American Journal of Molecular Biology
Vol.2 No.2(2012), Article ID:18948,8 pages DOI:10.4236/ajmb.2012.22015
Molecular cloning and characterization of fruit specific promoter from Cucumis sativus L.
![]()
Plant Molecular Biology Lab, Rajiv Gandhi Centre for Biotechnology, Poojappura, India
Email: *evsoniya@rgcb.res.in
Received 18 August 2011; revised 23 September 2011; accepted 4 October 2011
Keywords: Cucumber; Expansin; Fruit Specific Promoter; Transient Expression; Glucuronidase
ABSTRACT
The isolation and characterization of fruit specific promoters are critical for the manipulation of nutritional value and agronomic quality of fruits by genetic engineering and also opened a new era in edible vaccine technology. Expansins are proteins that induce loosening of individual plant cells by disrupting the non-covalent interactions between cellulose and hemicellulose microfibrils and hence have role in growth programs including fruit ripening. We report the identification of an expansin gene (CsExp) from Cucumis sativus that exhibits high levels of mRNA abundance and is specifically expressed in ripened fruit. The promoter region of CsExp also contains elements responsible for its fruit specific expression. Transient expression studies of the CsExp promoter were conducted with particle bombardment, followed by GUS histochemical assay and real time PCR. CaMV35S promoter was used as the positive control in all these experiments. Clear fruit specificity was observed for CsExp promoter in all the experiments. Thus CsExp promoter from Cucumber is a good candidate to target expression of the foreign genes to engineer fruit specific traits.
1. INTRODUCTION
Transgenic plant technology provides an indispensable and powerful tool for analysis of gene functions and stable expression of foreign genes [1-3]. In many cases tissue/organor developmental stage specific expression of transgenes in plants is highly desirable. In recent years, a large number of tissue or stage specific promoter sequences have been cloned and characterized [4]. In early 1980s some of the first plant transformation vectors were designed using NOS promoter to control the expression of selectable marker genes such as nptII (neomycin phosphor transferase) or APHII (aminoglycoside 3’-phosphotransferase) and DHFR (dihydrofolate reductase) [5- 7]. Many of the changes that occur during ripening process results from the induction of new enzymes activities [8] which in turn result from the activation of gene expression [9-11]. Regulation of the expression pattern of a particular gene can involve the specific binding of trans-acting factors to the cognate cis-elements, constituting a crucial step in transcriptional initiation. A number of fruit-specific genes that are activated during ripening have been isolated from tomato and other fruits [12, 13]. The promoters of fruit-specific genes would also be of great interest for use in strategies to manipulate fruit metabolism and produce valuable proteins such as antibody, biopharmaceuticals, and edible vaccines through methods of genetic engineering. However, the detailed mechanisms by which the expression of fruit specific genes is regulated are poorly understood, as many of the essential cis-elements have not been identified.
Expansins are plant cell-wall loosening proteins involved in cell enlargement and in a variety of other developmental processes in which cell-wall modification occurs [14]. In addition to these functions, a specific isoform of expansin plays a major role during fruit ripening. Expansin gene families have been identified in Tomato [15], Arabidopsis [16] and Cucumber [17] suggesting that divergent isoforms may act on different components of the wall, exhibit different developmental and environmental regulation or tissue specific expression. In the present study we describe the cloning and characterization of a fruit specific isoform of expansin gene promoter from cucumber and biolistic transient expression assay for rapid expression analysis.
2. MATERIALS AND METHODS
2.1. Plant Material
Mature seeds of Salad Cucumber (Cucumis sativus) were obtained from Agricultural College Vellayani, Thiruvananthapuram. Seeds were sterilized with 0.1% mercuric chloride solution for 5 minute, washed with sterile distilled water for several times and inoculated on hormonefree MS basal medium [18]. The seedlings obtained after 7 - 10 days were allowed to grow further under green house conditions and were used as the plant material in this study.
2.2. Total RNA Isolation, cDNA Synthesis and Amplification of Expansin Gene
Total RNA was isolated from 100 mg fruit tissue of C. sativus using TrizolTM reagent (Gibco BRL) according to the manufactures’ protocol. Five micrograms of total RNA were reverse transcribed at 42˚C using Moloney Murine Leukemia Virus Reverse Transcriptase (Promega) and 2.0 µg Oligo(dT)18 primer according to manufacturer’ recommendations. The expansin gene was amplified from cDNA by PCR using degenerate primers ExpF (5’-GSN CAY GCN ACN TTY TAY GGN G-3’) and ExpR (5’-YTG CCA RTT YTG NCC CCA RTT-3’). The PCR amplification was carried out with an initial denaturation at 94˚C for 5 min followed by 35 thermal cycles consisting of denaturation 94˚C for 30s; annealing at 58˚C for 90 sec; extension 72˚C for 1min followed by a final extension at 72˚C for 7 min. The product was cloned in pGEMT easy vector and sequenced.
2.3. Real Time PCR
Real Time PCR was carried out to determine the expression level of expansin in various tissues using ABI 7500 Real Time PCR system (Applied Biosystems). For that total RNA was isolated from the leaf, stem and fruit tissues (young and ripened) of greenhouse grown plants using TrizolTM Reagent as described earluer and respective cDNAs were prepared. The endogenous control used to normalize variance in the quantity of RNA and the amount of cDNA was elongation factor 1 α (ef1α) gene. Real time PCR was performed on an optical 96-well plate with SYBR® Green PCR Master mix (Applied Biosystems) using the primers ExpF and ExpR. Each reaction consists of 25 μl containing 2 μl of first strand cDNA, 12.5 µl of 2X SYBR® Green Master Mix and 0.2mM of forward and reverse primers and was run in triplicate. The thermal profile for all SYBR® Green PCRs was 50˚C for 2 min and 95˚C for 10 min, followed by 40 cycles of 95˚C for 10 s and 58˚C for 90 s.
2.4. Expansin Promoter Isolation and Analysis
The Universal Genome Walker Kit (BD Biosciences) was used to obtain promoter regions of C. sativus expansin gene. The genomic DNA was isolated from young leaves using the Gen Elute Plant DNA Extraction Kit (Sigma). The genomic DNA was digested with DraI, EcoR V, Pvu II and Stu I restriction enzymes to make a blunt end. Genome walker adaptors were ligated to the digests and the ligated products were then used as template to amplify promoter regions of expansin gene. Two gene specific primers GSP1 (5’-CAG ATG CGT CAC CAC CCC CAT AAA AAG-3’) and GSP2 (5’-GTT GGT GGC AGT GAC CCT AAT AGT TCC-3’) were designed for the amplification of the promoter region. The primary PCR was carried out using GSP1 and adapter specific primer AP1 (provided with the kit) using the following thermal conditions: 7 cycles of 94˚C for 30 s and 72˚C for 3 min followed by 32 cycles of 94˚C for 30s, 68˚C for 3 min and a final extension at 68˚C for 7min. Primary amplified product was diluted and used as the template for secondary PCR with GSP2 and adapter specific primer AP2 (provided with the kit) using the same conditions as mentioned for primary PCR. All PCR reactions were done using Advantage 2 polymerase mixture and relevant products were eluted and cloned ito pGEMT easy vector for sequencing. The sequence data was analyzed using the BLAST option available at NCBI (http://www.ncbi.nlm.gov/). Two nested primers ExpR1 (5’-GAA ATG CCA AAA CTT AGG ACC CC-3’) and ExpR2 (5’-AAG TGG GTT TGT AAG GTT GGA-3’) were designed to amplify the promoter of expansin alone. The cis-regulatory elements of the promoter were identified by PlantCARE database. Two primers PRF (5’-ATC ACR YGT CTC TGT GTT CAT CTG CTC CTC-3’) and PRR (5’-GAC CTC GAG ATT TCG CTC TTT ACT CCC TCC-3’) were designed for the amplification of the functional minimal promoter region and cloned into pGEMT easy vector for sequencing.
2.5. Vector Construction and Transient Expression Studies Using Particle Bombardment
The promoter less binary vector pCAMBIA1391Z harbouring GUS reporter gene was used to clone the isolated fruit specific promoter. The promoter fragment was sub-cloned in-frame into EcoR I site of pCAMBIA1391Z and the orientation was confirmed in transformed colonies by PCR using promoter specific forward primer PRF and GUS reverse primer (5’- GTA TCG GTG TGA GCG TCG CAG AAC-3’). To compare promoter activity, the binary vector pCAMBIA1301, where the GUS gene under the control of CaMV35S promoter was used as the positive control and promoter less binary vector pCAMBIA1391Z was used as the negative control. For the transient expression studies using particle bombardment, mature fruit and leaf tissues from Cucumber plants were used. Fruits were cut into 1 - 2 mm thin transverse slices and leaf segments or entire leaves were used for bombardment. The biolistic device used for the study was helium driven Biolistic®PDS-1000/He Particle Delivery System. The plasmid DNA was precipitated on to tungsten particles according to the protocol described with the instrument. Twenty five microliters of particle suspension was used for each bombardment. The system was set at 19 mm distance between the macrocarrier and the sample, and 23 mm distance between the macro carrier and the rupture disc. A vacuum of 5 mm Hg was applied and the particles were discharged when the helium pressure was 600 mm Hg.
2.6. GUS Assay
Following bombardment the explants were kept under dark conditions for 18 hours prior to the assay for GUS activity. Histochemical staining for GUS activity was performed using 5-bromo-4-chloro-3-indolyl-b-D-glucuronide (X-gluc) as a chromogenic substrate [19]. Tissues were vacuum infiltrated with X-Gluc reaction buffer (100 mM sodium phosphate buffer pH 7.2, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, 0.1% Triton X-100 and 1 mg/ml X-gluc) and incubated at 37˚C overnight. After incubation, chlorophyll was removed from green tissues by immersion in 70% ethanol, and tissue samples were observed.
3. RESULTS AND DISCUSSION
Recent advances in genetic engineering have provided the requisite tools to transform plants and express to foreign genes of agronomic importance in specific organ/ parts of plants. Plant genes that show tissue specificity, developmental specificity and a wide range of expression levels have been characterized [13]. The isolation and characterization of new fruit-specific promoters represents a key step in the development of biotechnological projects involving the expression of target genes in fruits of economical and edible crops. The fruit specific promoters will be useful to precisely limit expression of a desired gene in fruits at a particular stage of development, which has immense use in the case of edible vaccine production.
3.1. Isolation of Expansin Gene from C. sativus
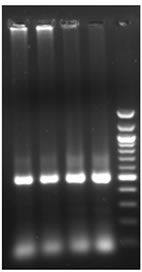
Total RNA was isolated from young leaves of cucumber and cDNA synthesis was carried out. RT-PCR using degenerate primers resulted in a single approximately 550 bp amplicon (Figure 1(a)). The amplified product was cloned into pGEM-T Easy vector, digested with EcoR I for confirmation and further sequenced. The sequence was analyzed at the nucleotide and protein level for its similarity to other reported expansin genes from the database. Interestingly the cucumber expansin isolated in this study exhibited a homology of 78% with Lycopersicon esculentum expansin (LeExp1), which is a fruit specific isoform [15,20]. The alignment of cucumber expansin with other fruit specific expansin genes of the database is shown in Figure 1(b).
3.2. Differential Expression of CsExp in Cucumber
To better understand function we investigated the expression pattern of isolated expansin gene in leaf, stem, young fruit and ripened fruits using real time PCR. This anlalyis confirmed that cucumber expansin gene showed a preferential expression in fruit as compared to leaves and stem (Figure 1(c)). In fruit tissue itself, the cucumber expansin showed a higher expression in ripened fruit in comparison with young ones.
3.3. Isolation and Analysis of 5’ Regulatory Regions of Cucumber Expansin Gene
The genomic DNA isolated from Cucumis sativus was digested with DraI, EcoR V, Pvu II and Stu I restriction enzymes and was used for the construction of the Genome Walker libraries. In nested type PCR on Genome Walker libraries, approximately 3 kb product was obtained Stu I library (Figure 1(d)), which was cloned in pGEMT easy vector and sequenced. The analysis of sequence data revealed that the 5’ region contained the cis-regulatory sequences and 3’ was similar to expansin gene. Analysis of this sequence using PlantCARE database identified the minimum region of promoter for successful fruit specific expression and this region was amplified using PRF and PRR primers (Figure 1(e)). The 1.5 kb promoter region along with the 5’ region of expansin gene was shown in Figure 2. Transcription start site was predicted using Softberry online tool. A number of potential regulatory motifs corresponding to known cis-elements of eukaryotic genes were found on PLACE signal scan search analysis of the sequence (Figure 2). A putative TATA box was found at the positions –29 to –25 bp upstream of the transcription start site, whereas a CAAT box was observed at positions –299 to –296 bp; these boxes function as basal promoter elements for transcription. Furthermore, regulatory elements with homology to those identified in fruit-specific or light-responsive (regulation) genes were found in the p CsExp region such as GATA box, I box, GCNT4 motif, TGTCAC motif, Skn motif etc. The fruit specific motif GCNT4 was observed at –438 to –442 bp, TGTCAC motif at –02 to –506 bp, Skn motif at –341 to –346 bp with respect to transcriptional start site. The region between –341 to –502 bp is required for the fruit specific expression. The G-box motif is currently one of the best characterized cis regulatory elements in plants and has been identified in diverse
 (a)
(a)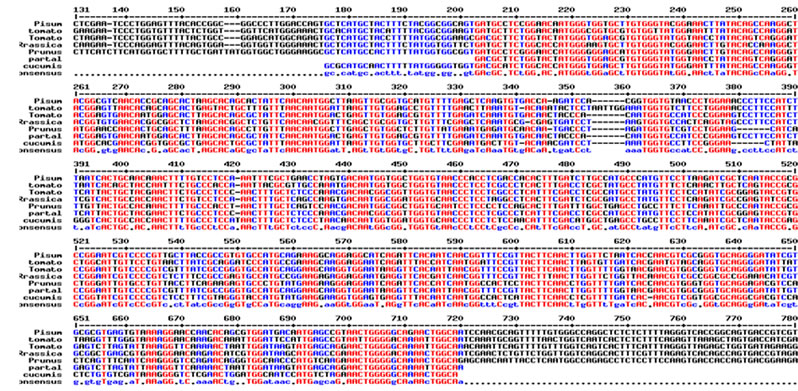 (b)
(b)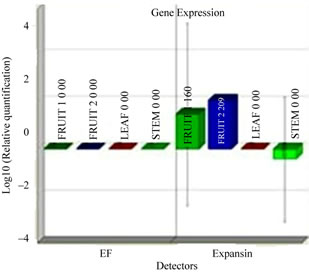 (c)
(c)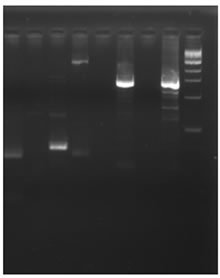 (d)
(d)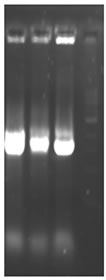 (e)
(e)
Figure 1. Isolation and characterization of expansin gene promoter for Cucumber. (a) Amplification of expansin gene from the cDNAs of cucumber. Amplified expansin gene (lanes 1-4) and 100bp ladder (lane 4); (b) Multiple alignment of expansin gene with other fruit specific isoforms from different plants; (c) Expression of expansin gene in different tissues of Cucumis sativus analyzed by Real time PCR. The expression of expansin gene in young fruits, expression of expansin gene in ripened fruits, expression of expansin gene in leaf tissues, and expression of expansin gene in stem; (d) Isolation of the promoter of expansin gene using BD Genome Walker kit. Ligated DNA amplified (Lanes 1-4-), negative control (lane 5), positive control (lane 6), lane 7-negative control, lane 8-positive control, and lane 9-1kb DNA ladder; (e) Cloning and amplification of the expansin promoter. Promoter region amplified (lanes 1-3) and 1 Kb DNA ladder (lane 4).
set of unrelated genes, including those controlled by visible, UV light [21], and ABA [22]. The G-box motif is an important factor for fruit specific expression. Most in vivo studies indicate that the G-box elements cannot act on their own but require the presence of additional cis acting elements for their function [23,24]. Thus it is very clear that the 1.5 kb promoter region of C. sativus expansin gene contians sufficient cis-elements required for the high and specific expression of the gene in fruit tissues.
3.4. The CsExp Promoter Directs the Expression of GUS in the Fruit Tissues
Transient expression studies using particle bomabardment were adopted for analyzing the activity of the CsExp promoter in leaves and fruit slices of Cucumis sativus. The transiently transformed expalnts were further used for GUS histochemical assay and real time PCR. For this the 1.5 kb promoter was cloned into the EcoR I site of the promoter less binary vector pCAMBIA1391Z which carries GUS as reporter gene. The schematic representation of the cloning of CsExp promoter to pCAMBIA1391Z is shown in Figure 3(a). The pCAMBIA- 1391Z-ExpPR would drive b-glucuronidase (GUS) gene expression under the control of expansin gene promoter while pCAMBIA1301 would drive b-glucuronidase (GUS) gene expression under the control of under the control of CaMV35S promoter. The main parameters that can be optimized in the biolistic transformation of the plant tissue are the amount of DNA used in each bombardment and the number of shots per sample [25-28]. In the present study, a very good expression was found; when 1 mg of the vector DNA was used as positive control.
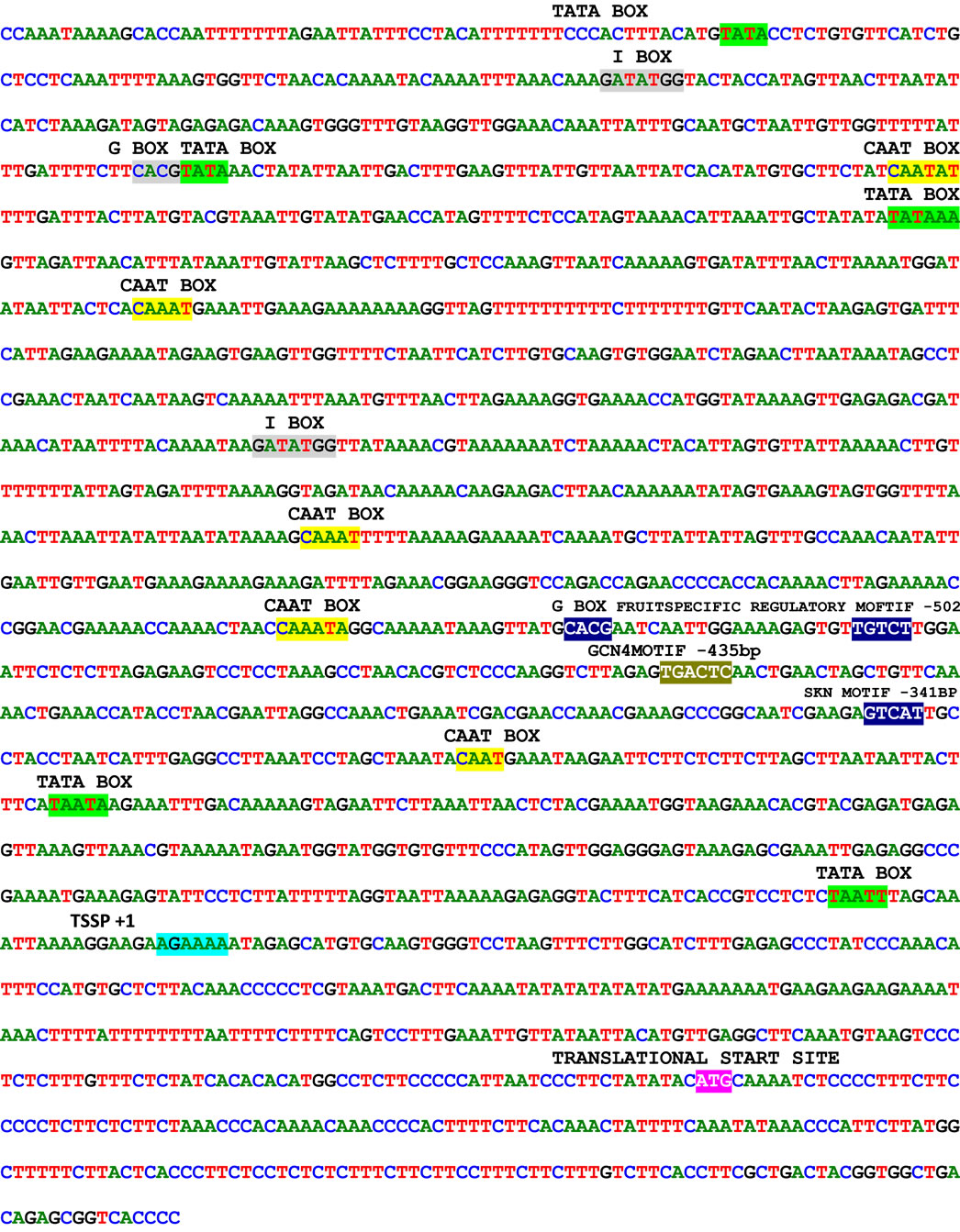
Figure 2. Promoter region of expansin gene from Cucumis sativus showing TATA box, CAAT box and fruit specific regulatory motifs. Predicted transcriptional start site is highlighted other predicted regulatory elements were shaded.
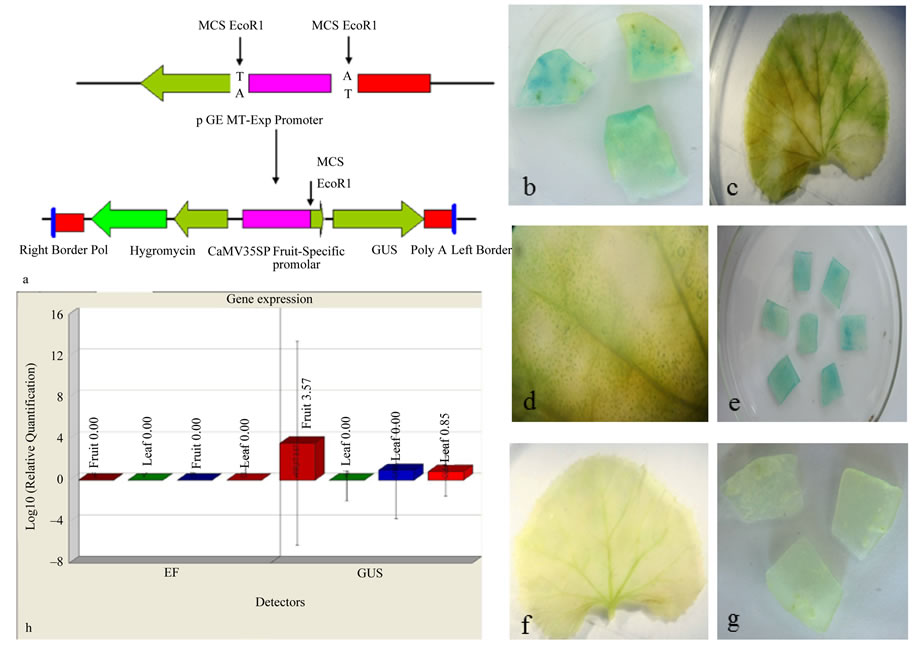
Figure 3. Transient expression studies of the fruit specific promoter using GUS reporter gene. a Schematic representation of cloning of the fruit specific promoter into the promoter less binary vector pCAMBIA1391Z; b Expression of GUS gene in fruit tissues bombarded with CaMV35S promoter; c Expression of GUS gene in leaf tissues bombarded with CaMV35S promoter; d Enlarged view of GUS gene expression in leaf bombarded with CaMV35S promoter; e Expression of GUS gene in fruit tissues bombarded with expansin gene promoter; f Expression of GUS gene in leaf tissues bombarded with fruit specific promoter; g Expression of GUS gene in fruit tissues bombarded with promoter less binary vector; h Real time PCR analysis of GUS transformants. The expression of GUS gene in fruit tissues bombarded with expansin promoter, expression of GUS gene in leaf tissues bombarded with expansin promoter, expression of GUS gene in fruit tissues bombarded with CaMV35S promoter, expression of GUS gene in leaf tissues bombarded with CaMV35S promoter.
When the binary vector carrying the expansin promoter was used for the bombardment, the fruit slices developed blue colour (Figure 3(e)) while in the leaf segments the colour was absent (Figure 3(f)). In the positive control experiments using pCAMBIA1301; where the GUS reporter gene under the control of CaMV35S promoter; blue coloured patches were observed both in the leaf and fruit slices (Figures 3(b)-(d)). In the negative control samples, the colouration were absent in both tissues (Figure 3(g)).
The expression analysis of expansin gene in different tissues after transient transformation was studied by real time PCR with EF gene as the endogenous control. The real time PCR analysis of the GUS transformants showed high level of expression in the fruit tissues under the control of expansin promoter while the positive control showed equal level of expression in all tissues (Figure 3(h)). The real time PCR method is very sensitive in detecting gene expression; it allowed quantification of relative expression and was more amenable to biological replication and statistical analysis. This approach is a powerful and sensitive means of studying the activity of a gene promoter as a function of cell development, or tissue and organ expression [29]. There are also several reports on the differential expression patterns of promoter activity based on the transient expression assay of these reporter genes [29-31]. In brief, the results from transient expression and real time PCR studies show that the expansin promoter isolated from Cucumis sativus displays a significant activity in cucumber fruits.
The isolation, characterization and functional analysis of organ speicific promoters, have become a key aspect in transgenic plant researches. They not only increase transgene expression in specific organs, but also can avoid unnecessary energy waste which results from the gene expression in other plant organs. As the promoter isolated in the present study is very much suitable for the high level expression in fruit tissues, it is very useful in the field of transgenic research for developing immunotherapeutic proteins including edible vaccines.
4. ACKNOWLEDGEMENTS
The authors would like to thank Kerala State Council for Science Technology and Environment, Indian Council of Medical Research for financial support.
REFERENCES
- Daniell, H., Muthukumar, B. and Lee, S.B. (2001) Marker free transgenic plants: Engineering the chloroplast genome without the use of antibiotic resistance genes. Current Genetics, 39, 109-110. doi:10.1007/s002940100185
- Fischer, R., Drossard, J., Commandeur, U., Schillberg, S. and Emans, N. (1999) Towards molecular farming in the future: Moving from diagnostic protein and antibody production in microbes to plants. Biotechnology and Applied Biochemistry, 30, 101-108.
- Wilke, D. (1999) Chemicals from biotechnology: Molecular plant genetics will challenge the chemical and the fermentation industry. Applied Microbiology Biotechnology, 52, 135-145. doi:10.1007/s002530051500
- Gupta, V., Khurana, R. and Tyagi, A.K. (2007) Promoters of two anther-specific genes confer organ-specific gene expression in a stage-specific manner in transgenic systems. Plant Cell Reports, 26, 1919-1931. doi:10.1007/s00299-007-0414-8
- Bevan, M.W., Barnes, W.M. and Chilton, M.D. (1983) A structure and transcription of the nopaline synthase gene region of T DNA. Nucleic Acid Research, 11, 369-385. doi:10.1093/nar/11.2.369
- Bevan, M.W. (1984) Binary agrobacterium vectors for plant transformation. Nucleic Acids Research, 12, 8711- 8721. doi:10.1093/nar/12.22.8711
- Herrera-Estrella, L., De Block, M., Messens, E., Harlensteens, J.P., Van Montagu, M. and Schell, J. (1983) Chimeric genes as dominant selectable markers in plant cells. EMBO Journal, 2, 987-995.
- Brady, C.J., McGlasson, B. and Speirs, J. (1987) The biochemistry of fruit ripening. In: Nevins, D. and Jones, R., Eds., Tomato Biotechnology, Alan R Liss Inc., New York, 279-288.
- Mansson, P.E., Hsu, D. and Stalker, D. (1985) Characterization of fruit specific cDNAs from tomato. Molecular and General Genetics, 200, 356-361. doi:10.1007/BF00425717
- Slater, A., Maunders, M.J., Edwards, K., Schuch, W. and Grierson, D. (1985) Isolation and characterization of cDNA clones for tomato polygalacturonase and other ripening related proteins. Plant Molecular Biology, 5, 137-147. doi:10.1007/BF00015677
- Biggs, M.S., Harriman, R.W. and Handa, A.K. (1986) Changes in gene expression during tomato fruit ripening. Plant Physiology, 81, 395-403. doi:10.1104/pp.81.2.395
- Chen, J.C., Jiang, C.Z., Gookin, T.E., Hunter, D.A., Clark, D.G. and Reid, M.S. (2004) Chalcone synthase as a reporter in virus-induced gene silencing studies of flower senescence. Plant Molecular Biology, 55, 521-530. doi:10.1007/s11103-004-0590-7
- Karaaslan, M. and Hrazdina, G. (2010) Characterization of an expansin gene and its ripening-specific promoter fragments from sour cherry (Prunus cerasus L.) cultivars. Acta Physiologiae Plantarum, 32, 1073-1084. doi:10.1007/s11738-010-0499-5
- Cosgrove, D.J. (2000) Loosening of plant cell walls by expansins. Nature, 407, 321-326. doi:10.1038/35030000
- Rose, J.K.C., Lee, H.H. and Bennett, A.B. (1997) Expression of a divergent expansin gene is fruit-specific and ripening regulated. Proceedings of the National Academy of Sciences of the USA, 94, 5955-5960. doi:10.1073/pnas.94.11.5955
- Cho, H.-T. and Cosgrove, D.J. (2002) Regulation of root hair initiation and expansin gene expression in Arabidopsis. The Plant Cell, 14, 3237-3253. doi:10.1105/tpc.006437
- Zhao, Q.X., Yuan, S., Wang, X., Zhang, Y., Zhu, H. and Lu, C.M. (2007) Restoration of mature etiolated cucumber hypocotyl cell wall susceptibility to expansin by pretreatment with fungal pectinases and EGTA in vitro. Plant Physiology, 147, 1874-1885.
- Murashige, T. and Skoog, S. (1962) A review medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiology, 15, 473-497. doi:10.1111/j.1399-3054.1962.tb08052.x
- Jefferson, R.A., Kavanagh, T.A. and Bevan, M.W. (1987) GUS fusions: B-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal, 6, 3901-3907.
- Anjanasree, K.N. and Bansal, K.C. (2003) Isolation and characterization of ripening-related expansin cDNA from tomato. Journal of Plant Biochemistry and Biotechnology, 12, 31-35.
- Hartmann, U., Valentine, W.J., Christie, J.M., Hays, J., Jenkins, G.I. and Weisshaar, B. (1998) Identification of UV/blue light response elements in Arabidopsis thaliana chalcone synthase promoter using a homologous protoplast transient expression system. Plant Molecular Biology, 36, 741-754. doi:10.1023/A:1005921914384
- Chandrasekharan, M.B., Bishop, K.J. and Tall, T.C. (2003) Module specific regulation of the beta-phaseolin promoter during embryogenesis. The Plant Journal, 33, 853- 866. doi:10.1046/j.1365-313X.2003.01678.x
- Foster, R., Izawa, T. and Chua, H. (1994) The bZIP proteins gather at ACGT elements. FASEB Journal, 8, 192- 200.
- Martinez-Hernadez, A., Lopez-Ochoa, L., Arguello-Astorga, G. and Herrera-Estrella, L. (2002) Functional properties and regulatory complexity of a minimal RBCS light-responsive unit activated by phyochrome and plastid signals. Plant Physiology, 128, 1223-1233. doi:10.1104/pp.010678
- Sanford, J.C., Klein, T.M., Wolf, E.D. and Allen, N. (1987) Delivery of substances into cells and tissues using a particle bombardment process. Particulate Science Technology, 5, 27-37. doi:10.1080/02726358708904533
- McElroy, D., Zhang, W., Cao, J. and Wu, R. (1990) Isolation of an efficient actin promoter for use in rice transformation. The Plant Cell, 2, 163-171.
- Ramli, Z. and Abdullah, S.N.A. (2003) Development of a transient promoter assay system for oil palm. Journal of Oil Palm Research, 15, 62-69.
- Webb, N.M., Mathews, H.V., Clendennen, S.K. and Kellogg J.A. (2000) Banana and melon promoters for expression of transgenic plants. CIPO Patent No. 2365259.
- Moriwaki, M., Yamakawa, T., Washino, T., Kodama, T. and Igarashi, Y. (1999) Organspecific expression of â-glucuronidase activity driven by the Arabidopsis heat-shock promoter in heat-stressed transgenic tobacco and Arabidopsis plants. Plant Molecular Biology, 28, 73-82.
- Crone, D., Rueda, J., Martin, K.L., Hamilton, D.A. and Mascarenhas, J.P. (2001) The differential expression of a heat shock promoter in floral and reproductive tissues. Plant, Cell and Environment, 24, 869-874. doi:10.1046/j.1365-3040.2001.00727.x
- Takahashi, T., Naito, S. and Komeda, Y. (1992) The Arabidopsis HSP8.2 promoter/GUS gene fusion in transgenic Arabidopsis plants: A powerful tool for the isolation of regulatory mutants of the heat-shock response. Plant Journal, 2, 751-761. doi:10.1111/j.1365-313X.1992.tb00144.x
NOTES
*Corresponding author.

