Advances in Biological Chemistry
Vol. 2 No. 4 (2012) , Article ID: 24647 , 6 pages DOI:10.4236/abc.2012.24045
Use of a naturally occurring codon bias for identifying topoisomerase mutations in ciprofloxacin resistant Escherichia coli using PCR and future prospects with other bacterial genera: A pilot study*
![]()
1Division of Infectious Diseases, Department of Pediatrics, The Saban Research Institute, Children’s Hospital Los Angeles, University of Southern California School of Medicine, Los Angeles, USA
2Tissue Culture and Drug Discovery Laboratory, Centre for Biotechnology, Anna University, Chennai, India
3Department of Microbiology, PSG Institute of Medical Sciences and Research, Coimbatore, India
Email: #skrishnan@chla.usc.edu
Received 12 September 2012; revised 14 October 2012; accepted 21 October 2012
Keywords: Escherichia coli Gyrase A; Codon Bias; Mutation; Ciprofloxacin Resistance; PCR
ABSTRACT
We developed a novel PCR method aimed at identifying and amplifying native codon sequences of mutation-prone amino acids in DNA gyrase implicated in quinolone resistance using a naturally occurring codon bias in E. coli DNA gyrase A.
1. INTRODUCTION
The bacterial DNA gyrase enzyme induces negative supercoils in the DNA during replication and is the target of action for quinolone drugs [1]. Escherichia coli ( E. coli) has 2 sub units, namely gyrase A (gyrA) and gyrase B (gyrB) that have molecular weights of 97 kDa and 90 kDa respectively [2]. GyrA interacts with DNA and cleaves it using its active site tyrosine residue (Tyr122) while GyrB has an ATPase active site. Recently, a small angle scattering X-ray study revealed the structure of GyrA and GyrB bound to DNA in E. coli [3]. Mutations in gyrase A (gyrA) lead to bacterial resistance to quinolone drugs. Substitution of serine at residue 83 (ser83) in gyrase A is the single most important reason for resistance to quinolones [4]. Mutations at ser83 of gyrA is the most frequent in Escherichia coli (E. coli) clinical isolates[5]. Mutation of ser83 to leucine (Leu) or tryptophan (Trp) confers high level resistance to quinolones while ser83 to Alanine (Ala) confers low level resistance [6]. Similarly, aspartate at residue 87 (asp87) is another important target for quinolone drugs. Mutation of asp87 in gyrase A to asparagine (Asp) or valine (Val) is common [7]. But in E. coli, the serine codon for primordial genes like ATP synthase and topoisomerase II is highly biased to TCN (where N can be A, T, G or C) than AGY (where Y can be C or T) [8]. Quinolone resistance is on the rise among E. coli clinical isolates [9]. Although regular disk diffusion techniques are employed to identify quinolone resistance [10] the molecular basis of quinolone resistance in E. coli attributed to mutations in topoisomerase II and presence of qnr genes could provide further understanding of the consistency of such resistance patterns.
PCR based detection of mutations that confer resistance, not only saves time when compared to disk diffusion methods, but also helps in identification of the nature of mutations that influence the resistance to antibiotics. Although various techniques for identifying mutations in gene sequences have been employed in microbiology laboratories around the world, they are not time and cost effective. We propose a new type of PCR using primers for amplification of native codon sequence or PAN PCR. By this method, only the native codon sequence is amplified in a PCR reaction while a mutated sequence will not. The naturally occurring codon bias in E. coli especially with respect to gyrase A forms the basis of this assay. This method was found to be quite efficient in identifying mutations in gyrA. Hence, identification of such codon biased sequences and design of PCR techniques targeting the mutations, not only in gyrase A but also in the target proteins for aminoglycosides, penicillins and macrolides.
2. MATERIAL AND METHODS
2.1. Bacterial Cultures and Disk Diffusion Experiments
52 non-repetitive E. coli strains were isolated from patients at the Department of Microbiology, PSG Institute of Medical Sciences and Research during 2008 and their antibiotic resistance profile for ciprofloxacin, ofloxacin, norfloxacin and nalidixic acid (HiMedia, India) was made using disk diffusion method as per CLSI guidelines. E. coli ATCC 25922 was used as a quality control strain for the experiments.
2.2. Primer Design for gyrA, parC and qnr
The primers for control gyrA and parC were designed as follows: gyrA-F, 5’-GACCTTGCGAGAAGAAAT TACAC-3’(position:7-28); gyrA-R,5’-GATGTTGGTTGCCATACCTACG-3’ (position 546 - 525). These sets of primers were used as internal control for the PAN PCR reactions. For analysis of the presence of the native codon sequence, the forward primer of gyrA was used with gyrAser83R, 5’-TCGTGTC ATAGACCGCCG-3’ (position 265-248) or gyrAasp87R (5’-GCGCCATGCGGACGATCGTGTC-3’). For analyzing plasmid-mediated quinolone resistance, we assessed the presence of qnr genes (qnrA1 to qnrA6, qnrB1 to qnrB6 and qnrS1 to qnrS2) using primer sequences published by Cattoir and others [11].
2.3. PCR Conditions
All the primers were purchased from Integrated DNA Technologies, USA. For the PCR reaction, the heat lysis method described previously [12] was followed. Briefly, 1 μL of supernatant containing template DNA was added to 9 μL of master mix containing 5 pM of forward and 5 pM each of reverse or PAN primers, 1 μL of 10× Taq buffer (GeneI, India) and 0.5 units of Taq DNA polymerase (GeneI, India).
The steps of PCR were as follows: Initial denaturation at 95˚C for 5 minutes, followed by 30 cycles of denaturation at 95˚C for 1 minute, annealing at 52˚C for gyrA and 47˚C for parC for 1 minute and 30 seconds, extension at 72˚C for 1 minute and a final extension at 72˚C for 5 minutes. PCR products were resolved on 1% agarose gels in 0.5× TBE buffer and stained with ethidium bromide for visualization.
3. RESULTS
In the disk diffusion method, 43 strains showed resistance to at least one of the four quinolone antibiotics (82.6%) and 9 strains were susceptible to all the four antibiotics. Among these 43 strains, 5 strains showed resistance only to nalidixic acid but were susceptible to the other three. The remaining 38 strains were resistant to all the four antibiotics (73%).
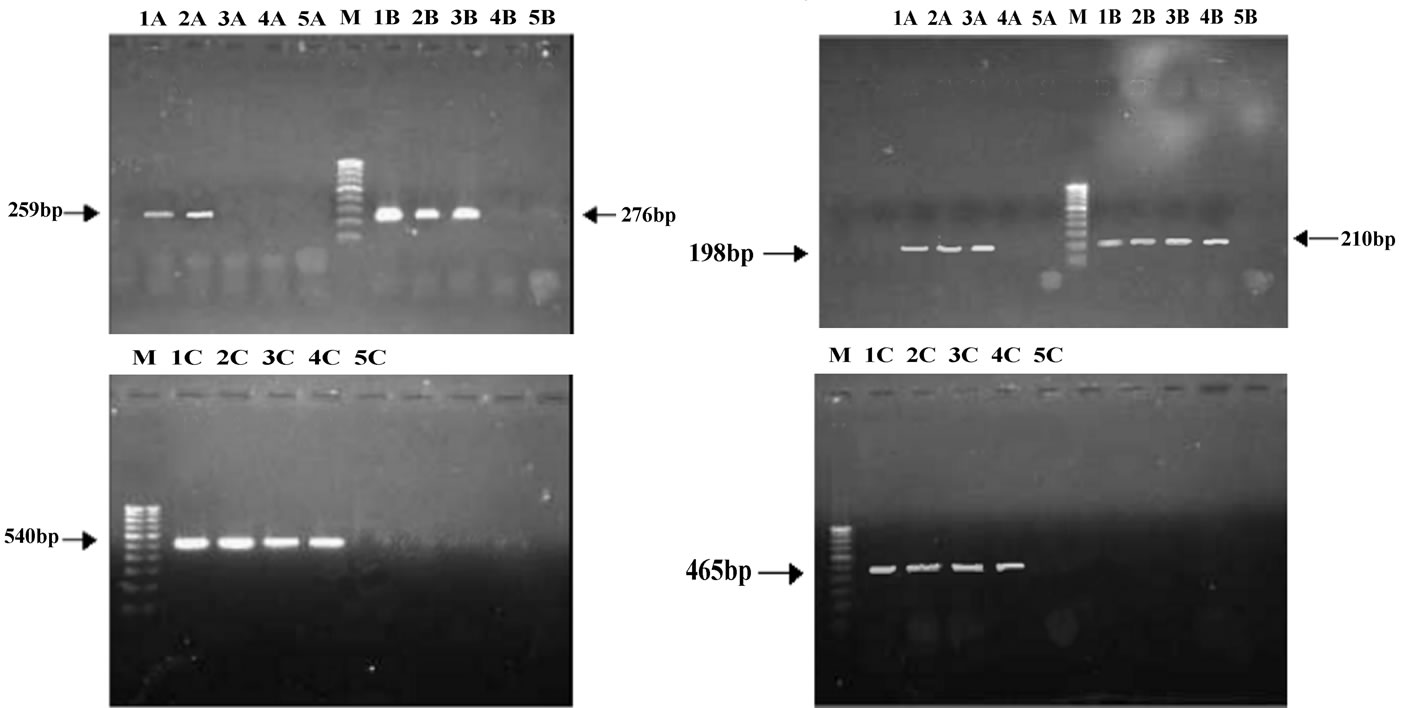
The resistance patterns observed by disk diffusion method extrapolated on to the PAN PCR results that exhibited specific mutation patterns, except for one strain, which was resistant to quinolones in the disk diffusion method but did not exhibit any mutations in gyrA. This could be due to presence of mutations in other genes like parE or aac6’-lb-cr or efflux pumps [13]. Among the resistant strains, 28 strains (68.4%) showed mutation in ser83 alone of gyrA while 2 strains showed mutation in both ser83 and asp87 of gyrA. PCR patterns of 4 representative strains are shown in Figure 1(a). One strain (isolate E5) showed mutation in ser80 of parC (Figure 1(b), lane 4A and Table 1). Of the remaining 9 resistant strains, 8 of them did not exhibit any mutations but harbored the qnrS or qnrB genes (4 of them had qnrS and the remaining 4 had qnrB) (Figure 1(c) and Table 1).
4. DISCUSSION
Resistance to antibiotics in E. coli has become a common feature in developing countries, especially in India, where amoxicillin and ciprofloxacin are the commonly prescribed antibiotics [14]. We developed a new method for detecting such mutations leading to antibiotic resistance, especially quinolones, using PCR based on naturally occurring codon bias and redundancy [8,15]. The efficiency of this method was also verified using some representative nucleotide sequences of gyrA from Escherichia coli genome maps available in the National Center for Biotechnology Information (NCBI) (Table 1). It was interesting to note that almost all of the strains had the same nucleotide sequence from bases 240 to 261 that comprise the ser83 and asp87 regions for gyrA. This observation is consistent with the codon bias (TCG for serine and GAC for aspartate) that was observed in E. coli [13]. Another interesting observation was that E. coli strain SMS-3-5 had mutations both at ser83 and asp87. As indicated in Table 2, the PAN primers would not amplify the gyrA sequence of this strain, if tested. We observed similar mutations (Ser to Leu and Asp to Asn) in the both the strains that possessed a double mutation when subjected to sequencing. This observation is also similar with earlier reports [6,7]. The enteropathogenic E. coli (EPEC) reference strain E2348/69 (O127: H6) was found to be ciprofloxacin sensitive in our earlier work [10]. As expected, we observed gyrA gene amplification using PAN PCR. Furthermore, the gyrA sequence of EPEC (Table 2) did not have any mutated sequences for ser83 or asp87 which is consistent with the sequence in NCBI which showed no mutations in ser83 and asp87 in EPEC’s gyrA sequence (Table 2). But we
 (a)
(a)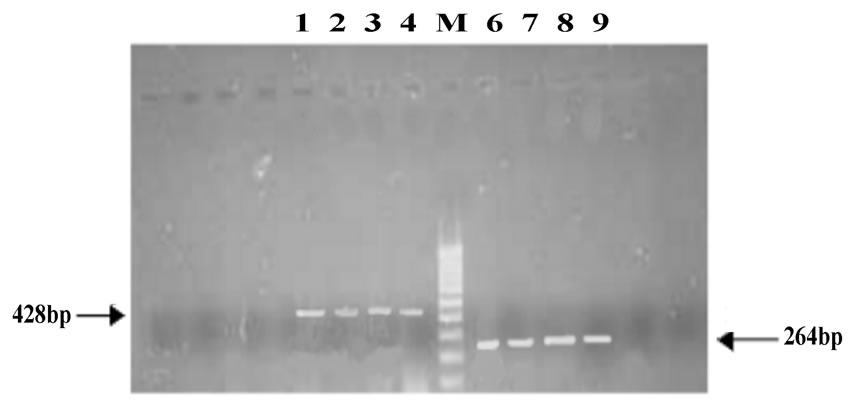 (b)
(b)
Figure 1. (a) Analysis of gyrA mutations by PCR; A: gyrA ser83; B: gyrA asp87; C: gyrA control; M: 100 base pair DNA ladder; 1: Quality control strain E. coli ATCC 25922; 2: E. coli A1 (No mutation); 3: E. coli A3 (mutation in ser83); 4: E. coli D4(mutation in both ser83 and asp87); 5: PCR negative control; (b) Analysis of parC mutations by PCR; A: parC ser80; B: parC glu84; C: parC control; M: 100 base pair DNA ladder; 1: Quality control strain E. coli ATCC 25922; 2: E. coli A1 (No mutation); 3: E. coli A3 (No mutation); 4: E. coli E5 (mutation in parC ser80); 5: PCR negative control; (c) Analysis of qnr genes by PCR; Lanes 1-4: qnrS positive strains E6, EHEC, Strain 1310 and Strain 932; Lanes 6-9: qnrB positive strains F6, F7, G1 and G2.
observed earlier that many of the EPEC clinical isolates were resistant to quinolones/fluoroquinolones. We subjected them to PCR analysis and found that they harbored mutations predominantly in ser83 codon.
Laboratories in developing countries seldom have facilities for DNA sequencing analysis. Apart from routine disk diffusion methods, molecular level analyses for the presence or absence of mutations at specific sites that confer resistance to the antibiotic has become imperative. Although DNA sequencing reveals the nature of mutations (i.e. Asp-87 to Asn or Ser-83 or Leu or Trp), its importance does not go beyond the mere fact that the substituted amino acid might possibly influence decrease in antibiotic affinity to the target site. On the other hand, techniques like PAN PCR, though would not reveal the amino acid substitution, can aid in identification of the presence of a mutation. Indeed, the bacterium’s natural selection process for codon sets is in fact advantageous, as in this case, to use them in diagnosis. In fact, our further search on gyrA sequences in other genera like Salmonella, Shigella and Helicobacter sp. further revealed that the ser83 and asp87 is mutated inherently in some of the strains. The PAN PCR would not amplify these sequences too. Hence, this method could also be used to identify quinolone/fluoroquinolone resistance across a wide range of genera. Further targets for antibiotic resistance in these genera could be analyzed for similar conserved codons and hence can serve to develop more
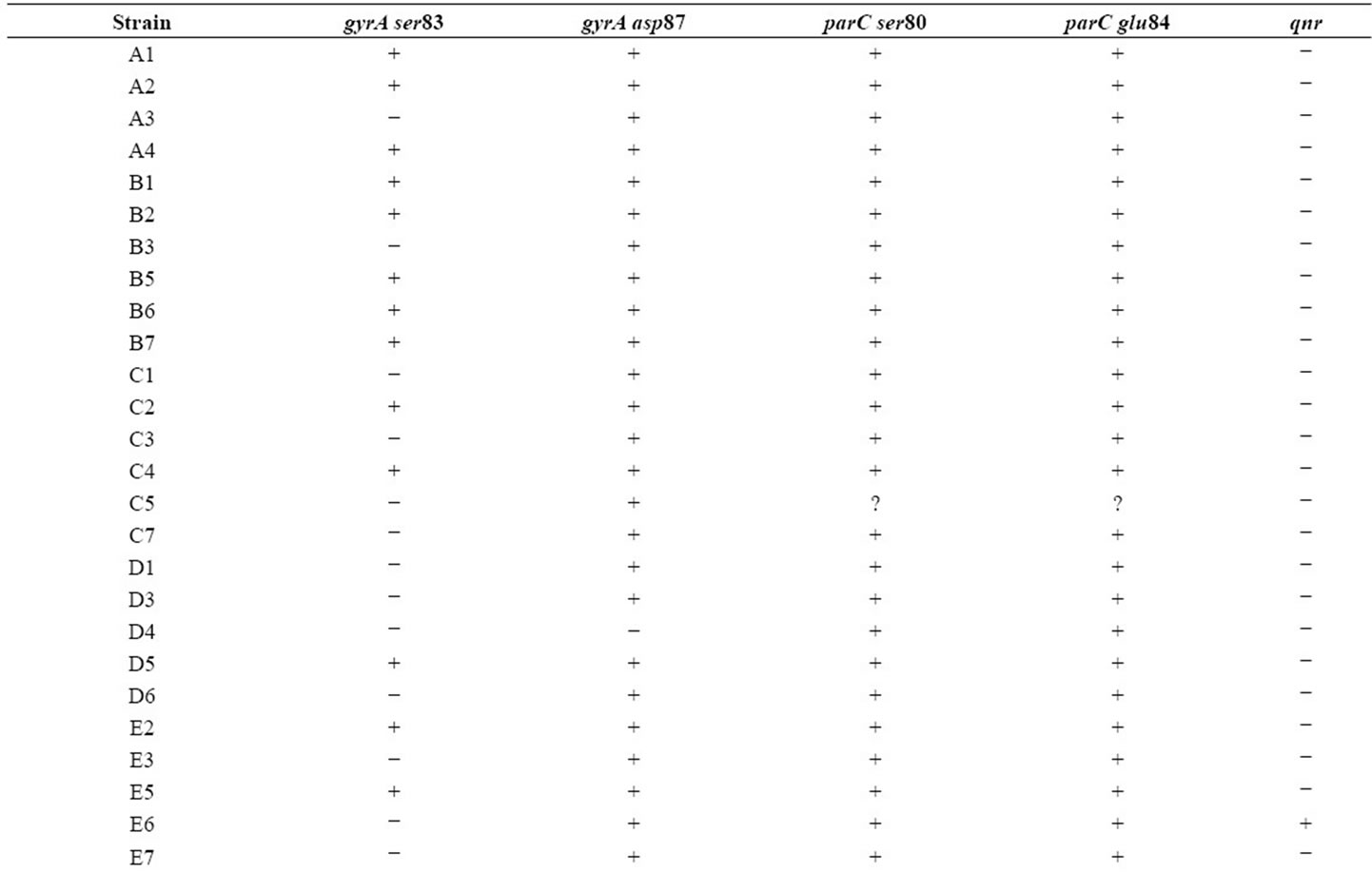

Table 1. Mutation patterns observed in E. coli strains. “+” indicates amplification (presence of native sequence) and “−” indicates lack of amplification (presence of mutation) for gyrA and parC sequences. For qnr genes, “+” indicates presence of the gene while “−” indicates absence.
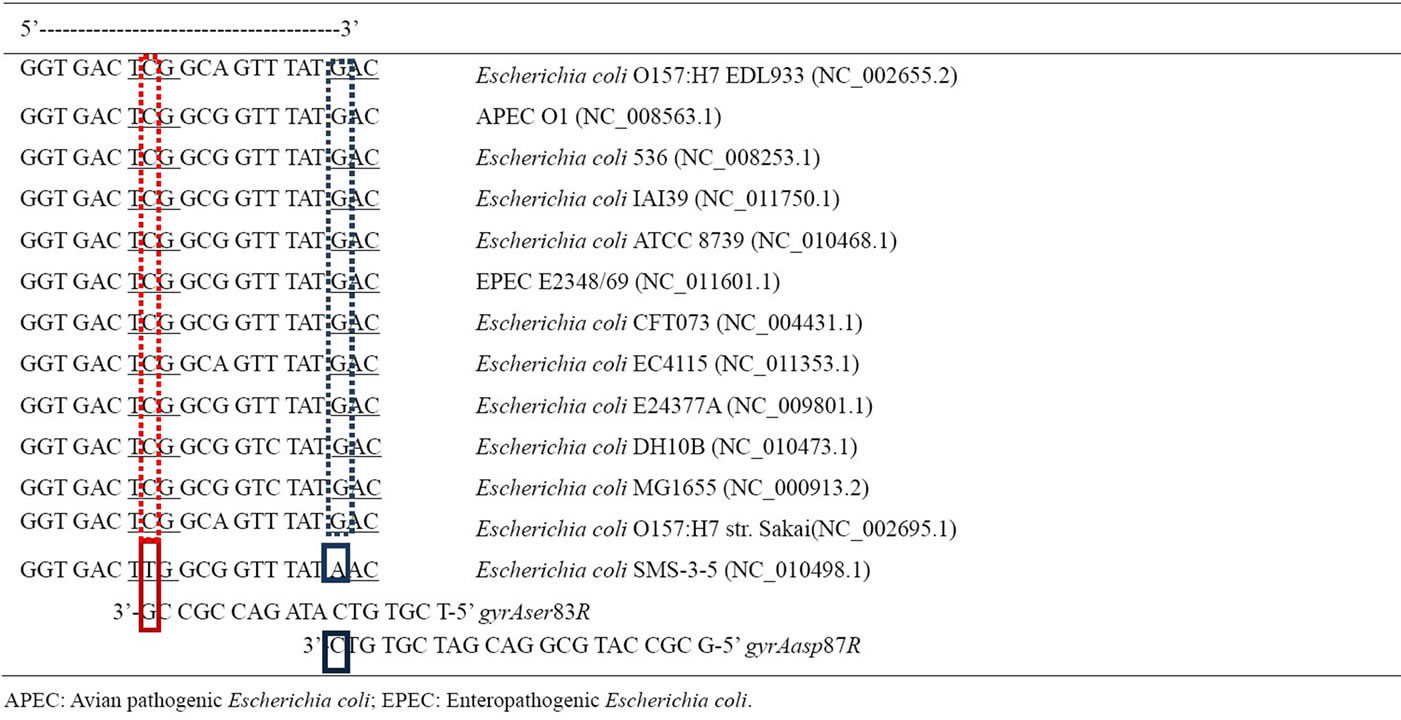
Table 2. Prediction of PAN PCR efficacy with representative E. coli pathogens: Nucleotide sequence of gyrA of different Escherichia coli strains from National Center for Biotechnology Information (NCBI). The NCBI sequence reference number is provided within brackets. Nucleotides 240-261 are represented here. Serine codon (TCG) is at positions 247-249 while Aspartate (GAC) is at positions 259-261. Reverse primers are aligned with putative complementary sequences. Red and blue solid boxes are the primer annealing bases from which DNA polymerase extends the complementary strand from gyrAser83R and gyrAasp87R respectively. Strain Escherichia coli SMS-3-5 exhibits mutations in both ser83 and asp87 due to base mismatch between 3’ terminal base of primer and the DNA sequence. PAN PCR primers would not amplify this sequence and is indicative of quinolone resistance. The remaining 11 representative pathogenic E. coli strains have exact complementary base pairs for ser83 and asp87 highlighted as dashed red and blue boxes respectively. PAN PCR primers would amplify these sequences and is indicative of quinolone susceptibility.
detection methods for multiple antibiotic resistance in bacteria. PAN PCR can also reduce the time required for detection of quinolone resistance and help in preventing the indiscriminate usage of quinolone drugs. Clinically, it can open avenues for exploration of other amino acids in gyrase A that can be targets other than these mutationprone amino acids for developing anti-bacterials. We [10] and others [16] have explored the same, where we have independently reported the importance of Arg-121 in the QRDR region as a pivotal amino acid for ciprofloxacin binding. A small molecule that was found to be highly potent against enteropathogenic E. coli was also found to be targeting Arg-121, apart from interacting with other amino acids like Methionine (Met-120), Alanine (Ala- 117) and Aspartate (Asp-82) [10].
To conclude, PAN PCR helps in the identification of mutations, though not substitutions, but nevertheless reveals the need for targeting other regions in gyrase A, not only for quinolones, but for other prospective antibacterials. It also encourages researchers to search and target more of such native codon sequences that are conserved within and across many other bacterial genera and the mutation of which, might result in antibiotic resistance or increased virulence.
5. ACKNOWLEDGEMENTS
The authors thank Muralikrishnan, Department of Microbiology, PSG Institute of Medical Sciences and Research for E. coli culture maintenance during the study. The Council for Scientific and Industrial Research (CSIR), Government of India funded this work.
![]()
![]()
REFERENCES
- Reece, R.J. and Maxwell, A. (1991) DNA gyrase: Structure and function. Critical Reviews in Biochemistry and Molecular Biology, 26, 335-375. doi:10.3109/10409239109114072
- Collin, F., Karkare, S. and Maxwell, A. (2011) Exploiting bacterial DNA gyrase as a drug target: Current state and perspectives. Applied Microbiology and Biotechnology, 67, 479-497. doi:10.1007/s00253-011-3557-z
- Baker, N.M., Weigand, S., Maar-Mathias, S. and Mondragon, A. (2011) Solution structures of DNA-bound gyrase. Nucleic Acids Research, 39, 755-766. doi:10.1093/nar/gkq799
- Oppegaard, H. and Sorum, H. (1996) Cloning and nucleotide sequence of the DNA gyrase gyrA gene from the fish pathogen Aeromonas salmonicida. Antimicrobial Agents and Chemotherapy, 40, 1126-1133.
- Ruiz, J. (2003) Mechanisms of resistance to quinolones: Target alterations, decreased accumulation and DNA gyrase protection. Journal of Antimicrobial Chemotherapy, 51, 1109-1117. doi:10.1093/jac/dkg222
- Gruger, T., Nitiss, J.L., Maxwell, A., Zechiedrich, E.L., Heisig, P., Seeber, S., Pommier, Y. and Strumberg, D. (2004) A mutation in Escherichia coli DNA gyrase conferring quinolone resistance results in sensitivity to drugs targeting eukaryotic topoisomerase II. Antimicrobial Agents and Chemotherapy, 48, 4495-4504. doi:10.1128/AAC.48.12.4495-4504.2004
- Barnard, F.M. and Maxwell, A. (2001) Interaction between DNA gyrase and quinolones: Effects of alanine mutations at GyrA subunit residues Ser(83) and Asp(87). Antimicrobial Agents and Chemotherapy, 45, 1994-2000. doi:10.1128/AAC.45.7.1994-2000.2001
- Diaz-Lazcoz, Y., Henaut, A., Vigier, P. and Risler, J.L. (1995) Differential codon usage for conserved amino acids: Evidence that the serine codons TCN were primordial. Journal of Molecular Biology, 250, 123-127. doi:10.1006/jmbi.1995.0363
- Kahali, S., Sarkar, B., Rajendran, K., Khanam, J., Yamasaki, S., Nandy, R.K., Bhattacharya, S.K. and Ramamurthy, T. (2004) Virulence characteristics and molecular epidemiology of enteroaggregative Escherichia coli isolates from hospitalized diarrheal patients in Kolkata, India. Journal of Clinical Microbiology, 42, 4111-4120. doi:10.1128/JCM.42.9.4111-4120.2004
- Subramanian, K., Selvakkumar, C., Vinaykumar, K.S., Goswami, N., Meenakshisundaram, S., Balakrishnan, A. and Lakshmi, B.S. (2009) Tackling multiple antibiotic resistance in enteropathogenic Escherichia coli (EPEC) clinical isolates: A diarylheptanoid from Alpinia officenarum shows promising antibacterial and immunomodulatory activity against EPEC and its lipopolysaccharideinduced inflammation. International Journal of Antimicrobial Agents, 33, 244-250. doi:10.1016/j.ijantimicag.2008.08.032
- Cattoir, V., Poirel, L., Rotimi, V., Soussy, C.J. and Nordmann, P. (2007) Multiplex PCR for detection of plasmidmediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. Journal of Antimicrobial Chemotherapy, 60, 394-397. doi:10.1093/jac/dkm204
- Qiang, Y.Z., Qin, T., Fu, W., Cheng, W.P., Li, Y.S. and Yi, G. (2002) Use of a rapid mismatch PCR method to detect gyrA and parC mutations in ciprofloxacin-resistant clinical isolates of Escherichia coli. Journal of Antimicrobial Chemotherapy, 49, 549-552. doi:10.1093/jac/49.3.549
- Cesaro, A., Bettoni, R.R., Lascols, C., Merens, A., Soussy, C.J. and Cambau, E. (2008) Low selection of topoisomerase mutants from strains of Escherichia coli harbouring plasmid-borne qnr genes. Journal of Antimicrobial Chemotherapy, 61, 1007-1015. doi:10.1093/jac/dkn077
- Pandey, M., Khan, A., Das, S.C., Sarkar, B., Kahali, S., Chakraborty, S., Chattopadhyay, S., Yamasaki, S., Takeda, Y., Nair, G.B. and Ramamurthy, T. (2003) Association of cytolethal distending toxin locus cdtB with enteropathogenic Escherichia coli isolated from patients with acute diarrhea in Calcutta, India. Journal of Clinical Microbiology, 41, 5277-5281. doi:10.1128/JCM.41.11.5277-5281.2003
- Karlin, S., Mrazek, J. and Campbell, A.M. (1998) Codon usages in different gene classes of the Escherichia coli genome. Molecular Microbiology, 29, 1341-1355. doi:10.1046/j.1365-2958.1998.01008.x
- Madurga, S., Sanchez-Cespedes, J., Belda, I., Vila, J. and Giralt, E. (2008) Mechanism of binding of fluoroquinolones to the quinolone resistance-determining region of DNA gyrase: Towards an understanding of the molecular basis of quinolone resistance. Chembiochem, 9, 2081- 2086. doi:10.1002/cbic.200800041
ABBREVIATIONS
Ala: Alanine;
Arg: Arginine;
Asp/asp: Aspartate;
DNA: Deoxyribonucleic acid;
E. coli: Escherichia coli;
Leu: Leucine;
Met: Methionine;
NCBI: National Center for Biotechnology Information;
PCR: Polymerase Chain Reaction;
Ser/ser: Serine;
Trp: Tryptophan;
Val: Valine.
NOTES
*The authors declare no competing interests.
#Corresponding author.

