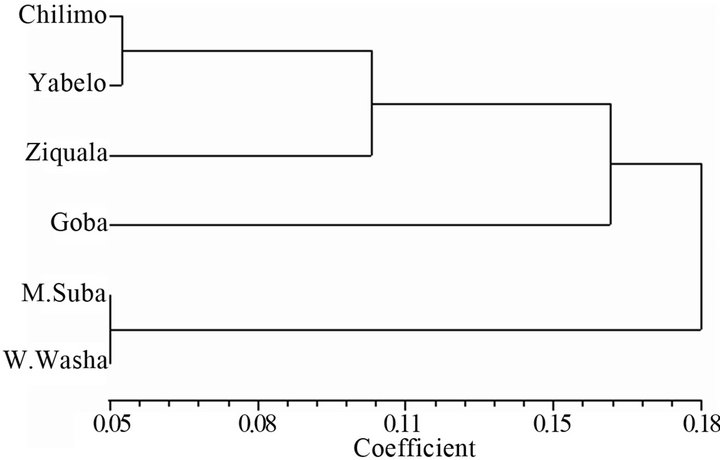
Figure 4. UPGMA dendrogram based on pair wise Nei’s unbiased genetic distances.
morphism of the five microsatellite loci [17] in J. przewalskii reported polymorphism for locus Jc-32 which failed to show amplification in J. procera. Conversely, [18] failed to observe amplification record in J. przewalskii at locus Jc-37 which was found highly polymorphic in J. procera and they got monomorphism in J. przewalskii at locus Jc-31 which was polymorphic in J. procera. This might be an indication that J. procera is genetically divergent not only from J. communis but also from other species within the genus Juniperus. Reference [18] also failed to produce any successful amplification with J. przewalskii at the locus Jc-16.
The double loci amplification by primer pair Jc-16 in both J. procera and J. communis (control) is in accordance with the report of [28] who observed multiple binding sites of the primer in her further investigation though [17] reported it as successful microsatellite primer. The frequency of microsatellite regions in the genome increases with the genome size of an organism [29-31]. Hence, primer sets designed for a single target microsatellite region may encounter more than one region in large genomes, thereby possibility of amplifying additional non target regions [31]. Conifers in general are reported to have large genome with a highly repetitive DNA that has made difficulty for recovery of microsatellite markers from their genome [14,32-34]. Accordingly, the experiences of single primer pair amplifying multiple loci in conifer genomes have also been commonplace [14,31,35].
4.2. Population Genetic Variation
J. procera is diocious and hence it is obligatorily outcrossing wind pollinated species [10,11,27]. The high within population variation in this study is likely attributable to these mating features of the species which was also reported for many species having similar reproductive biology [5,36-40). In addition, the strong gene migration of the species via seed which is mediated by long traveling birds [11,13] likely pronounced the within population genetic variation by narrowing down the variation among populations [39,41].
The lowest pairwise genetic distance which was realized between Menagesha-Suba and Wef-Washa may confirm the historical record that claims Menagesha-Suba as plantation of wildlings from Wef-Washa back in 15th century [42,43]. These two populations were also the only undifferentiated pairs in AFLP analysis of the six populations [44]. The low differentiation between Chilimo and Yabelo, however; is contrary to the one reported by [44] between which the authors computed the highest differentiation. The result in this study is likely due to the allele size 155 bps at Jc-31 which was shared between the two populations at higher frequency. The allele sizes 155 and 156bps at Jc-31 seem to have been evolved through insertion and deletions process that has gone in the flanking region of the locus [45]. Therefore, by chance, the two populations (Chilimo and Yabelo) probably have made relatively same rate of evolution in favor of the allele size 155 bps which can be considered as Homoplasy [46,47].
4.3. Heterozygosity and Fixations
The relatively and consistently higher heterozygosity at both of the loci for the Goba population might indicate the strong reproductive contact of this population with other populations. The Goba population is a sub population of an extensive mega Juniper population over the Bale Mountains [48,49]. Nevertheless, this population was found with inferior gene diversity of all the populations investigated through AFLP analysis, which was assumed to suffer from bottleneck associated with sever disturbance [44]. Bottlenecks have little effect on heterozygosity [50-52]. This likely has enabled the Goba population in maintaining its superiority in heterozygosity in this study while the population was reported with lower gene diversity at AFLPs [44].
On the other hand, the higher fixation (FIS = 0.584) computed for Chilimo might be an indication of the higher inbreeding [53,54] in this population which might be resulted from limited geneflow [55-57]. Chilimo was also reported with lower gene diversity among the central highland populations based on AFLP analysis [44]. Chilimo is located in the western margin of Juniper habitats in Ethiopia [58].
4.4. Geographic Isolations and Private (Endemic) Allele Concentrations
The reason for higher concentration of endemic alleles in the south eastern populations (Goba and Yabelo) may somehow be related to the exclusive isolation of these populations from the rest by the Great Rift Valley as a physical barrier to gene flow. Reference [44] also reported that these two populations were genetically divergent from the other populations. Yet, the two south easterners exist at substantially long geographic distance apart each other which likely enabled them to harbor large number of endemic alleles independently. The Bale Mountains, where the Goba population belongs are known to have high concentration of endemic faunas and floras resulting from its unique ecology [49,59] experiencing temperature extremes between −15˚C to +26˚C [60]. The unique ecological features of the area may also contribute for change of certain genetic factors in the non-endemic organisms as well which perhaps have resulted in high level of private allele in Goba population. The Yabelo population harbored a total of three endemic alleles of which two were at locus Jc-37 and the other at Jc-31. The high allelic frequency (>10%) of the endemic allele with the size of 164 bps (locus Jc-37) in Yabelo may indicate the uniqueness of this population. Yabelo is recognized as lowland Juniper population in Ethiopia and it was reported for having unique seed morphology with a higher germination rate [61,62]. The two endemic alleles observed in Chilimo population may also be attributed to the geographic position of the population, which is at the western margin of the ecological zones for the Juniper forests in Ethiopia [58].
4.5. Conservation Implication of the Genetic Structure
Although most of the variations resided within the populations, due mainly to the mating system and gene flow mechanism of the species, the significant differentiation among the populations entails the level of variation of genetic information harbored in each population. Furthermore, the high private allele concentration for the geographically isolated populations clearly suggested that these populations deserve a special attention for genetic conservation. Earlier [61,62] reported the Juniper at Yabelo having unique morphological and physiological traits signifying the need of special conservation attention of this population. Contrary to [44] who reported low diversity in Goba population, the present study scored both the highest heterozigocity and private alleles in this population. This, as stated above, may emanate from bottleneck problems that have little effect on heterozygosity but likely be associated with the excessive disturbances of the population. Therefore, appropriate steps must be taken to put in place a proper genetic resources conservation strategy before such populations, particularly those with high concentration of private alleles are genetically eroded as their private alleles might not be regained from anywhere else once they are lost. The higher fixation score in Chilimo population seem to be due to selective logging of the elite genotypes with higher diameter [63,64] and due to its geographic isolation with the other populations [58]. In order to avoid genetic erosion and inbreeding depression, this population may require further genetic enrichment involving the introgression of new genetic backgrounds into the existing population by inter-planting of seedlings from other populations.
5. CONCLUSION
The less transferability of the microsatellites in J. communis to J. procera elucidates the genome structure differences of the two species. It is likely that the genome structure of J. procera has evolved in response to its current unique habitats, which may make it divergence not only from J. communis but also from other species of the genus. Though only two polymorphic loci were involved in this study for the assessment of J. procera populations in Ethiopia, results were able to generate precisely valid information relevant for conservation strategy set up and history reconstruction. However, we suggest more microsatellite markers either specifically developed for the species or those that could be transferred from related taxa to be involved for detail investigation and generate reliable data in further studies.
6. ACKNOWLEDGEMENTS
The German Academic Exchange Service (DAAD) is appreciated for financial support to the first author. We thank, Olga Artes, Oleksandra Dolynska and Gerold Dinkel for technical support during the laboratory work. We also thank Gemechu Keneni (Dr.) for editing the manuscript and Demeke Nigussie for his assistance in plotting the sample sites on Ethiopian map.
REFERENCES
- Keiper, F.J., Hayden, M.J., Park, R.F. and Wellings, C.R. (2003) Molecular genetic variability of Australian isolates of five cereal rust pathogens. Mycological Research, 107, 545-556. doi:10.1017/S0953756203007809
- Stefenon, V.M., Gailing, O. and Finkeldey, R. (2008) The role of gene flow in shaping genetic structures of the subtropical conifer species Araucaria angustifolia. Plant Biology, 10, 356-364. doi:10.1111/j.1438-8677.2008.00048.x
- Finkeldey, R. and Hattemer, H.H. (2007) Tropical forest genetics. Springer-Verlag, Berlin, Heidelberg, 315 pp. doi:10.1007/978-3-540-37398-8
- Dow, B.D. and Ashley, M.V. (1996) Microsatellite analysis of seed dispersal and parentage of saplings in bur oak, Quercus macrocarpa. Molecular Ecology, 5, 120- 132. doi:10.1111/j.1365-294X.1996.tb00357.x
- Streiff, R., Labbe, T., Bacilieri, R., Steinkellner, H., Glossl, J. and Kremer, A. (1998) With in population genetics structure in Quercus rubor L. and Quercus petaea (Matt.) Liebl. Assessed with izozymes and microsatellites. Molecular Ecology, 7, 317-328. doi:10.1046/j.1365-294X.1998.00360.x
- Collevatti, R.G., Grattapaglia, D. and Hay, J.D. (2001) Population genetic structure of the endangered tropical tree species Caryocar brasiliense, based on variability at microsatellite loci. Molecular Ecology, 10, 349-356.
- Mottura, M.C., Finkeldey, R., Verga, A.R. and Gailing, O. (2005) Development and characterization of microsatellite markers for Prosopis chilensis and Prosopis flexuosa and cross-species amplification. Plant Molecular Biology Reporter, 22, 251-258. doi:10.1007/BF02773135
- Pandey, M. (2005) Development of microsatellites in sycamore maple (Acer pseudoplatanus L.) and their application in population genetics. Ph.D. Dissertation, Institute of Forest Genetics and Forest Tree Breeding, GeorgAugust University of Göttingen, Göttingen.
- Pohjonen, V. and Pukkala, T. (1992) Juniperus procera Hocht. ex Endl. in Ethiopian forestry. Forest Ecology and Management, 49, 75-85. doi:10.1016/0378-1127(92)90161-2
- Negash, L. (1995) Indigenous trees of Ethiopia: Biology, uses and propagation techniques. SLU Rprocentralen, Umea, 285 pp.
- Adams, R.P. (2004) Juniperus of the world: The genus Juniperus. Trafford Publishing, Victoria BC, Canada, 275 pp.
- Kerfoot, O. (1975) Origin and speciation of the Cupressaceae in Sub-Sahara Africa. Boissiera, 24, 145-150.
- Adams, R.P., Demeke, T. and Abulfatih, H.A. (1993) RAPD DNA Fingerprints and terpenoids: Clues to past migrations of Juniperus in Arabia and east Africa. Theoretical and Applied Genetics, 87, 22-26. doi:10.1007/BF00223738
- Berube, Y., Ritland, C. and Ritland, K. (2003) Isolation, characterization, and cross-species utility of microsatellites in yellow cedar (Chamaecyparis nootkatensis). Genome, 46, 353-361. doi:10.1139/g03-014
- Nakao, Y., Itwata, H., Mutsumota, A., Tsumura, Y. and Tomaru, N. (2001) Highly polymorphic microsatellite markers in chmaecyparis obtusa. Canadian Journal of Forest Research, 31, 2248-2251. doi:10.1139/x01-145
- Mlangeni, E.T. (2005) Genetic diversity and population structure in the East African Pencil Cedar, Juniperus procera (Cupressaceae). M.Sc. Thesis, University of Oslo, Oslo.
- Michalczyk, I.M., Sebastiani, F., Buonamici, A., Cremer, E., Mengel, C., Ziegenhagen, B. and Vendramin, G.G. (2006) Characterization of highly polymorphic nuclear microsatellite loci in Juniperus communis L. Molecular Ecology Notes, 6, 346-348. doi:10.1111/j.1471-8286.2005.01227.x
- Zhang, Q., Yang, Y.Z., Wu, G.L., Zhang, D.Y. and Liu, J.Q. (2008) Isolation and characterization of microsatellite DNA primers in Juniperus przewalskii Kom (Cupressaceae). Conservation Genetics, 9, 767-769. doi:10.1007/s10592-007-9387-y
- Opgenoorth, L. (2009) Identification and characterization of microsatellite marker in the tetraploid Juniperus tibetica Kom. using next generation sequencing. Conservation Genetic Resources, 1, 253-255. doi:10.1007/s12686-009-9062-3
- Bekele, A.T. (2007) Useful trees and shrubs for Ethiopia: Identification, propagation and management for agricultural and pastoral community. World Agroforestry Center, Nairobi, 559 pp.
- Garzulia, M. (2006) Threatened, endangered and vulnerable tree species: A comparison between FRA 2005 and the IUCN red list. Working Paper 108/E, FAO, Forestry Department, Rome, Italy.
- Borghesio, L., Giannetti, F., Ndang’ang’a, K. and Shimelis, A. (2004) The present conservation status of Juniperus forests in the South Ethiopian Endemic Bird Area. African Journal of Ecology, 42, 137-143. doi:10.1111/j.1365-2028.2004.00511.x
- IUCN (2009) Juniperus procera: Red list of threatened species. Conifer Specialist Group Report, 1998.
- Nei, M. (1978) Estimation of Average heterozygosity and genetic distance from a small number of individuals. Genetics, 89, 583-590.
- Yeh, F., Yang, R.C. and Boyle, T. (2000) POPGEN (version 1.32), microsoft windows-based freeware for population genetic analysis. http://www.ualberta.ca/~fyeh/pr01.htm
- Rohlf, F.J. (1998) NTSYSpc. Numerical taxonomy and multivariate analysis system. Version 2.0, User Guide: Exeter Software, Setauket, New York, 31 pp.
- Farjon, A. (2005) A monograph of Cupressaceae and Sciadopitys. Royal Botanic Gardens, Kew, 648 pp.
- Michalczyk, I.M. (2008) Application of DNA marker systems to test for genetic imprints of habitat fragmentation in Juniperus communis L. on different spatial and temporal scales. Ph.D. Dissertation, Department of Conservation Biology, Philipps-University of Marburg, Marburg.
- Primmer, C.R., Raudsepp, T., Chowdhary, B.P., Moller, A.P. and Ellegren, H. (1997) Low frequency of microsatellites in the avian genome. Genome Research, 7, 471- 482.
- Hancock, J.M. (1999) Microsatellites and other simple sequences: Genomic context and mutational mechanism, In: Goldstein, D.B. and Schlötterer, C., Eds., Microsatellites: Evolution and applications, Oxford University Press, New York, 1-9.
- Garner, T.W.J. (2002) Genome size and microsatellites: The effect of nuclear size on amplification potential. Genome, 45, 212-215. doi:10.1139/g01-113
- Fischer, D. and Bachman, K. (1998) Microsatellite enrichment in organisms with large genomes (Allium cepa L.). BioTechniques, 24, 796-802.
- Elsik, C.G. and Williams, C.G. (2001) Low-copy microsatellite recovery from a conifer genome. Theoretical and Applied Genetics, 13, 1189-1195. doi:10.1007/s001220100725
- Bogunic, F., Muratovic, E., Brown, S.C. and SiljakYakovlev, S. (2003) Genome size and base composition of five Pinus species from the Balkan region. Plant Cell Reports, 22, 59-63. doi:10.1007/s00299-003-0653-2
- Fisher, P.J., Richardson, T.E. and Gardner, R.C. (1998) Characteristics of singleand multi-copy microsatellites from Pinus radiate. Theoretical and Applied Genetics, 96, 969-979. doi:10.1007/s001220050828
- Billington, H.L. (1992) Effect of population size on genetic variation in a dioecious conifer. Conservation Biology, 5, 115-119. doi:10.1111/j.1523-1739.1991.tb00394.x
- Hamrick, J.L., Godt, M.J.W. and Sherman-Broyles, S.L. (1992) Factors influencing levels of genetic diversity in woody plant species. New Forests, 6, 95-124. doi:10.1007/BF00120641
- Hamrick, J.L. and Godt, M.J.W. (1996) Effects of life history traits on genetic diversity in plant species. Transaction of the Royal Society of London Series B, 351, 1291-1298. doi:10.1098/rstb.1996.0112
- Weidema, I.R., Magnussen, L.S. and Philipp, M. (2000) Gene flow and mode of pollination in a dry-grassland species, Filipendula vulgaris (Rosaceae). Heredity, 84, 311-320. doi:10.1046/j.1365-2540.2000.00669.x
- Gerard, J., Oostermeijer, B. and de Knegt, B. (2004) Genetic population structure of the wind-pollinated, dioecious shrub Juniperus communis in fragmented Dutch heathlands. Plant Species Biology, 19, 175-184. doi:10.1111/j.1442-1984.2004.00113.x
- White, T.L., Adams, W.T. and Neale, D.B. (2007) Forest genetics. Oxford University Press, Oxford. doi:10.1079/9781845932855.0000
- Demissew, S. (1988) The floristic composition of the Menagesha state forest and the need to conserve such forests in Ethiopia. Mountain Research and Development, 8, 243-247. doi:10.2307/3673454
- Eshetu, Z. (2002) Historical C3–C4 vegetation pattern on forested mountain slopes: Its implication for ecological rehabilitation of degraded highlands of Ethiopia by afforestation. Journal of Tropical Ecology, 18, 743-758. doi:10.1017/S0266467402002481
- Sertse, D., Gailing, O., Eliades, E.G. and Finkeldey, R. (2011) Anthropogenic and natural causes influencing genetic structure of Juniperus procera. Hochst. ex. Endl. in the Ethiopian highlands. Genetic Resource and Crop Evolution, 58, 849-859. doi:10.1007/s10722-010-9623-z
- Lee, S.L., Tani, N., Ng, K.K.S. and Tsumura, Y. (2004) Characterization of 15 polymorphic microsatellite loci in an endangered tropical tree Hopea bilitonensis (Dipterocarpaceae) in Peninsular Malaysia. Molecular Ecology Notes, 4, 147-149. doi:10.1111/j.1471-8286.2004.00593.x
- Sanderson, M.J. and Hufford, L. (1996) Homoplasy: The recurrence of similarity in evolution. Academic Press, San Diego.
- Donoghue, M.J. and Ree, R.H. (2000) Homoplasy and developmental constraints: A model and example from plants. American Zoologist, 40, 759-769. doi:10.1668/0003-1569(2000)040[0759:HADCAM]2.0.CO;2
- Malcolm, J. and Evangelista, P.H. (2005) The range and status of Mountain Nyala: Case study in Bale Mountains, Ethiopia, 42 pp. www.ethiopianwolf.org/publications
- Umer, M., Lamb, H.F., Bonnefill, R., Le´zin, A.M., Tierceline, J.J., Gibert, E., Cazet, J.P. and Watrin, J. (2007) Late pleistocene and holocene vegetation history of the Bale Mountains, Ethiopia. Quaternary Science Reviews, 26, 2229-2246. doi:10.1016/j.quascirev.2007.05.004
- Allendorf, F.W. (1986) Genetic drift and the loss of alleles versus heterozygosity. Zoo Biology, 5, 181-190. doi:10.1002/zoo.1430050212
- Knapp, E.E. and Connors, P.G. (1999) Genetic consequences of a single-founder population bottleneck in Trifolium amoenum (Fabaceae). American Journal of Botany, 86, 124-130. doi:10.2307/2656961
- Keller, L.F., Jeffery, K.J., Arcese, P., Beaumont, M.A., Hochachka, W.M., Smith, J.N.M. and Bruford, M.W. (2001) Immigration and the ephemerality of a natural population bottleneck: Evidence from molecular markers. Proceedings: Biological Sciences, 268, 1387-1394. doi:10.1098/rspb.2001.1607
- Wright, S. (1931) Evolution in Mendelian populations. Genetics, 16, 97-159.
- Wright, S. (1951) The genetical structure of populations. Annals of Eugenics, 15, 323-354.
- Suter, M., Schneller, J.J. and Vogel, J.C. (2000) Investigations into the genetic variation, population structure, and breeding systems of the fern Asplenium trichomanes subsp. Quadrivalens. International Journal of Plant Science, 161, 233-244. doi:10.1086/314258
- Franceschinelli, E.V., Jacobi, C.M., Drummond, G.M. and Resende, M.F.S. (2006) The genetic diversity of two Brazilian Vellozia (Velloziaceae) with different patterns of spatial distribution and pollination biology. Annals of Botany, 97, 585-592. doi:10.1093/aob/mcl007
- Collevatti, R.G., Leite, K.C.E., de Miranda, G.H.B. and Rodrigues, F.H.G. (2007) Evidence of high inbreeding in a population of the endangered giant anteater, Myrmecophaga tridactyla (Myrmecophagidae), from Emas National Park. Genetics and Molecular Biology, 30, 112- 120. doi:10.1590/S1415-47572007000100020
- Bekele, T. (1994) Phytosociology and ecology of a humid afromontane forest on the central plateau of Ethiopia. Journal of Vegetation Science, 5, 87-98. doi:10.2307/3235642
- Kingdon, J. (1990) Island Africa: The evolution of Africa’s rare animals and plants. Princeton University Press, Princeton.
- [61] Hillman, J.C. (1988) The Bale Mountains National Park area, southeast Ethiopia, and its management. Mountain Research and Development, 8, 253-258. doi:10.2307/3673456
- [62] Mamo, N., Mihretu, M., Fekadu, M., Tigabu, M. and Teketay, D. (2006) Variation in seed and germination characteristics among Juniperus procera populations in Ethiopia. Forest Ecology and Management, 225, 320- 327. doi:10.1016/j.foreco.2006.01.026
- [63] Tigabu, M., Fjellström, J., Odén, P.C. and Teketay, D. (2007) Germination of Juniperus procera seeds in response to stratification and smoke treatments, and detection of insect-damaged seeds with VIS + NIR spectroscopy. New Forests, 33, 155-169. doi:10.1007/s11056-006-9020-9
- [64] Wodemariam, T.G. (1998) Diversity of woody plants and avi fauna in a dry afromontane forest: On the central platue of Ethiopia. Master of Science Thesis, Swedish University of Agricultural Science, Skinnskatteberg.
- [65] Shumi, G. (2009) The structure and regeneration status of trees and shub species of Chilimo forest-ecological sustainability indicators for participatory forest management (PFM) in Oromia Ethiopia. Master of Science Thesis, University of Dresden, Dresden.
APPENDIX
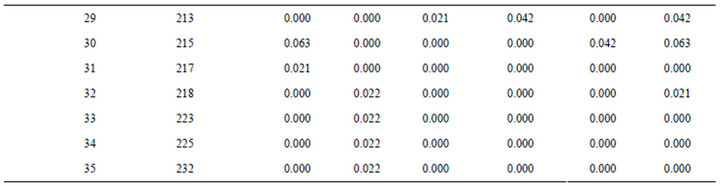
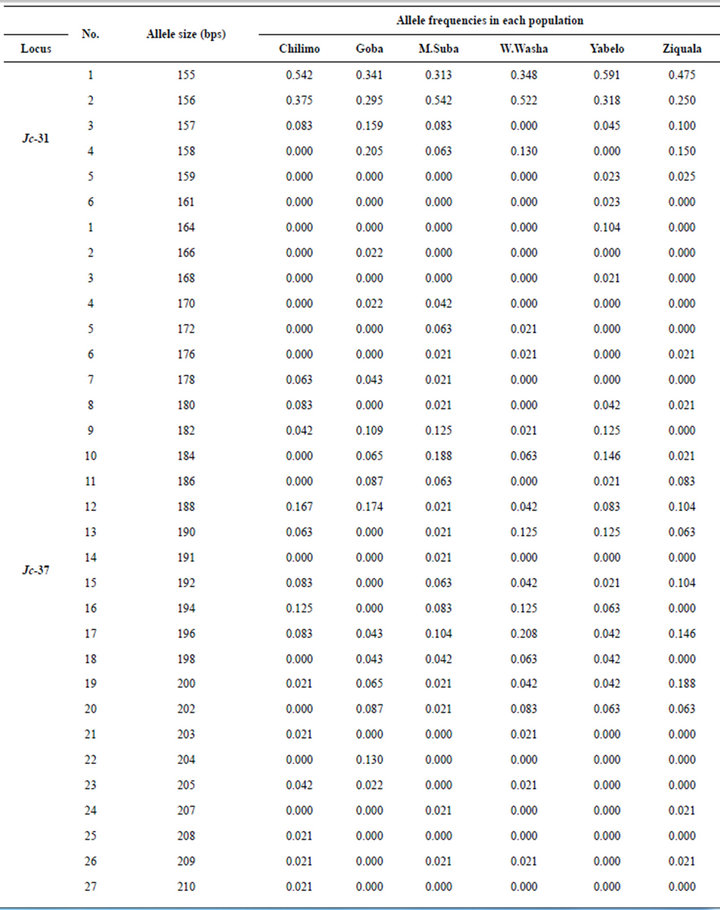
Allelic frequencies at microsatellite loci Jc-31 and Jc-37.
NOTES
*Corresponding author.

