Open Journal of Genetics
Vol. 2 No. 4 (2012) , Article ID: 26313 , 11 pages DOI:10.4236/ojgen.2012.24027
Transcriptome analysis of the Tityus serrulatus scorpion venom gland
![]()
Departamento de Biologia Geral, Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, Belo Horizonte, Brazil
Email: ekalapo@icb.ufmg.br
Received 20 October 2012; revised 21 November 2012; accepted 29 November 2012
Keywords: Scorpions; Antimicrobial Peptides; Neutoxins; Venom Glands; Brazilian Yellow Scorpion
ABSTRACT
The Tityus serrulatus scorpion is considered the most dangerous scorpion in Brazil and is responsible for several cases of human envenomation annually. In this study, we performed transcriptome profiling of the T. serrulatus venom gland. In addition to transcripts with housekeeping functions, such as those related to protein synthesis, energy supply and structural processes, transcripts from thirty-five families of venom peptides or proteins were identified. These transcripts included three new complete sequences of toxins and more than a dozen putative venom gland proteins/peptides. The venom gland transcriptome profile was verified by comparison with the previously determined proteomic profile. In conclusion, this transcriptome data provides novel insights into the putative mechanisms underlying the venomous character of T. serrulatus. The collected data of scorpion transcripts and proteins/peptides described herein may be an important resource for identifying candidate targets of molecular therapies and preventative measures.
1. INTRODUCTION
In 2007, the World Health Organization (WHO) officially designated human envenomation by scorpion sting as a neglected public health issue and recommended urgent international action with a focus on the tropical regions where these venomous animals are abundant. In Brazil, scorpion stings have been recognized a serious public health threat for many decades; yet, the annual incidence of accidental stings remains high, with more than 50,000 cases reported in 2010 [1]. The most frequent culprit among the cases of human envenomation by Brazilian scorpions is the Tityus serrulatus of the Buthidae family [2,3]. As such, this species (known commonly as the Brazilian yellow scorpion) has been the subject of extensive research efforts to isolate and characterize its toxins and other venom components for potential clinical benefit [4-13].
Analyses of T. serrulatus venom have revealed a complex mixture of functionally diverse molecules, including neurotoxins, proteases, hyaluronidases, and hypotensins [3-14]. Of these, the neurotoxins are believed to play a key role in the pathogenesis of scorpionism through their interactions with sodium channels and potassium channels (as reviewed by Cologna et al. [3]). However, to gain a more in-depth and comprehensive understanding of the molecular components of the T. serrulatus venom gland, the gene expression profiling is necessary.
Transcriptomic analyses of the scorpion venom gland have been performed recently for species belonging to various scorpion families, including Buthidae (T. stigmurus and Centruroides noxius), Scorpiopidae (Scorpiops margerisonae and S. jendeki), Scorpionidae (Pandinus cavimanus and Heterometrus petersii), Iuridae (Hadrurus gertschi), and Liochelidae (Opisthacanthus cayaporum) [15-26]. Transcriptomes have provided important insights into the biological processes that are taking place inside venom gland cells [15,18,19,26], as well as identified new genes and expression patterns that form the basis of functional genomics studies [27,28]. Such data for the venom glands of different species represents a resource of potential toxinological and clinical relevance [29,30].
In the current study, the molecular complexity of T. serrulatus venom was investigated by transcriptome profiling of the venom gland. This approach identified the main cellular components and revealed new putative venom constituents, some of which may be candidate targets of new therapeutic strategies to help promote the health of sting victims.
2. MATERIAL AND METHODS
2.1. Library Construction
A cDNA library was constructed from active venom glands of 60 T. serrulatus scorpions that had been milked two days prior to the RNA extraction, as previously described [31].
2.2. Expressed Sequence Tag (EST) Sequencing, Data Processing, and Bioinformatic Analysis
For large-scale DNA sequencing (EST generation), random clones were grown in antibiotic selective medium for approximately 18 h. The plasmid DNA was then isolated using the standard alkaline lysis method and sequenced on an ABI 3130 sequencer with reagents from the BigDye Sequencing Kit (Applied Biosystems Inc., Foster City, CA, USA) and the standard M13 forward or reverse primers.
The resultant trace files of the sequenced clones were applied to the Phred program for base calling and quality scoring using a Phred score cut-off value of 20 [32]. The nucleotide sequences corresponding to vector, adaptors, and Escherichia coli DNA, and any short transcripts (<100 pb) were removed by the SeqClean program [33] (http://compbio.dfci.harvard.edu/tgi/software). The final sequences were deposited in GenBank under accession IDs JK731601-JK732954. The TGICL program [34] was used to assemble high-quality ESTs into contigs (overlapping ESTs that together represent a consensus sequence). Any ESTs without significant similarity to any other ESTs were classified as singlets. Considering the diversity of scorpion toxins, those clusters putative to encode venom peptides were re-examined manually to pick out individual different isoforms. The electropherogram of each putative isoform was visually inspected to confirm the sequencing quality in the polymorphic region.
To annotate the contigs, the set of contigs plus singlets (collectively designated as “uniques”) were searched against the UniProt database [35] (March 2011) using the BLASTx [36] (e-values < 1 × 10–10) and nr database using BLASTx (e-values < 1 × 10–10) in the BLAST2GO program [37]. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis [38] was also performed. The toxin nomenclature was assigned according to the recommendations of King et al. [39] and Cologna et al. [3].
Amino acid sequence of the transcripts was deduced using the open reading frame (ORF) Finder program (http://www.ncbi.nlm.nih.gov/projects/gorf/). Subsequenty, the Seed Server (unpublished, Guedes et al.) was used to search for venom component orthologs. Briefly, the Seed Server methodology uses a protein of interest to group homologues by means of Seed Linkage software [40] and UniRef50 Enriched KEGG Orthology (UEKO) clusters built with the procedure described by Fernandes et al. [41]. The sequences used for alignment analysis were retrieved from the UniProt database and the alignment was performed by webPrank [42] and visualised by the Jalview program [43]. Peptide signal prediction was carried out for putative venom components by using the SignalP 4.0 program [44].
3. RESULTS
3.1. EST Sequencing and Clustering
A total of 1629 high-quality ESTs were generated from the T. serrulatus venom glands. The average length of these ESTs was 421 bp. TGICL-based clustering of the 1629 ESTs yielded 185 contigs (= 1171 ESTs). The average length of these contigs was 625 pb. Four-hundred-and-fifty-eight ESTs showed no significant similarity to any other ESTs in the database and were identified as singlets.
3.2. T. serrulatus Venom Gland Transcript Profile
The BLASTx searches against the UniProt database indicated that among the 643 uniques (185 contigs and 458 singlets), 54% (348 uniques representing 1259 ESTs) encoded precursors of known proteins. The remaining 46% (295 uniques representing 370 ESTs) had e-values >1 × 10–10 and were thus designated as having no match. These no-match transcripts may represent new proteins or peptides.
Cellular localisation and function analyses classified the uniquesand ESTs-deduced products into 10 categories; the distributions of which are shown in Figures 1(a) and (b). The most frequently represented functional categories among the cellular components in T. serrulatus venom gland were: Protein synthesis and processing, structural function, and energy supply.
3.3. Housekeeping Genes
When considering only the ESTs that presented similarity with sequences in the database, about 40% of ESTs were related to cellular components. KEGG analysis (corroborated with manual annotation; Table 1) indicated that the most frequently represented pathways were also involved in protein synthesis and processing (ribosome and protein processing in the endoplasmic reticulum), structural processes (cardiac muscle contraction—important to venom release; regulation of actin cytoskeleton), and energy supply (oxidative phosphorylation). The automated KEGG pathway analysis and manual annotation method yielded similar but not identical results, likely due to the different rules of each approach used to categorize the transcript products.
By manual annotation, the most frequently represented functional categories were protein synthesis and pro cessing (37 ESTs coding ribosomal proteins), eukaryotic
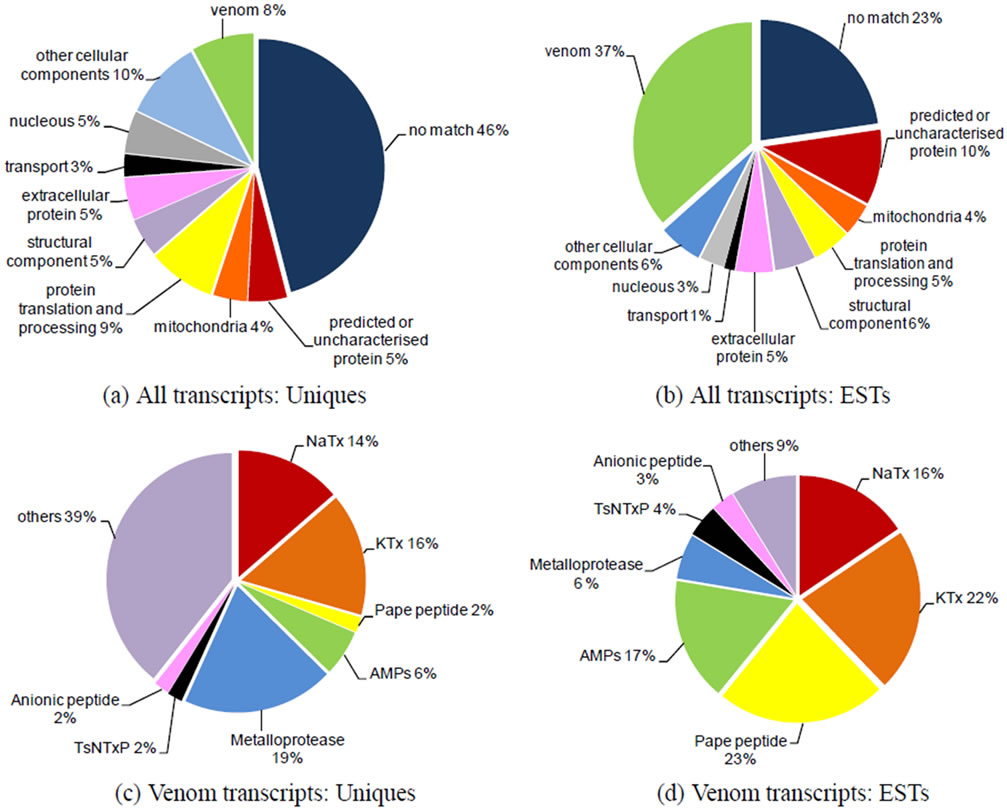
Figure 1. Molecular characteristics of the T. serrulatus venom gland transcriptome. (a) Relative proportion of each category in the uniques. “Venom” includes transcripts encoding toxins and other secreted components previously described in scorpion venom. “Mitochondrial” and “Nuclear” categories comprise ESTs coding conserved proteins located in these cellular organelles. “Protein translation and processing” contains transcripts encoding, for instance, ribosomal protein, disulfide-isomerase, and other proteins related to protein synthesis. “Structural components” includes mainly cytoskeleton proteins, such as actin, myosin, and tubulin. “Transport” comprises transcripts encoding proteins involved in intracellular trafficking, such as the copper transport protein. “Predicted or uncharacterized protein” includes ESTs similar to previously described sequences that have no functional assessment. “Extracellular” comprises transcripts encoding extracellular proteins that are, for instance, found in the extracellular matrix, such as fibronectin. “Other cellular components” includes ESTs encoding cellular components that were not included in any of the other categories; (b) Distribution of ESTs in the same categories described in (a); (c) Relative proportion of uniques encoding different classes of venom components; (d) Abundance of transcripts in the main peptide/protein families of T. serrulatus venom from Table 2.
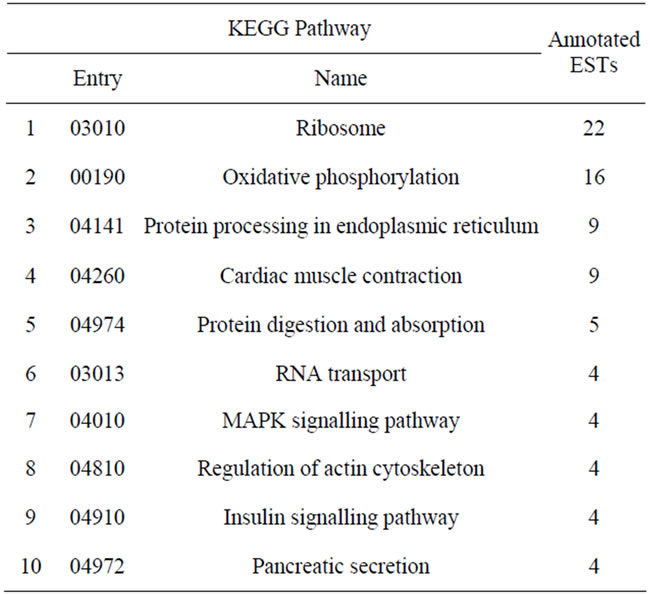
Table 1. The most well represented KEGG pathways in the T. serrulatus transcriptome.
translation initiation (11 ESTs), elongation factors (seven ESTs), disulfite isomerases (four ESTs), and heat shock proteins (HSPs; four ESTs). Seven ESTs encoded peptides that were related to degradation through the ubiquitin-proteasome system. These transcripts might play a role in ensuring that misfolded or otherwise abnormal proteins are recognised and eliminated, or may regulate various molecular pathways in the venom gland, as suggested by Rendón-Anaya et al. [26].
Several transcripts expressed in the venom gland of T. serrulatus were identified as structural components, which may be involved in the maintenance of gland structure and/or contractile activity that mediates venom release. In particular, transcripts were found that encoded actin (16 ESTs), troponin (11 ESTs), myosin (eight ESTs), alpha tubulin (seven ESTs), paramyosin (15 ESTs), and five other ESTs specifically related to cytoskeleton organisation.
The synthesis of venom is considered an energetically costly process. About 5% of the ESTs (8% of uniques) aligned with mitochondrial proteins encoded by nuclear DNA. The majority of these transcripts were related to energy production; in particular transcripts were found that encoded cytochrome c oxidase (17 ESTs), NADHubiquinone oxidoreductase (15 ESTs), ATP synthase (11 ESTs), and cytochrome b (6 ESTs).
3.4. ESTs Related to Venom Components
Five-hundred-and-ninety-four of the ESTs comprised 51 clusters and coded for 35 different families of peptides or proteins related to venom components (Table 2). Except for the incomplete sequences (indicated by asterisks in Table 2), all transcripts had signal peptides. The most abundant and diverse venom transcripts (Figures 1(d)-(e)) encoded neurotoxins (sodium channel toxins or NaTxs and potassium channel toxins or KTxs). In addition, a wide variety of transcripts encoded secreted peptides and proteins. The components that were abundantly expressed included the Pape peptide, similar to bradikinin-potentiating peptide or BPP (UniProt ID: P86821), antimicrobial peptide (AMP), metalloproteases (zinc metalloproteases [UniProt ID: P85842] plus antareases [UniProt ID: P86392]), non-toxic protein NTxP (TsNTxP; UniProt ID: O77463), anionic peptide and hypotensin (UniProt IDs: P84189 and P86824); however, phospholipase A2, hyaluronidase (UniProt ID: P86821), allergen (UniProt ID: P85840) and some cysteine-rich peptides were less frequently expressed.
3.5. New Venom Components of T. serrulatus
Although T. serrulatus venom has been extensively studied, the current transcriptome analysis revealed at least three complete sequences of new potential toxins and more than a dozen new venom components (Table 2). The complete sequence of some of these are detailed below and presented in Figures 2-5. It is important to note that still other new venom components may be represented by those sequences that were designated as nomatch (see the Discussion for more detail).
3.5.1. Sodium Channel Toxins
Three of the known major NaTxs of T. serrulatus venom, Ts1, Ts2 and Ts3, were found in the current transcripttome analysis. Although the Ts5 toxin was not found in this study, a probable new NaTx that is similar to Ts5 (P45659; identity = 86%) was found (designated as Ts17; U-BUTX-Ts1a). Figure 2(a) shows the alignment of the Ts17 sequence with Ts3 (UniProt ID: P01496), Ts5, and other orthologs. In addition, five ESTs matched significantly with the U1-buthitoxin-Hj1a (identities = 63%), a predicted NaTx derived from Hottentotta judaicus, and the T. serrulatus sequence was named Ts18 (U-BUTX-Ts1b) (Figure 2(b)).

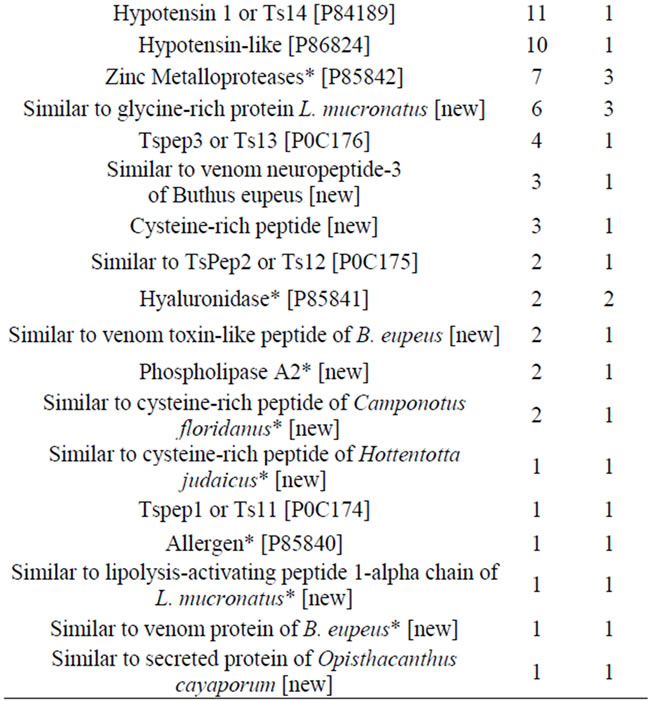
Table 2. ESTs and contigs of T. serrulatus venom components.
3.5.2. Potassium Channel Toxin
Among the KTxs identified in T. serrulatus venom, Ts6, Ts7, Ts8, Ts15 and Ts16 had been previously identified. However, the Ts9 was not found in this study. The potassium channel toxin beta-KTx 2 (UniProt ID: P69940) had been previously identified by proteomic analysis and deposited as peptide fragment [30]; however, our analysis identified its precursor (designated as Ts19; UBUTX-Ts1c) and its orthologs (Figure 2(c)).
3.5.3. Other components
The category “other components” included non-neurotoxic venom components that had been previously described for T. serrulatus as well as potential new venom components harbouring signal peptides and/or showing similarity to venom constituents of other scorpion species (Table 2). However, more evidence is needed to confirm the occurrence of these protein and/or peptide in T. serrulatus venom. It is possible that the expression of some of these secreted components is restricted to the venom gland (for example, secreted components in connective tissue of this organ). Here, we summarize the data of new sequences for highly-expressed transcripts, including the Pape peptide, AMPs, anionic peptides, and hypotensins.
Second only to the neurotoxins, the Pape peptide was the most highly expressed transcript in the T. serrulatus venom gland transcriptome. The Pape peptide (UniProt ID: P86821) was previously identified by proteomic analysis and deposited as a peptide fragment [30]. The complete sequence of the peptide precursor and its orthologs are shown in Figure 3. While the signal peptide is highly conserved among these peptides, the N-terminal region is moderately conserved and the C-terminal region shows very little conservation.
Sequences of AMPs and anionic peptides were abundant in the venom gland transcriptome of T. serrulatus, indicating that these components may play important roles in the function of this organ. The complete se-
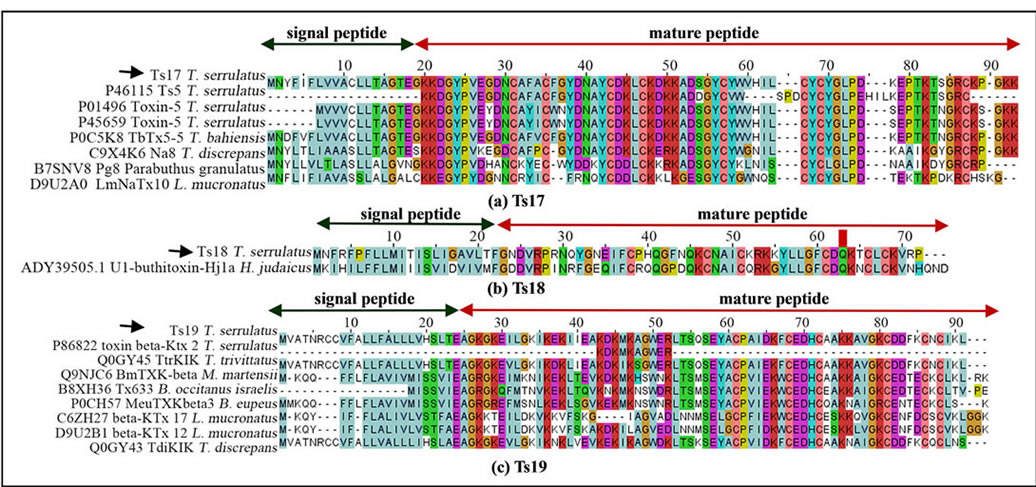
Figure 2. Sequence alignments of NaTxs ((a) and (b)) and KTx (c). (a) The translated sequence of Ts17 and orthologs. For comparison, the paralogous sequences of Ts3 and Ts5 are also presented. The UniProt ID and the corresponding scorpion species are indicated; (b) The Ts18 from the T. serrulatus cDNA library and the related sequence of U1-buthitoxin-Hj1a from Hottentotta judaicus, a predicted NaTx [22]; (c) Complete sequence of Ts19 (KTxs) and ortholog alignment. The UniProt IDs and corresponding scorpion species from genus Tityus, Mesobuthus, Lynchas, Buthus are indicated. The deposited fragment of the potassium channel toxin beta-KTx 2 is presented for comparison. The signal peptide (green arrow) and cleavage sites are shown for Ts17 (between 19 - 20 amino acid resides), Ts18 (between 21 - 22 amino acid resides) and Ts19 (25 - 26 amino acid resides). Black arrows indicate sequences identified in the current study. Dots represent gaps introduced to improve alignment.
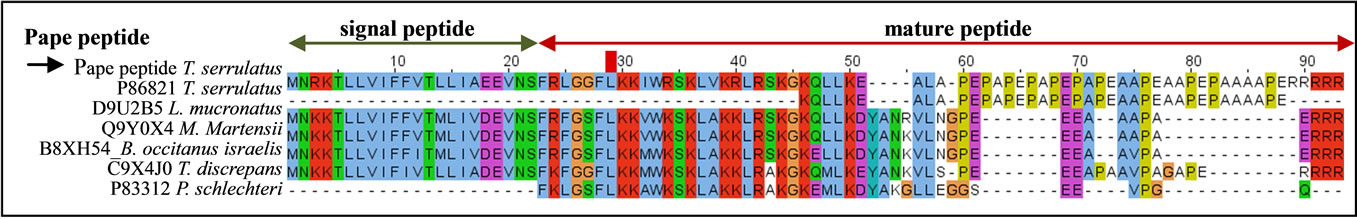
Figure 3. Alignment of the Pape peptide with its orthologs. The black arrow indicates the sequence identified in the current study and P86821 is the Pape peptide previously identified. The signal peptide (green arrow) and the cleavage site (between 22 - 23 amino acid resides) are shown. The UniProt ID and the corresponding scorpion species are indicated. Dots represent gaps introduced to improve alignment.
quences of these venom components are shown in Figure 4. Three different sequences coded an AMP highly similar to the putative AMP of T. costatus (Figure 4(a)). In addition, two ponericin-like sequences were found and presumed to be antimicrobial peptide (Figure 4(b)). In this study, 18 ESTs encoded the same sequence of anionic peptide (Figure 4(c)). Anionic peptides have been previously reported as highly expressed and conserved among the Buthidae scorpion species [17,23]. Although the function of these peptides remains unknown [45], some researchers have suggested that they might play antimicrobial activity [46] or an important role in pH balance, since neurotoxins are basic peptides [18,22].
Hypotensins were identified in T. serrulatus venom gland transcriptome. These random-coiled linear peptides are characterized by the bradykinin-potentiating peptide amino acid signature [13]. Of the three sequences in the UniProt database (hypotensin-1, UniProt ID: P84189; hypotensin-2, UniProt ID: P84190; hypotensin-like peptide, UniProt ID: P86824), the complete sequence of hypotensin-like and hypotensin-1 precursors were identified (Figure 5).
4. DISCUSSION
4.1. Transcriptome Profiling Has Provided Comprehensive
Information of venom glands, accelerate the discovery of their peptides and proteins [47]. Currently, 12 studies in the publicly available literature have reported data of the venom glands from various scorpion species [15-26]. The study described herein not
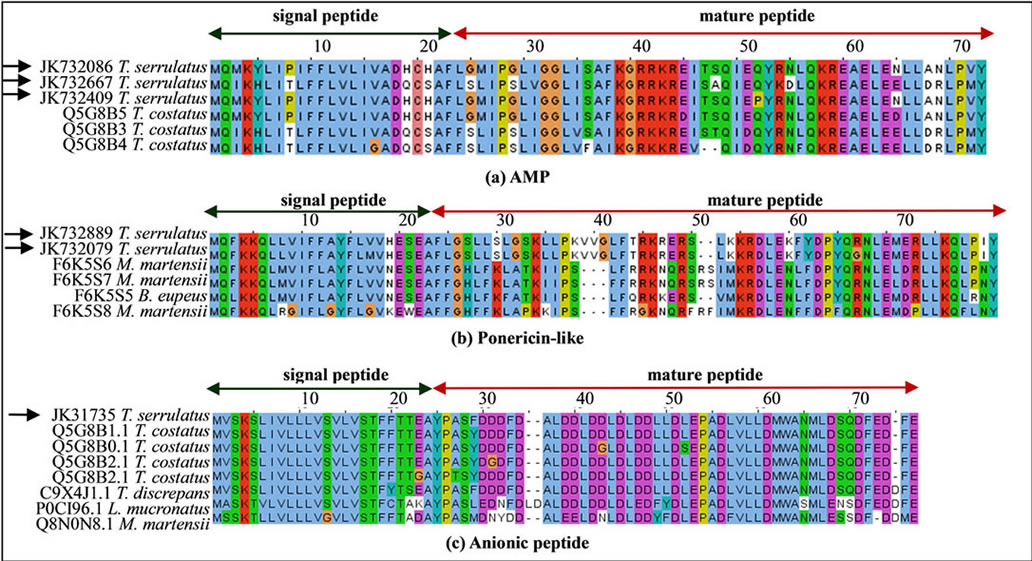
Figure 4. Sequence alignment of putative antimicrobial peptides and orthologs. (a) Three sequences of AMP exist in the Buthidae scorpions T. serrulatus and T. costatus; (b) Two Ponericin-like peptide sequences, presumed to be an antimicrobial peptide, were found in T. serrulatus transcriptome; (c) Sequence of anionic peptide found in T. serrulatus and alignment with its relatively conserved orthologs from other Buthidae scorpions. The signal peptide (green arrow) and cleavage sites are shown for AMP (between 22 - 23 amino acid resides), Ponericin-like peptide (between 23 - 24 amino acid resides) and anionic peptide (24 - 25 amino acid resides) Black arrows indicate sequences identified in the current study.
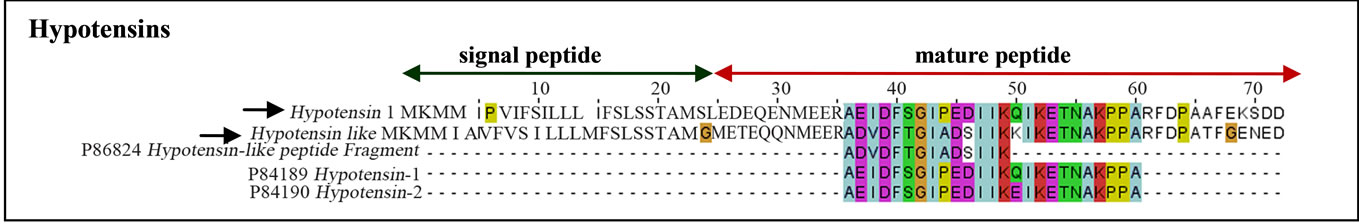
Figure 5. Sequence of hypotensins found in T. serrulatus. Black arrows indicate sequences identified in the current study. The signal peptide (green arrow) with the cleavage site between 24 and 25 amino acid residues is shown.
only provides the first transcriptome data for the T. serrulatus venom gland but also the largest transcript catalogue for scorpion venom glands obtained by Sanger sequencing to date. This work also focused on the discovery of new genes (using the EST approach) which when added to the larger databases will allow for better anchoring of transcriptomic short reads generated by most of the next-generation sequencing (NGS) platforms.
In this study, 35 different peptide families, coded by 594 ESTs, were identified as related to venom components. Previous proteomic analysis of T. serrulatus venom [14,30] has identified many toxins, in addition, venom components that have since been isolated and studied individually [4-9,13]. The Animal Toxin Annotation Program (UniProt) [48], which annotates the secreted proteins in animal venoms, has already identified 32 peptides/proteins in T. serrulatus venom. The current transcriptome analysis identified three precursors of potential new toxins (Ts17, Ts18, and Ts19) and more than a dozen new venom components in T. serrulatus. However, this list is certainly not exhaustive, since a large number of sequences without matches (295 uniques representing 370 transcripts) were found. We intend to continue to explore the entire dataset to identify other new venom components and to verify the biological function and clinical relevance in envenomation for any promising factors.
Some of the venom components previously identified were not identified by the current transcriptome analysis, such as Peptide T (UniProt ID: Q9TWR4), Ts5 (UniProt ID: P45659) and alpha amylase (UniProt ID: P85843). However, the absence of venom components at the transcript level is not a surprising observation. In fact, a previous study in C. noxius indicated that the powerful pyrosequencing platform, producing over three million reads, was unable to detect the entire panel of known toxins [26]. Some hypotheses to explain the transcript absence are: 1) The transcriptomic analysis performed did not provide complete coverage; 2) Toxin genes were down-regulated at the time of RNA extraction or had undergone microRNA-mediated degradation during processing, as suggested by Rendón-Anaya et al. [26]; and 3) the venom protein/peptide had undergone post-translational modifications that significantly differentiated it from the intact form, as described by Pimenta et al. [14].
Considering the previously reported protein/peptide compositions of T. serrulatus venom [3,14,30], we expected to find to a high level of neurotoxin expression. Indeed, NaTxs, the main agents responsible for the toxic effects of T. serrulatus envenomation [49], presented high expression; in particular, this was observed for Ts1 and Ts2. In the current study, NaTx represented about 6% of the total transcripts. However, transcriptome analysis of another scorpion Brazilian species’, T. stigmurus, venom gland [25], showed lower expression levels of NaTxs (1.3% of the total transcripts).The higher expression of NaTxs in T. serrulatus may be related to the higher lethality of this scorpion, as compared to T. stigmurus [25]. KTxs also represented a high number of ESTs in T. serrulatus, representing about 8% of the total transcripts. Specifically, Ts8 and Ts19 were responsible for the higher expression level. However, the overall expression level of KTxs in T. serrulatus was lower than in T. stigmurus, for which KTxs represented 13.5%. The differences in expression level of NaTxs and KTxs between T. serrulatus and T. stigmurus may be speciesspecific or reflect the differences of transcriptional profiles for active and resting venom glands. The T. stigmurus cDNA library was performed for resting venom glands [25], while the current study used active venom glands.
Another important finding from the current study is the robust expression of some particular components, such as the Pape peptide, AMPs, anionic peptide, and metalloproteases. Although further analysis is required to uncover the precise functions of these venom components in the active venom gland of T. serrulatus, their observed abundance suggests an important role in the biological function of this species’ venom gland. Indeed, the Pape peptide and a peptide similar to Ponericin-L1 and Ponericin-L2 were identified in the previous proteomic study of T. serrulatus venom performed by Rates et al. [30]. The current transcriptome analysis identified precursors of both peptides. Interestingly, the Pape peptide represented 8.5% of the total transcripts. This peptide was similar to the BPP found in other scorpion species and Parabutoporin, an antimicrobial peptide identified in Parabuthus schlechteri (Figure 3). Despite its higher expression level, the function of the Pape peptide in T. serrulatus venom remains unknown. Besides the Ponericin-like peptide, sequence similar to AMP was found. Various AMPs have been previously identified in the venom of several scorpion species [15,18,19] and appear to be highly expressed in the S. jendeki [18], H. petersii [20], Isometrus maculatus [23] and T. stigmurus [26] species. While no consensus has yet been reached about the precise functions of AMP in scorpion venom, it is theorized they may act as protectants against bacterial infection or potentiators of neurotoxin action [50]. The demonstrated antibacterial activities of AMPs in animals, plants, and insects have indicated their potential for use as antimicrobial agents [46,50-52]. Hence, the AMPs described in the current study might represent potential candidates for anti-infective drugs.
Morgenstern et al. [22] showed a higher abundance of proteases and metalloproteases in the resting venom gland of the Buthidae scorpion. All animals used in the present study had been milked prior to the mRNA extraction, yet the expression level of metalloproteases was as high as some of the NaTx and KTx sequences. Prosdocimi et al. [53] found a similar high expression of metalloproteases (astacin family) in the Gasteracantha cancriformis spider’s spinning gland transcriptome, and suggested that these proteins may play a role in the remodelling processes of silk fibre deposition. A higher expression of these proteases was also observed in the L. mucronatus venom, suggesting that these proteins might play a central role in scorpion venom as well [21]. Indeed, several antarease members of the metalloprotease family have been purified from the T. serrulatus venom gland and shown to selectively cleave the essential SNARE protein within mammalian pancreatic tissue; it has been suggested that this function may be responsible for the pancreatitis that develops in some patients following scorpion envenomation [54]. Alternatively, the metalloproteases may play a specific non-toxic role in the scorpion’s venom gland. A large number of fragments derived from larger peptides have been found by the proteomic studies of venom glands [14,30]. It appears that different fragments derived from the same peptide might have distinct biological functions [55]. Hence, we speculate that the protease-mediated fragmentation may act to exponentially increase the diversity of venom peptides and their biological targets.
The scorpion’s venom is mainly used for prey capture and defence. The venom gland is a complex organ where a large number of substances are synthesized. Transcriptomic tools have accelerated the description of venom components, greatly expanding the publicly available sequence databases. A large diversity of venom components were found using this approach: NaTxs, KTxs, scorpines, calcines, AMPs, BPPs, anionic peptides, proteases, glycine-rich peptides, phospholipases, lectins, hypotensins [15-26]. In the current transcriptome study— the first of its kind to examine the active venom gland of T. serrulatus—several of these components were found as well as several new components, including potential toxins, AMPs, and cysteine-rich peptides. Another important finding was the surprisingly high transcription levels of some non-neurotoxic components, such as the Pape peptide, AMPs, anionic peptides, metalloproteases, and hypotensins. Overall, this analysis provides a novel and more comprehensive insight into the venom arsenal of T. serrulatus. These data may act as an important resource for future investigations of the evolution of the scorpion venom arsenal, envenomation mechanisms, and discovery of bioactive peptides and proteins.
5. CONCLUSION
This work represents a step towards better understanding the gene expression profile of the active T. serrulatus venom gland, expanding our knowledge of its peptide and protein content. The transcriptome analysis revealed at least three new precursors of toxins and more than a dozen potential venom components. Besides the more common toxins (NaTx and KTx), some transcripts, such as the Pape peptide, metalloproteases, AMPs, anionic peptides, and hypotensins, presented high expression level. The functions of these components may help to advance our understanding of the biological and molecular processes of the venom gland. The gene expression profile of the venom gland agreed with the activated state of this organ and revealed the activities of protein synthesis and processing and energy supply. Some potential bioactive proteins/peptides (such as Ts17, Ts18, Ts19, and AMPs) described in this work may be an important resource for the investigation and characterization of molecules applicable in pharmaceutical research and biotechnology.
6. ACKNOWLEDGEMENTS
We wish to thank the Brazilian National Council for Research (CNPq), FAPEMIG and CAPES (Edital Toxinologia 063/2010), for financial support. The FAPEMIG scholarship awarded to Érika Ramos de Alvarenga is also gratefully acknowledged. The authors also are grateful to Dr. J. Miguel Ortega and Raphael L. M. Guedes for help in the bioinformatics analysis.
REFERENCES
- Brazilian Ministry of Health (2011) http://portal.saude.gov.br/portal/saude/profissional/visualizar_texto.cfm?idtxt=31519
- Chippaux, J.P. and Goyffon, M. (2008) Epidemiology of scorpionism: A global appraisal. Acta Tropica, 107, 71-79. doi:10.1016/j.actatropica.2008.05.021
- Cologna, C.T., Marcussi, S., Giglio, J.R., Soares, A.M. and Arantes, E.C. (2009) Tityus serrulatus scorpion venom and toxins: An overview. Protein and Peptide Letters, 16, 920-932. doi:10.2174/092986609788923329
- Possani, L.D., Alagón, A.C., Fletcher Jr, P.L. and Erickson, B.W. (1977) Purification and properties of mammalian toxins from the venom of Brazilian Scorpion Tityus serrulatus Lutz and Mello. Archives of Biochemistry and Biophysics, 180, 394-403. doi:10.1016/0003-9861(77)90053-4
- Arantes, E.C., Prado, W.A., Sampaio, S.V., Giglio, J.R. (1989) A simplified procedure for the fractionation of Tityus serrulatus venom: Isolation and partial characterization of TsTX-IV, a new neurotoxin. Toxicon, 27, 907-916. doi:10.1016/0041-0101(89)90102-5
- Sampaio, S.V., Arantes, E.C., Prado, W.A., Riccioppo Neto, F. and Giglio, J.R. (1991) Further characterization of toxins T1IV (TsTX-III) and T2IV from Tityus serrulatus scorpion venom. Toxicon, 29, 663-672. doi:10.1016/0041-0101(91)90058-Y
- Martin-Eauclaire, M.F., Céard, B., Ribeiro, A.M., Diniz, C.R., Rochat, H. and Bougis, P.E. (1994) Biochemical, pharmacological and genomic characterisation of Ts IV, an alpha-toxin from the venom of the South American scorpion Tityus serrulatus. FEBS Letters, 342, 181-184. doi:10.1016/0014-5793(94)80496-6
- Sampaio, S.V., Coutinho-Netto, J., Arantes, E.C., Marangoni, S., Oliveira, B. and Giglio, J.R. (1996) Isolation of toxin TsTX-VI from Tityus serrulatus scorpion venom. Effects on the release of neurotransmitters from synaptosomes. Biochemistry and Molecular Biology International, 39, 729-740. doi:10.1080/15216549600201811
- Guatimosim, S.C., Prado, V.F., Diniz, C.R., Chávez-Olórtegui, C. and Kalapothakis, E. (1999) Molecular cloning and genomic analysis of TsNTxp: An immunogenic protein from Tityus serrulatus scorpion venom. Toxicon, 37, 507-517. doi:10.1016/S0041-0101(98)00187-1
- Novello, J.C., Arantes, E.C., Varanda, W.A., Oliveira, B., Giglio, J.R. and Marangoni, S. (1999) TsTX-IV, a short chain four-disulfide-bridged neurotoxin from Tityus serrulatus venom which acts on Ca2+-activated K+ channels. Toxicon, 37, 651-660. doi:10.1016/S0041-0101(98)00206-2
- Petricevich, V.L., Hernández Cruz, A., Coronas, F.I. and Possani, L.D. (2007) Toxin gamma from Tityus serrulatus scorpion venom plays an essential role in immunomodulation of macrophages. Toxicon, 50, 666-675. doi:10.1016/j.toxicon.2007.06.001
- Mendes, T.M., Dias, F., Horta, C.C., Pena, I.F., Arantes, E.C. and Kalapothakis, E. (2008) Effective Tityus serrulatus anti-venom produced using the Ts1 component. Toxicon, 52, 787-793. doi:10.1016/j.toxicon.2008.08.005
- Verano-Braga, T., Rocha-Resende, C., Silva, D.M., Ianzer, D., Martin-Eauclaire, M.F., et al. (2008) Tityus serrulatus hypotensins: A new family of peptides from scorpion venom. Biochemical and Biophysical Research Communications, 371, 515-520. doi:10.1016/j.bbrc.2008.04.104
- Pimenta, A.M., Stöcklin, R., Favreau, P., Bougis, P.E. and Martin-Eauclaire, M.F. (2001) Moving pieces in a proteomic puzzle: Mass fingerprinting of toxic fractions from the venom of Tityus serrulatus (Scorpiones, Buthidae). Rapid Communications in Mass Spectrometry, 15, 1562-1572.
- Schwartz, E.F., Diego-Garcia, E., Rodríguez de la Vega, R.C. and Possani, L.D. (2007) Transcriptome analysis of the venom gland of the Mexican scorpion Hadrurus gertschi (Arachnida: Scorpiones). BMC genomics, 8, 119-119. doi:10.1186/1471-2164-8-119
- Kozminsky-Atias, A., Bar-Shalom, A., Mishmar, D. and Zilberberg, N. (2008) Assembling an arsenal, the scorpion way. BMC Evolutionary Biology, 8, 333-333. doi:10.1186/1471-2148-8-333
- D’Suze, G., Schwartz, E.F., García-Gómez, B.I., Sevcik, C. and Possani, L.D. (2009) Molecular cloning and nucleotide sequence analysis of genes from a cDNA library of the scorpion Tityus discrepans. Biochimie, 91, 1010-1019.
- Ma, Y., Zhao, R., He, Y., Li, S., Liu, J., Wu, Y., et al. (2009) Transcriptome analysis of the venom gland of the scorpion Scorpiops jendeki: Implication for the evolution of the scorpion venom arsenal. BMC genomics, 10, 290-290. doi:10.1186/1471-2164-10-290
- Silva, E.C., Camargos, T.S., Maranhão, A.Q., Silva-Pereira, I., Silva, L.P., Possani, L.D., et al. (2009) Cloning and characterization of cDNA sequences encoding for new venom peptides of the Brazilian scorpion Opisthacanthus cayaporum. Toxicon, 54, 252-261. doi:10.1016/j.toxicon.2009.04.010
- Ma, Y., Zhao, Y., Zhao, R., Zhang, W., He, Y., Wu, Y., et al. (2010) Molecular diversity of toxic components from the scorpion Heterometrus petersii venom revealed by proteomic and transcriptome analysis. Proteomics, 10, 2471-2485. doi:10.1002/pmic.200900763
- Ruiming, Z., Yibao, M., Yawen, H., Zhiyong, D., Yingliang, W., Zhijian, C., et al. (2010) Comparative venom gland transcriptome analysis of the scorpion Lychas mucronatus reveals intraspecific toxic gene diversity and new venomous components. BMC genomics, 11, 452-452. doi:10.1186/1471-2164-11-452
- Morgenstern, D., Rohde, B.H., King, G.F., Tal, T., Sher, D. and Zlotkin, E. (2011) The tale of a resting gland: Transcriptome of a replete venom gland from the scorpion Hottentotta judaicus. Toxicon, 57, 695-703. doi:10.1016/j.toxicon.2011.02.001
- Ma, Y., He, Y., Zhao, R., Wu, Y., Li, W. and Cao, Z. (2011) Extreme diversity of scorpion venom peptides and proteins revealed by transcriptomic analysis: Implication for proteome evolution of scorpion venom arsenal. Journal of Proteomics, 75, 1563-1576. doi:10.1016/j.jprot.2011.11.029
- Diego-García, E., Peigneur, S., Clynen, E., Marien, T., Czech, L., Schoofs, L., et al. (2012) Molecular diversity of the telson and venom components from Pandinus cavimanus (Scorpionidae Latreille 1802): Transcriptome, venomics and function. Proteomics, 12, 313-328. doi:10.1002/pmic.201100409
- Almeida, D.D., Scortecci, K.C., Kobashi, L.S., AgnezLima, L.F., Medeiros, S.R., Silva-Junior, A.A., et al. (2012) Profiling the resting venom gland of the scorpion Tityus stigmurus through a transcriptomic survey. BMC Genomics, 13, 362. doi:10.1186/1471-2164-13-362
- Rendón-Anaya, M., Delaye, L., Possani, L.D. and HerreraEstrella, A. (2012) Global transcriptome analysis of the Scorpion Centruroides noxius: New toxin families and evolutionary insights from an ancestral scorpion species. PLoS One, 7, e43331. doi:10.1371/journal.pone.0043331
- Adams, M.D., Kelley, J.M., Gocayne, J.D., Dubnick. M., Polymeropoulos, M.H., Xiao, H., et al. (1991) Complementary DNA sequencing: Expressed sequence tags and human genome project. Science, 252, 1651-1656. doi:10.1126/science.2047873
- Takasuga, A., Hirotsune, S., Itoh, R., Jitohzono, A., Suzuki, H., Aso, H., et al. (2001) Establishment of a high throughput EST sequencing system using poly(A) tail-removed cDNA libraries and determination of 36 000 bovine ESTs. Nucleic Acids Research, 29, e108-e108. doi:10.1093/nar/29.22.e108
- Zhang, B., Liu, Q., Yin, W., Zhang, X., Huang, Y., Luo, Y., et al. (2006) Transcriptome analysis of Deinagkistrodon acutus venomous gland focusing on cellular structure and functional aspects using expressed sequence tags. BMC genomics, 7, 152-152. doi:10.1186/1471-2164-7-152
- Rates, B., Ferraz, K.K., Borges, M.H., Richardson, M., De Lima, M.E. and Pimenta, A.M. (2008) Tityus serrulatus venom peptidomics: Assessing venom peptide diversity. Toxicon, 52, 611-618. doi:10.1016/j.toxicon.2008.07.010
- Kalapothakis, E., Jardim, S., Magalhães, A.C., Mendes, T.M., De Marco, L., Afonso, L.C., et al. (2001) Screening of expression libraries using ELISA: Identification of immunogenic proteins from Tityus bahiensis and Tityus serrulatus venom. Toxicon, 39, 679-685. doi:10.1016/S0041-0101(00)00194-X
- Ewing, B., Hillier, L., Wendl, M.C., Green, P. (1998) Basecalling of automated sequencer traces using phred. I. Accuracy assessment. Genome Research, 8, 175-185.
- SeqClean. http://compbio.dfci.harvard.edu/tgi/software
- Pertea, G., Huang, X., Liang, F., Antonescu, V., Sultana, R., Karamycheva, S., et al. (2003) TIGR gene indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics, 19, 651-652. doi:10.1093/bioinformatics/btg034
- Consortium, T.U. (2010) The universal protein resource (UniProt) in 2010. Nucleic Acids Research, 38, D142-D148. doi:10.1093/nar/gkp846
- Altschul, S., Madden, T., Schäffer, A., Zhang, J., Zhang, Z., Miller, et al. (1997) Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research, 25, 3389-3402. doi:10.1093/nar/25.17.3389
- Conesa, A., Götz, S., García-Gómez, J., Terol, J., Talón, M. and Robles, M. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 21, 3674-3676. doi:10.1093/bioinformatics/bti610
- Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H. and Kanehisa, M. (1999) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research, 27, 29-34. doi:10.1093/nar/27.1.29
- King, G.F., Gentz, M.C., Escoubas, P. and Nicholson, G.M. (2008) A rational nomenclature for naming peptide toxins from spiders and other venomous animals. Toxicon, 52, 264-276. doi:10.1016/j.toxicon.2008.05.020
- Barbosa-Silva, A., Satagopam, V., Schneider, R. and Ortega, J.M. (2008) Clustering of cognate proteins among distinct proteomes derived from multiple links to a single seed sequence. BMC Bioinformatics, 9, 141. doi:10.1186/1471-2105-9-141
- Fernandes, G.R., Barbosa, D.V.C., Prosdocimi, F., Pena, I.A., Santana-Santos, L., Coelho Junior, et al. (2008) A procedure to recruit members to enlarge protein family databases—the building of UECOG (UniRef-Enriched COG Database) as a model. Genetics and Molecular Research, 7, 910-924. doi:10.4238/vol7-3X-Meeting008
- Löytynoja, A. and Goldman, N. (2010) webPRANK: A phylogeny-aware multiple sequence aligner with interacttive alignment browser. BMC Bioinformatics, 11, 579-579. doi:10.1186/1471-2105-11-579
- Waterhouse, A.M., Procter, J.B., Martin, D.M.A, Clamp, M. and Barton, G.J. (2009) Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics, 25, 1189-1191. doi:10.1093/bioinformatics/btp033
- Petersen, T.N., Brunak, S., von Heijne, G. and Nielsen, H. (2011) SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nature Methods, 8, 785-786. doi:10.1038/nmeth.1701
- Diego-García, E., Batista, C.V., García-Gómez, B.I., Lucas, S., Candido, D.M., Gómez-Lagunas, F., et al. (2005) The Brazilian scorpion Tityus costatus karsch: Genes, peptides and function. Toxicon, 45, 273-283. doi:10.1016/j.toxicon.2004.10.014
- Harris, F., Dennison, S.R. and Phoenix, D.A. (2009) Anionic antimicrobial peptides from eukaryotic organisms. Current Protein and Peptide Science, 10, 585-606. doi:10.2174/138920309789630589
- Rodríguez de la Vega, R.C., Schwartz, E.F. and Possani, L.D. (2010) Mining on scorpion venom biodiversity. Toxicon, 56, 1155-1161. doi:10.1016/j.toxicon.2009.11.010
- Jungo F., Estreicher A., Bairoch A., Bougueleret L. and Xenarios I. (2010) Animal toxins: How is complexity represented in databases? Toxins, 2, 262-282. doi:10.3390/toxins2020261
- Kalapothakis, E. and Chávez-Olórtegui, C. (1997) Venom variability among several Tityus serrulatus specimens. Toxicon, 35, 1523-1529. doi:10.1016/S0041-0101(97)00017-2
- Carballar-Lejarazú, R., Rodríguez, M., de la Cruz HernándezHernández, F., Ramos-Castañeda, J., Possani, L., ZuritaOrtega, et al. (2008) Recombinant scorpine: A multifunctional antimicrobial peptide with activity against different pathogens. Cellular and Molecular Life Science, 65, 3081-3092. doi:10.1007/s00018-008-8250-8
- Pimenta, A.M. and De Lima, M.E. (2005) Small peptides, big world: Biotechnological potential in neglected bioactive peptides from arthropod venoms. Journal of Peptide Science, 11, 670-676. doi:10.1002/psc.701
- Dai, C., Ma, Y., Zhao, Z., Zhao, R., Wang, Q., Wu, Y., et al. (2008) Mucroporin, the first cationic host defense peptide from the venom of Lychas mucronatus. Antimicrobial Agents and Chemotherapy, 52, 3967-3972. doi:10.1128/AAC.00542-08
- Prosdocimi, F., Bittencourt, D., da Silva, F.R., Kirst, M., Motta, P.C. and Rech, E.L. (2011) Spinning gland transcriptomics from two main clades of spiders (order: Araneae) insights on their molecular, anatomical and behavioral evolution. PLoS One, 6, e21634. doi:10.1371/journal.pone.0021634
- Fletcher, P.L., Fletcher, M.D., Weninger, K., Anderson, T.E. and Martin, B.M. (2010) Vesicle-associated membrane protein (VAMP) cleavage by a new metalloprotease from the Brazilian scorpion Tityus serrulatus. The Journal of Biological Chemistry, 285, 7405-7416. doi:10.1074/jbc.M109.028365
- Diego-García, E., Abdel-Mottaleb, Y., Schwartz, E.F., de la Vega, R.C., Tytgat, J. and Possani, L.D. (2008) Cytolytic and K+ channel blocking activities of beta-KTx and scorpine-like peptides purified from scorpion venoms. Cellular and Molecular Life Sciences, 65, 187-200. doi:10.1007/s00018-007-7370-x

