Journal of Cancer Therapy
Vol.5 No.9(2014), Article ID:49182,18 pages
DOI:10.4236/jct.2014.59094
Neoplastic-Like CELL Changes of Normal Fibroblast Cells Associated with Evolutionary Conserved Maternal and Paternal Genomic Autonomous Behavior (Gonomery)
Kirsten H. Walen
CROMOS, Richmond, CA, USA
Email: kwalencromos@gmail.com
Copyright © 2014 by author and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 18 June 2014; revised 15 July 2014; accepted 11 August 2014
Abstract
The present comparative review discusses conservation of early evolutionary, relic genetics in the genome of man, which determine two different mechanistic reductive division systems expressed by normal, human diploid cells. The divisions were orderly and segregated genomes reductively to near-diploid daughter cells, which showed gain of a proliferative advantage (GPA) over cells of origin. This fact of GPA expression is a fundamental requirement for initiation of tumorigenesis. The division systems were responses to a carcinogen-free induction system, consisting of short (1 - 3 days) exposures of young cells to nutritional deprivation of amino acid glutamine (AAD). In recovery growth (2 - 4 days) endo-tetra/ochtoploid cells and normal diploid metaphase cells demonstrated chromosomal reductive divisions to respectively heterozygous and homozygous altered daughter cells. Both division systems showed co-segregating whole complements, which for reduction of the diploid metaphases could only arise from gonomeric-based autonomous behavior of maternal and paternal (mat/pat) genomes. The timely associated appearance with these latter divisions was fast growing small-cells (1/2 volume-size reduced from normal diploidy), which became homozygous from haploid, genomic doubling. Both reductive divisions thus produced genome altered progeny cells with GPA, which was associated with pre-cancer-like cell-phenotypic changes. Since both “undesirable” reductive divisions expressed orderly division sequences, their genetic controls were assumed to be “old genetics”, evolutionarily conserved in the genome of man. Support for this idea was a search for evidential material in the evolutionary record from primeval time, when haploid organisms were established. The theory was that endopolyploid and gonomery-based reductive divisions relieved the early eukaryotic organisms from accidental, non-proliferative diploidy and polyploidy, bringing the organism back to vegetative haploid proliferation. Asexual cycles were common for maintenance of propagating haploid and diploid early unicellular eukaryotes. Reduction of accidental diploidy was referred to as “one-step meiosis” which meant gonomeric-based maternal and paternal genomic independent segregations. This interpretation was supported by exceptional chromosomal behaviors. However, multiple divisions expressing non-disjunction was the choice-explanation from evolutionists, which today is also suggested for the rarer LL-1 near haploid leukemia. These preserved non-mitotic mechanistic divisions systems are today witnessed in apomixes and parthenogenesis in many animal phyla. Thus, the indications are the modern genome of man harbors, relic-genetics from past “good” evolvements assuring “stable” proliferation of ancient, primitive eukaryotes, but with cancer-like effects for normal human cells.
Keywords: Cytogenetics, Pathologic Cytology, Endomitosis, Division Skewedness, Pathological Mitosis, Metaphase Rosettes, Homozygous, LOH, Growth Pattern, Nutrition, Amino Acid

1. Introduction
1.1. Goals and Aims
The hypothesis of man as a genetic carrier of relic genes originated from observations of endopolyploid and diploid mechanistic divisions to para-diploid and haploid progeny genomes showing gain of a proliferative advantage (GPA) in normal, human, diploid cell populations. Increase in cell fitness is the fundamental requirement in a cancer initiation process, and occurred in a nutritional, carcinogen-free induction system. Ergo, man could be in control (by lifestyle) of his own destiny regarding cancer development or not. The question became: how and when in evolutionary time did these undesirable, genomic reducing systems enter man’s genome? Is it a recent evolutionary divergence for instance linked to transition from unicellular to multicellular organisms, or was it established in the primeval time when haploid organisms prevailed? Answers to these questions and how GPA was genetically determined, may well be instrumental in discovery of molecular ways that can foster a vaccine development for cancer incidence reduction.
1) General early evolutionary happenings for ancient unicellular organism In the book “The Touchstone of Life” [1] , the first successful existence of a self-replicating, informational DNA molecule with evolutionary development to primitive organisms is discussed. Old (“relic”) genetics and epigenetics were mentioned as sources for cell-to-cell communication in evolutionary advanced multicellular organisms. This fact together with ongoing Encode’s findings of “deeply weird” genes that “can make more than one protein” (e.g., peculiar interon-exon spliceosomes), was proposed to be evolutionary dragged along as “baggage” [2] [3] . This possibility suggested that other “left-over”, past good unicellular reproductive mechanisms, could occupy the genome of man. Early shared features in mitosis between prokaryotes (Archaea, Bacteria) and early eukaryotes, are actin and RecA-family recombinant proteins plus several others, and for meiosis the specific proteins, DMC1 and Rec8 are shared [4] -[6] . But operational meiosis was not a feature of the very early, primitive eukaryotes that had gained polyploid status in response to environmental induced genomic damage. For survival with procreative ability this damage had to be repaired accurately, which was claimed could only be done by meiotic recombination [7] . Molecular data from such primitive eukaryotes and from prokaryotes suggest that a type of meiosis was always present as a continuum evolvement of eukaryotes from prokaryotes. Sexual reproduction in Bacteria by a transformation process was suggested to be evolutionary transferred to eukaryotes, which therefore, possessed the ability to repair genomic damage accurately by the presence of recombinant meiotic gene products. Other views claimed simultaneous evolvement of mitosis and meiosis, or that meiosis originated from mitosis which only required four mitotic changes: 1) homologous chromosome pairing, 2) capacity for recombination, 3) suppression of sister-chromatid separation in meiosis I (M-I), and 4) cohesion between sister chromatids maintenance until M-II [5] .
Eukaryotes with closed divisions inside the nuclear membrane were considered to be more primitive than those with variously “open” divisions [8] [9] . Bernstein and Bernstein [7] argued that survival with procreative ability of ancient eukaryotes stemmed from their capacity of meiotic recombination (pairing of homologs) for repair of accidental, acquired genomic damage. Human cells showed mitotic repair exchanges associated with genome reductive divisions of polyploidy [10] -[14] . Accidental re-replication of the primitive haploid genome (genomic doubling) to diploidy, may not have been with the greatest fidelity, leading to genomic damage in need of repair [15] . These authors argued that accurate repair was achievable by exchanges between original, pristine (damage-free) template DNA strands and damaged strands from faulty replication. This suggestion puts evolvement of meiosis from diploidy into the evolutionary “future”, which was in agreement with other evolutionists’ way of thinking [16] [17] . They, from existing cytology-observations claimed that polyploidy was the route to genomic repair mechanisms, and that such polyploid, unicellular eukaryotes underwent genome reductive processes. For the present presentation this latter suggestion is crucial, because it brings into the ongoing debate (see above) endo-polyploidy from endomitosis (the megakaryocyte type). For in vitro cells endotetraploidy with 4-chromatid chromosomes (diplochromosomes/pairs of sister chromosomes), the “Hurst-Nurse-type” recombinant repair of genomic damage was evident from tritiated thymidine labeling, and from chiasma-terminalization in anaphase [13] [18] . Such mitotic, chiasmatic exchanges between sister-chromatids of diplochromosomes (4n/8C) is also part of the disease picture for the prototype “cancer-model” Blooms’ syndrome [19] . There is also the possibility that cells in early evolutionary time did not undergo accurate DNA repair processes, because of faulty repair mechanisms, giving rise to mutational variation with evolutionary adaptive value in the development of new species [20] [21] .
2) Cytogenetics of two mechanistic genome reductive divisions The present pictorial display (figures) cannot give full justice to the details and consequences of these reductive divisions, which produce genomic altered para-diploid (transformed) cells expressing gain of a proliferative advantage (GPA) [10] -[14] . Therefore, the following summary of cellular happenings must precede present experimental result with the knowledge that GPA is essential for cancer initiating events. Endomitosis, the megakaryocyte-type, is increasingly mentioned in cancer-related literature concerned with aneuploidy as a hallmark in tumorigenesis. The endomitotic reductive divisions were found to be a response in a carcinogen-free induction system that was applied to young normal, human diploid cells. The important for the documentation of both types of mechanistic division sequences were experiments designed for in situ observations of growing cell populations. This was achieved by growing normal, human cells directly on microscopy slides/coverslips with harvest of attached cells without chromosomal spreading of metaphases, and not solution-based slide-making. Such preparations showing endo-tetraploid reduction division revealed the following chromosomal details: co-segregating whole complements in both prophase and anaphase, change to star-like (rosette-like) morphology of segregating genomes, and perpendicular orientation to the cytoskeleton axis of prophase and endo-tetraploid segregating genomes. The reduction of normal diploid metaphases to haploidy also showed: co-segregation of maternal and paternal (mat/pat) genomes from each other (i.e., gonomery), proliferative capacity from genomic doubling to homozygosity, change to star-like mitotic figures, and 1/2 cell-volume reduction to small-cells. Other details were absence of apoptotic cells, and both types of reductive divisions occurred without any indications of activated mitotic checkpoint arrest controls. The perpendicular orientation of the divisions relative to the cytoskeleton axis freed cells from cell contact inhibition with result of cell polarity change/loss in multilayered growths.
At this point in the present presentation it seems appropriate with an evaluation of presently ongoing, preferred techniques/methodology in cancer research. The present cytogenetic/morphology approach has over the last two decades mostly been substituted by molecular chemistry and the machines for PCR, SNP, and flow cytometry. The latter device can in short time scan a large cell population for presence of diploid and polyploid cells, but a full cycle of mitosis becomes over-stepped. Furthermore, with its latest imaging capability, specific gene activity can be monitored (up-down regulations) in for instance the change from diploidy to endopolyploidy. Such data together with the methodologies of comparative genomic hybridization (details of karyotypic changes) and antibody fluorescence analyses (gene-specific activity) are today’s preferred experimental tools in cancer research (exclusive of mouse cancer model studies). The question is whether these approaches supply maximum information when considering a possible tumorigenic initiation process from endopolyploidy with 4-chromatid chromosomes. Firstly, flow cytometry cannot count chromatids and distinguish between diplochromosomes (#46) and regular tetraploidy (#92), and 2n/4C and 4n/4C cells can also not be distinguished from each other, and neither can such cells be assigned with confidence to cell cycle phase (G1 versus G2), because diplochromosomes segregate bichromatid, 4C cells into G1 of the cell cycle, rendering such division products incapable of entering normal mitosis. These are serious flaws, because two types of tetraploidy with very different division sequences are not acknowledged in the cancer research community. The ongoing cancer-experimental methodology has yet produced mutated cells expressing GPA with origin from normal, human cell populations. And furthermore, how can experimental use of already cancer-transformed cell lines reveal initiating processes (e.g., HCT116, p53+/+). Clearly, present cancer research related molecular machinery will not detect present types of GPA before; in situ microscopy of mitosis (prophase to telophase/interphase) is included in the experimental approach.
3) Presentation of pictorial illustrations of genome reductive divisions of endopolyploidy and diploid metaphases
1.2. Materials and Methods
Throughout data collection for previous publications [10] -[14] , the two primary cell strains L-645 and WI-38 were used, firstly, associated with dysfunctional telomeres at early senescence, and secondly, changed to young cell propagation (at about 20 population doublings) in a carcinogen-free induction system of both reductive divisions. The inducing system consisted of short-time (2 - 4 days) amino acid glutamine deprivation (AAD – Eagles’ basal medium + 2% calf serum (fbs), pen/strep) followed by recovery in complete medium (Eagles’ + 10% fbs + pen/strep + 3 mmol/100 glutamine) showed in recovery growth all aspects of cellular events associated with both types of genome reductive divisions. The details of this methodology for the present results were as follows: flask cultures (25 cc) seeded with 1 to 1.5 million cells (both cell strains) at 18th and 19th passages allowed 1-day growth in complete medium, washed 2× with Hanks balanced salt solution, and then were exposed to medium without glutamine. Such flask cultures were left untouched for 2, 3, and 4 days, before cell detachment from trypsin/versene treatments. One and two well chamber, glass slides were seeded with 3 - 4000 from each detached cell population such that for each AAD exposure day there were harvested slides with 1, 2, 3, 4 days growths. Harvesting was done by fixation in Carnoys’ fixative (3 parts methyl alcohol:1 part acidic acid). The fixed preparations for in situ cytogenetic/morphology studies were stained/not stained with 1% Giemsa, and subjected to microscopy photography, including phase contrast. This simple procedure has the added advantage that different fixatives can be used for immune-fluorescent and genomic hybridization studies and, with whatever manipulation desired. The older type of chromosomal staining, Giemsa and Feulgen, gives far superior, crisp, cell and chromosomal staining results than the today’ preferred methods of DAPI or Propidium Iodide that gives “fuzzy” images (made worst by stamp-sized journal pictures). Other positive aspects of this simple AAD methodology are the ease with which it can be repeated, and that the cost of these old, staining methods are only a small fraction of the cost for the newer methods. (New toys are not necessarily an improvement upon older ways, which is supported by today’s maintenance of crystal violetor fuchsin-based haematoxylin in cancer diagnostic pathology).
2. Results
Summarily the highlights of the meiotic-like (ML) divisions of endo-tetraploid cells (4n/8C) with diplochromosomes (Figure 1(A), Figure 1(B), see legend) contain 46-4-chromatid chromosomes (Figure 2(B)) from “glued” together sister-pairs (Figure 1(F)—4n/8C), a first division (ML-1) splits the pairs apart with co-segregation of whole complements from each other to 4n/4C cells (Figures 2(C)-2(F)). The co-segregating process was marked by change to characteristic star-like morphology (rosette-like figures) (Figures 2(C)-2(E)), which were in a perpendicular orientation to the cytoskeleton axis (Figure 1(C)). The resulting chromosomal groups at telophase showed chromosomal condensation associated with cytokinesis (Figure 2(F)). Compared to normal mitotic anaphase (Figure 2(A)) these star-like divisions to 4n/4C cells did not show segregation-individuality of single chromosomes on a spindle apparatus. This change for sister chromosomal segregation (bichromatid chromosomes) is clearly demonstrated in Figure 2(I) showing a polar and a side view of one division figure. The possible explanation of synchronized divisions from binucleated cells is excluded by chromatin bridges being double, which indicates segregations of bichromatid whole complement from each other (Figure 1(B), Figure 1(C) & Figure 2(H)). A very important fact is that is that co-segregation of whole complements is into G1-phase of the cell cycle (see Discussion). These 4n/4C cells from reductive division of 4n/8C cells rarely went through an immediate second division (ML-2) to 2n/2C para-diploid cells (Figure 1(B), Figure 1(D), Figure 1(E) & Figure 2(F), Figure 2(G)). These genome altered cells (transformed) showed in extended passage-growth
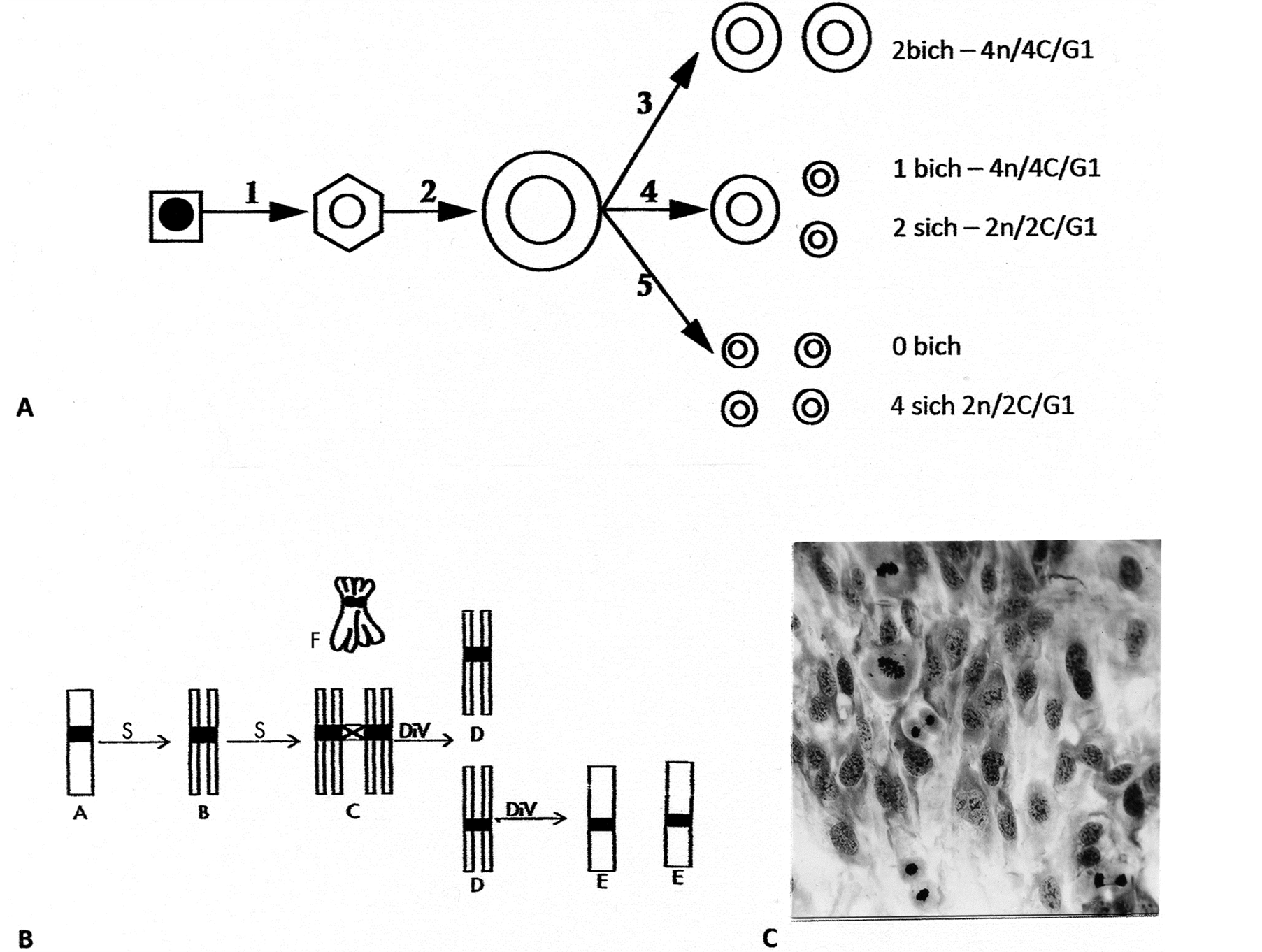
Figure 1. Line drawings of endotetraploid genomic reductive division to para-diploid cells. (A) Interphase happenings following two consecutive S-periods that by for example, flow cytometry (FC) can be separated into tetraploid cells (4n/8C) from step 2 and reduction to cells (4n/4C) from step 3 and one product from step 4. The remaining 6 small, cell-products, the para-diploid cells (2n/2C), would be missed by FC in a normal diploid cell population; (B) Line drawings of chromatid segregation-sequences during the reduction division: The two S-periods produce 4-chromatid chromosomes by two sister bichromatid chromosomes being cohesed together (diplochromosomes). This complex has three centromere regions: synthetically an old (C-cross) and two younger (black) centromere regions. The old centromere region resolves first and separates the sisterpairs from each other with results of cells containing bichromatid chromosomes in G1 (D-cells). Rarer divisions of these D-cell products give rise to the para-diploid cells (see text); (C) A polyploid cell-area in a normal cell population exposed to glutamine deprivation (see text) shows 3 divisions in a perpendicular orientation relative to their own cytoskeleton cell axis and to surrounding cells. Giemsa stain, enlarged: (C) 396×.
increased growth rate of morpho-logically changed cells, forming a changed growth pattern (Figure 3(A), Figure 3(B)) from normal fibroblast-pattern of striation. This unusual mechanistic cell-division system is an unprecedented discovery with base in genomic damage leading to endo-polyploidization. That nutritional deprivation of one single amino acid can lead to genomic damage of normal cells is unquestionably supported by presence in the recovery-growth of cells with chromosome breakage and various segregating chromosomal abnormalities (Figures 2(J)-(M)).
The details of the second genome reductive process showed halving/near-halving (asymmetric) of diploid metaphase rosette figures (Figures 3(C)-(E)) into two haploid groups of chromosomes which was followed by chromosomal condensation and cytokinesis (Figures 3(F)-(H)) [14] . These occurrences of haploidization, also with star-like morphology in mitotic proliferation (Figures 3(C)-(E)), gave rise to a small, 1/2 cell-volume reduced diploid cell-type. The haploid genomes from gonomeric-based maternal and paternal (mat/pat) genomic, independent segregations apparently doubled to homozygous cells, (which awaits confirmation by sequence array haplotyping). The “half-size” of these small-cells was retained during mitosis, i.e., size reduction was permanent (Figures 2(C)-(E)). Of note is the clear indication of induced genomic damage by presence of DNA-
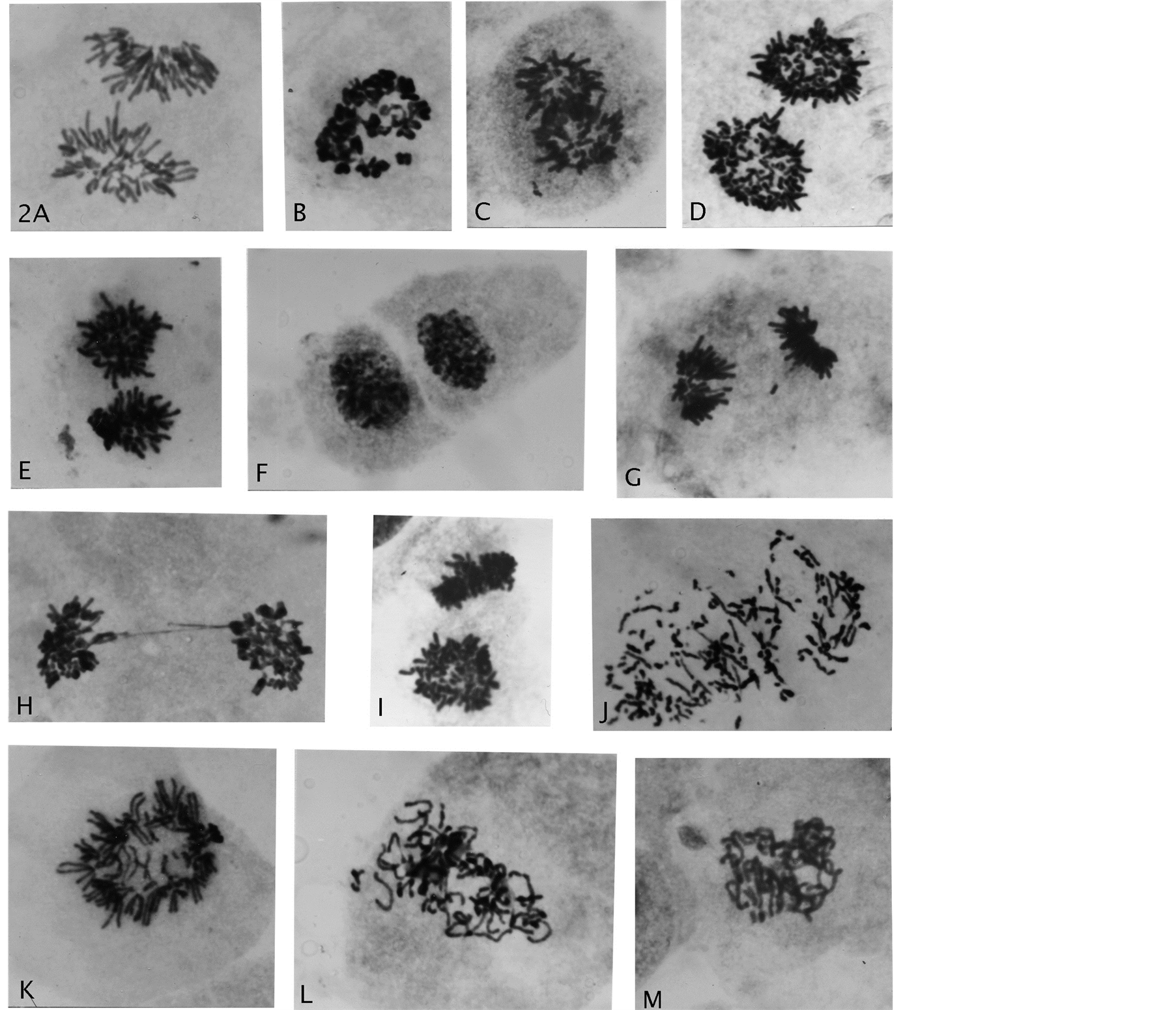
Figure 2. Genome reductional division of endotetraploid cells. (A) A normal anaphase cell; (B) A rosette with diplo-chromosomes; (C)-(E) Segregation of whole bichromatid complements with star-like morphology to telophase nuclei separated by cytokinesis; (F) (G) Rarer division of one endo-derived 2n/4C telophase product into 2n/2C chromosomal groups; (H) Telophase double chromatin bridge; (I) Polar and side view of star-like complements; (J)-(M) Anaphase with chromosomal breakage, premature chromatid separations, early and late prophase genomic separations. Giemsa stain, all enlarged 2475×.
repair sister chromatid exchange-figures in anaphase (Figures 3(I)-(L)). To this authors’ knowledge, these specific chromosomal configurations in diploid cells, leaves no doubt about chiasmata-terminalization, which is a first-time demonstration. In extended passage-growth the small-cells showed streaming growth (hyperplasia-like) (Figure 3(M)) of uniformed-sized nuclei, possessing multiple small nucleoli (Figure 3(N)). Increase in proliferation rate (GPA) was immediate upon “birth” of these cells, which very likely was a result from their homozygous constitution expressing loss of heterozygosity for tumor suppressor genes. Thus, the lesson is that early effects from AAD is not easily detected: the innocently “normal looking” rosette rings (Figure 4(A)) with orderly chromosomal arrangements for mitotic fidelity, may be hiding potential gonomery with haploid sets capable of autonomous behavior for very undesirable division-consequences (Figure 4(B), Figure 4(C)) in normal human cells. These latter figures do not show metaphase rosette behavior for normal mitotic segregation, but indicate asymmetric gonomeric segregations (unpubl.).
3. Discussion
1) Reductive endopolyploidy creating a cancerous potential
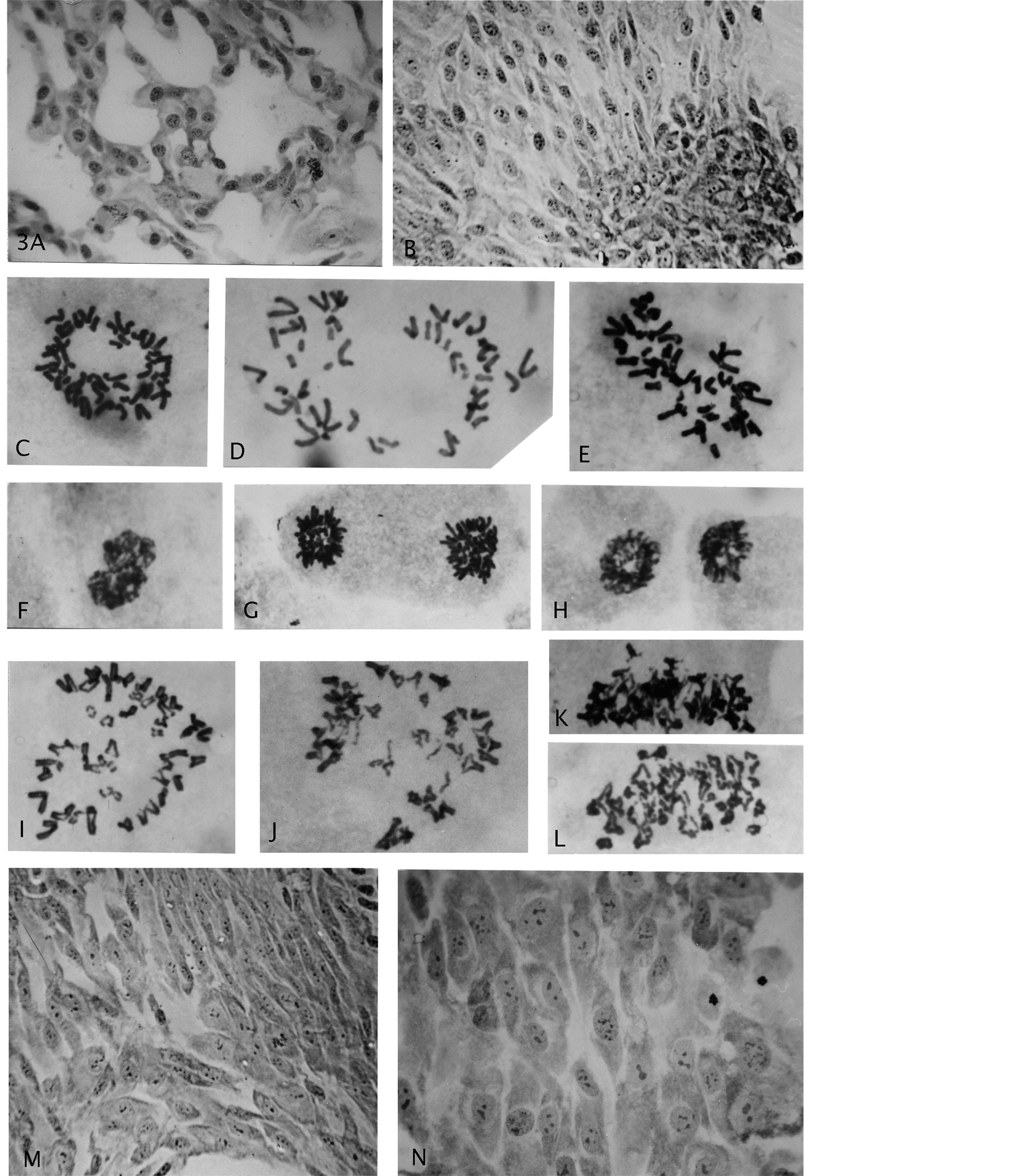
Figure 3. (A) (B) Growth of endo-derived para-diploid cells with roundish nuclei and a heaped, multi layered, focal growth; (C) A diploid normal rosette figure; (D)-(H) Segregated haploid groups of chromosomes (gonomery) and resultant diploid star-like, size-reduced mitotic figures (compare Figure 2(A)); (I)-(L) Examples of cells showing sister chromatid exchanges; (M) (N) Gonomeric-derived cell growth (haploidization) from conversion to diploidy by genomic doubling, showing nuclear size uniformity and multiple small nucleoli. Giemsa stain, enlargements: (A) (B) (M) (N) 619×, (G)-(L) 2475×.
Previous and present results clearly demonstrate that the genome of man contains old “evolutionary” genetics, determining orderly, genome-reductive behavior for two different division-systems. They both gave rise to transformed cells that had gained pre-cancer-like increased fitness associated with altered cell-phenotypes. The surprise is that these changes were immediately expressed by the different progenies, and importantly, can be
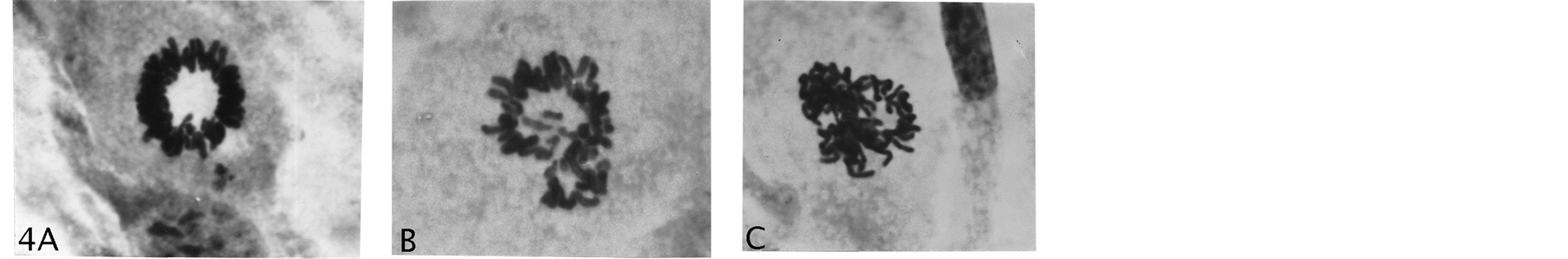
Figure 4. (A) A normal rosette figure; (B) (C) Examples of asymmetric gonomeric-based chromosomal distributions with potentials for cells with LOH. Giemsa stain, (A) (B) (C) 2475×.
produced at will in the present AAD, carcinogen-free inducing-system. Any type of a cellular system that can give rise to GPA from normal cells is suspect of being a cancer-related initiation process, and the question is whether in vivo pre-cancers show for example, endopolyploid involvement in their origin. Yes, Barrett’ esophagus disease, which is associated with tobacco use, obesity and gastroesophageal reflux has several positive pointers [22] . This disease has increased risk for change into esophageal adenocarcinoma, and has been (is) in the forefront as a model system for studies on prevention and therapeutic cure. Recently pre-cancerous lesions were described as “4N (G2-tetraploid)” from flow cytometry-DNA measures. This description is confusing/ -misleading without C-values, and one can only guess that the description means either G2-4n/8C or G1-4n/4C cells, with both conditions expected to instigate mitotic divisions. Surprisingly, the expected mitoses were not present in these lesions, although the molecular machinery was indicated [22] . The most likely explanation from this latter fact is endo-tetraploid (46-4n/8C) reduction-division to 46-4n/4C cells, which will be in G1 of the cell cycle. Bichromatid chromosomes (normal metaphase chromosomes) in G1 renders such cells incompetent to enter regular mitosis. But, 4n/4C cells in G1 can enter into S-periods with genomic doubling to 4-chromatid diplochromosomes (4n/8C, endo-tetraploidy). Such cells are vulnerable for transient endo-reductive divisions back to 4n/4C cells in G1, which with repeated such cycling would result in growth-enlargement of Barrett’ esophagus pre-cancerous lesions. Additionally, it is known that mutations in EAC (esophageal adenocarcinoma) were present in matched Barrett’ esophagus, which points to a common precursor cellular event, here suggested to be transient endo-tetraploid reductive division.
Interestingly, this type of endo-G1-cycling appears to have support from so-called “pathological mitosis” which “--- reflect cell-cycle disorder, ---” and that “--- dysplasia originates from undetected nuclear defects” [23] . Single division DNA-measurements for meta-interphase cells demonstrated: 4C+/−; 5C exceeding values, 6.1C; 8.4C, and anaphase division figures showed asymmetric segregations: 2.0 - 1.9; 2.0 - 2.0; 2.9 - 4.6; 2.5 - 2.9 in ulcerative colitis and in other dysplasia with creation of significant aneuploidy. The author avoided implication of endo-polyploidy simply, because of believes in endo-polyploidy being a proliferative dead end.
2) Consequences and peculiarities of low level endo-polyploidy Above it was pointed out that methodologies in cancer research are insufficient for discoveries of present type, suggested cancer-initiating cellular events. Endo-reductive cycling with the division in a perpendicular orientation to the cytoskeleton axis (Figure 1(C)) lead to freedom from cell contact inhibition for the two daughter cells. In extended growth such cells showed cell polarity change, especially clearly demonstrated in abnormal multilayered growth [24] [25] . Another important peculiarity is absence of an S-period between ML-1 division and an immediately following (rarer) ML-2 division, which leads to 2n/2C para-diploid (transformed) cells. The chromosomal 4-chomatid complex as outlined (Figure 1(B)) is held together by 3 centromere/-kinetochore regions, which from their time of synthesis is one old (C cross) and two younger regions (C black). The old region holds the pairs of sister chromosomes together whereas the newer regions holds together the two chromatids of single sister chromosomes. The essence of the endo-polyploid “reductive-power” is in the asynchronous dissolution of these regions with separation of the old centromere-kinetochore region in the first meiotic-like division (ML-1), followed by separation of the newer regions ones in the second division (ML-2). These somatic cell divisions are peculiarly similar in some details to meiosis (meiotic-like) as for instance [Figure 3 & Figure 4 above] of Wilkins and Holiday [5] . Moreover, in tumor progression associated with depolyploidization to lower ploidy-levels the meiotic specific gene-products DMC1, Rec8 and MOS were up-regulated [26] . The uniqueness of this meiotic-like endo-two-step division-system is illustrated from fibroblastic growth showing the perpendicular orientation of co-segregating whole complements in the first division (Figure 1(C)). This is a peculiarity expressed for the unicellular, radiolaren Aulacantha (see below). Modest asymmetric first-divisions would contribute to aneuploidy, perhaps facilitating evolutionary probability for development of organismal-diversity [20] [21] . But as will be clear in the following, there were other different reductive divisions of higher levels of polyploidy for the more primitive unicellular organisms [17] .
3) Literature knowledge/acceptance of reductive endo-polyploid division It is increasingly understood that G2/M cells (normal metaphase chromosomes) with genomic damage in a repair process (gamma-H2AX foci) can enter an S-period by mitotic slippage [27] with result of genomic doubling to endotetraploidy. The earlier literature associated the tetraploid-condition with formation of 46, 4-chromatid diplochromosomes (4n/8C) i.e., from endo-reduplication [10] [28] . Presently there is increasing mentioning of endoreplication, but its chromosomal structural consequence (4-chromatid chromosomes) as distinct from regular, genomic doubling to 92 chromosomes lacks understanding regarding a non-mitotic division [29] -[32] . Even when encountered in cancer-cytogenetics, there appears to be no curiosity about their division-behavior, which was a major interest for mouse ascites tumor cells some decades ago [33] . These divisions are orderly to daughter cells, which have inherited endo-division special traits as for instance co-segregation of complete complements. Such traits became incorporated into the daughter’ innate mitotic machinery best expressed by prophase cell’ ability to segregate complete genomes directly into anaphase [12] . If for example, the acknowledged presence of tetraploid diplochromosomal cells had been considered with division-results, the claim of a “transformed cell phenotype” from presence of trisomy 8 sole-abnormality would be radically different [34] . Their working cell population (10 mm dishes) was genotypically mixed from diplochromosomal reductive divisions back to “transformed para-diploidy”, recently supported by the finding that “--- the tetraploid (from diplochromosomes) descendants were more transformed based ---” in two assay systems than diploid cells [30] [35] . Special division-systems that can bypass active, mitotic checkpoint control (p53-positive cells) have (so far) not been considered in the origin of the so-called transformed cell-phenotype. Since this latter process has recently been shown to occur for young cells with long telomeres [13] [14] it is very important that pre-cancer-like cell changes are recognized with origin from an endo-polyploid mechanistic cell division-system.
Another reason hampering general recognition of diplochromosomal genome reductive divisions is as mentioned superficial, mimicry of conventional 2-step meiosis. The sister pairs of chromosomes stays closely cohesed from prophase to metaphase showing four chromatids, which can give the appearance of “meiotic” pairing of homologous chromosomes. Cleveland [36] [37] was guilty of such interpretations from his cytology-observations of primitive, flagellate, symbionts in the cockroach [17] and remarked: “--- At prophase synapsis begins, and soon the plainest tetrads I have ever seen are formed --- followed by --- chromosomes going to the poles as dyads”. He did not observe meiosis-II with four products and remarked further: “--- divisions resembled normal mitosis”. This confusion is easily solved by the demonstration of a haploid number of tetrads for real meiosis, whereas diplochromosomal divisions show the diploid number (46) of sister-pairs (“tetrads”) of chromosomes.
The cytological presentations from unicellular eukaryotes is also problematic in that there too, is an omission of a distinction between the regular and the endomitotic type of polyploidy [8] [9] . But most frustrating is that present-day referrals to publications explaining/hypothesizing the origin of cancer-aneuploidy without mentioning endo-polyploid DNA-replication systems with consequences [38] -[40] . Diagrams of regular tetraploid (92 chromosomes) cells exhibiting various mitotic segregation mistakes is the going trend, but becomes inadequate information considering possible cell-population-dynamics when normal cells are induced to undergo genomic changes (unpubl.). Compared to present attitude to endopolyploidy, ancient unicellular organisms were on the contrary often considered to be endo-tetraploid from endomitosis [5] [9] [15] [16] , the megakaryocyte-type [41] . Such special replication produce cells with occurrence of anaphase A-only and back to a failed mitosis, but with retained capacity for return to an S-period, doubling the genome to 46, 4-chromatid diplochromosomes (pairs of sister chromosomes). Multiple cycles of endomitosis lead to polyteny (many threads), in-capable of division, as also is characteristic of highly polyploidy, differentiated cells. For the early eukaryote, low and high polyploid levels existed as phases in their life cycles, such that the purpose of establishment of a reductive polyploid division-system would be to bring the organism back to haploid/diploid vegetative life [15] [16] . On their already established mitosis, the endomitotic reductive segregations would be chaotic, because mitosis is built for 2-chromatid chromosomal segregation and not for 4. Both Kondrashov [16] and Haig [17] therefore, argued for pre-existence of a depolyploidization system, before presence of meiosis and sexual cycles. They considered that primeval, stressful, environmental conditions caused damage to haploid genomes in G2 (bichromatid chromosomes) and would be in need of a DNA-repair system, that likely involved endomitotic polyploidization. Accurate repair by recombination-events associated with depolyploidization may still be the way to haploid vegetative life as described for “chromidia” production [42] . Thus, it appears that DNA-damage-associated endo-polyploidization is an ancient evolutionary phenomenon, which has genome reductive capacity long before presence of repair by paired homologous regions. Endomitosis to high polyploidy of eukaryotes living today (e.g., unicellular radiolaren Aulacantha, see below) may therefore, have been an early primitive, adapted change before meiosis came on the scene. Importantly, these considerations do not negate existence of other types of mechanisms for genome reduction (see below—haploidization).
In this regard there is possibility that diplochromosomes can express instability and “fall-apart” into components (singleand bichromatid units) and proceed through anaphase to aneuploid daughter cells [38] -[40] . There are two ways that endo-replication can occur: either by consecutive S-periods [43] or by endomitosis, the megakaryocyte type [41] . As mentioned, the latter genomic doubling process goes through anaphase-A followed by chromosomal condensation and returns to endotetraploid interphase cells (4n/4C). It is not known to what extent anaphase-A separate/weaken the cohesion between the 3 centromere regions of diplochromosomes (Figure 1(B)), especially in repeated endomitotic replication as discussed for Barrett’ esophagus. But in either case the first meiotic-like (ML-1) division has probability of abnormal aneuploid chromosomal distribution, the whole chromosomal loss-type [44] that however, can be hidden by compensated uniparental disomy (UPD) [45] . These potentially, unstable divisions may also be associated with breakage [13] [14] with possibility for chromosomal rearrangements that can lead to segmental UPDs [46] . Certainly, if diplo-chromosomal structure can be modified into units there would be provisions for genomic diversity leading to clonal selection of cells with GPA from loss of heterozygosity (LOH). But, why would these atypical ML-1 divisions from endomitosis not show arrest by SAC? Because these “endo-divisions” were already primed for perpendicular segregation relative to the cytoskeleton axis, which avoids spindle-assembly-checkpoint (SAC) activation (see below Aulacantha). The cytoskeleton partakes in mitotic cytokinesis and its fibrils reorganize in accord with the development of the spindle apparatus, maintaining the cytoskeleton “axial” orientation [47] .
4) Gonomery in the evolutionary record Now the pressing question is how the non-mitosis-related reductive endo-division is connected to gonomery? Among the ancestral, diploid eukaryotes, microsporidia from molecular data: “--- emerged earlier than any other protistan groups” [9] , and together with red algae showed endo-polyploid genome reductive behavior to haploidy. But, an example is first presented for high level “endoreduplicated genomes” undergoing depolyploidization that would be difficult to disqualify, because of careful quantitative DNA-analyses [48] . It is from the colonic alga Eudorina californica (family, Volvocaceae) with closed nuclear divisions, where in vegetative reproduction the conidial cells re-replicated their genomes 64 times the haploid genome. This was followed by genome reduction by cleavage (amitosis) to 64, free swimming haploid “plakeas”. The reduction to proliferative haploidy cannot be anything else than gonomery-based “mat and pat” genomic segregation from each other. Other examples of reproductive-associated amitosis of high endopolyploidy are from Euglena, dinoflagelates and the amoeba Paratetramitus jugosus—seen as “--- segregation of multiple genomes” [9] [17] [42] [48] [49] . The amoeba example with high level endopolyploidy reduced to haploidy by multiple-fission (amitosis). Strangely, these amitotic reductions led to accurate genome segregations to volume-reduced small offspring cells. (It is not known whether cleavage, fission and fragmentation of nuclei are the same processes, and is for convenience grouped into amitosis.) A likely explanation for gonomery is the organization of endomitotic mat and pat genomes to each other. High endo-polyploidy (16n/32C & 32n/64C) for human primary cells either reduced genomic contents by amitosis to multinuclear cells (MNCs), or by sequential, amitotic partitioning of diploid-sized nuclei (i.e., nuclear budding) from the large, mother nucleus, that was capable of compartmentalized mitoses in mother-cell cytoplasm [50] [51] . Nuclear size differences in MNCs indicated diploidy and haploid segregations associated with genomic doubling as observed for rat embryonic cells (see below).
But interestingly, before leaving endo-polyploid reductive division it should be known that Eudorina (Volvocaceae family) showed division-behavior supporting an evolvement from the Chlamydomona alga, which interpretively performed “meiotic”, two-step reduction-division of tetraploid cells in the absence of an intervening DNA-synthetic period [16] [17] [48] . These divisions with available cytology can equally well be from reductive endopolyploidy, which is supported from more detailed chromosomal analyses of the unicellular radiolaren Aulachanta scolymantha [9] [52] . The division events were: segregations to genome reduced cells in the absence of centrioles and a spindle apparatus, and co-segregating genomes were in a perpen-dicular orientation relative to the cell’s cytoskeleton axis (see above: perpendicular ML-1 division). Raikov [9] wrote: “--- chromosomes (were) not on plate as in regular mitosis, but perpendicular to it”, and D’Amato [28] classified the segregation as “parallel” as opposed to axial (see consequences: cell polarity change [25] ). In oral cancer-cells the division-axis was skewed relative to the cytoskeleton axis suggesting, cancer-cell derivation from endo-polyploidy, which gave rise to offspring cells without the use of a normal spindle apparatus [53] . Whether endo-tetraploid cells (4n/8C) can express gonomeric-based reductive divisions is preliminary indicated by 46 diplo-chromosomes undergoing bipolar segregation with two groups of near-23 diplo-chromosomes (unpubl). This happening suggests that 23 mat-diplo-chromosomes separated from 23 pat-diplochromosomes, which is likely if such mat and pat genomes (4n/4C each) were arranged opposite to each other in metaphase rosette figures (see below for diploid arrangements). Interpretively, such segregations would lead to homozygous tetraploid (4n/4C) cells in G1 (see Barrett’ esophagus) with cycling probability from GPA-associated with loss of tumor suppressor genes.
The absence of centrioles figures strongly in the evolutionary record from ancient Archaea with membraneattached nucleoid fission to eukaryotes lacking a spindle apparatus (microsporidia, Trichomonas, ameoeba) [9] [32] [54] . Divisions lacking centrioles were even expressed in phylogenetic “high-up” snakes and wallaby PtK-1 cells, the latter cells showed that microtubules “--- did not come to a focal point” [55] [56] . Absence of centrioles is also a feature in evolutionary advanced, fission yeast [57] , which is often a chosen organism in cancer-cell experimentation. These primitive divisions with absence of a normal spindle apparatus, were found to be associated with the presence of actin, myosin and motor proteins which were suggested to form a moving “matrix carpet” in non-mitotic divisions [58] [59] . The reception of these ideas have been rather cool [58] , but rethinking is necessary, because these ideas may give clues to the absence of arrest by SAC of endopolyploid perpendicular divisions [25] . Thus, the absence of centrioles in multi-cellular organisms is an indication of evolutionary conservation of an ancient cell-division-system, indicated to be connected to division-perpendicularity and SAC-avoidance.
The next obvious question is whether there are cytological occurrences (indications) of gonomery for unicellular organisms claimed to undergo primitive “one-step meiosis” by a single reduction-division from diploidy to haploidy? This search, starting from Cleveland’ [36] symbiotic flagellates showed that the chromosomes gathered together in two rings from end-to-end attachments. Why the formation of two rings? The most likely answer is mat and pat genomic rings, which supposedly must have been established when diploid cells originated from haploidy. Although, there are many claims of one-step meiosis in different eukaryotic taxa, the more difficult to “explain away” is for example, from the dinoflagellate Crypthecodinium cohnii [17] . Gametogenic cysts divided into two daughter cells, and zygotic cysts into four daughter cells, all daughters being haploid. Similarly Eimeria tenella showed haploid gametes performing syngamy (fertilization) with diploid segregations back to haploidy without chromosomal replication. In mirosporidia (oldest eukaryotes) haploid cells fused to binucleated cells in which the nuclei with close contacts went through several cycles of synchronized, individual, haploid mitoses before syngamy [17] .
5) Gonomery in multicellular organisms and in normal human cells Gonomeric-based mat and pat genomic segregations is an acknowledge occurrence in obligate parthenogenesis of endopolyploidy in most animal phyla where the genome reductive process can end with haploidy for various male-organisms (Crustacea, Isopoda, Lepidoptera, wasp, aphids and honeybees) [60] . For the crustacean Copepoda, gonomery is well established with links to “chromatin diminution” [61] . And, the latest example of gonomery is from haploid proliferation from unfertilized human egg-cells, demonstrating stem-cell characteristics [62] . Principally, this latter phenomenon is similar to human terratomas (significant change to carcinomas) from fertilization of an empty egg-cell with an X-bearing sperm, found to be homozygous diploid cells from genomic doubling of the haploid genome [63] [64] . Evolutionary scientists found it difficult to accept reductive divisions of diploidy to haploid cells (i.e., “one-step meiosis”) before the advent of meiosis. In fungi (yeast) these events are seen as parasexual issues [65] [66] , which however, were suggested to have been acquired subsequent to meiosis-presence in eukaryotes [5] . And, Cleveland’ [37] cytological studies of symbiotic, flagellates in cockroaches, claiming alternation between diploid vegetative life and a haploid “sexual” phase was disqualified as ambiguous cytology [17] . But when one-step-meiosis was acknowledged, the assumed mechanistic route from diploidy to haploidy was believed to be a series of mitoses with gradual chromosome loss from non-disjunction [16] , —not unlike today’s suggestion for certain leukemias (see below). However, the most recent study of diploid yeast cells showed: “--- rapid conversion to haploid --- DNA content ---” [66] , and likewise rapid, for human-cell-haploidization (Figure 3(C)-(H)) from a haploid grouping process (Figure 3(D), Figure 3(E)) of chromosomes in normal metaphase cells [13] [14] . Such rosette-figures segregated near-haploid (~23) genomes by a halving-process of the chromosomal rings, which proceeded with telophase nuclei performing cytokinesis, and further to mitosis from genomic doubling (assumed as for terratomas) to homozygous cells. Similarly, chromosome painted rosettes in metaphase showed human, mat and pat genomes in opposite positions in the ring of chromosomes [67] [68] . Gonomery-based mat and pat unit-identity with segregations from each other in mitosis was shown for early embryonic mouse cells, but with reduction to 10% in later divisions [69] . The basic origin of gonomery from ovum and sperm was shown for a primitive hermaphroditic tubularian for which the haploid numbers of chromosomes were in specific order, forming linier arrays from end-to-end attachments. Following syngamy (fertilization), a large rosette figure was formed with mat and pat complements arranged opposite to each other [70] .
Stern [71] in his critical review on somatic cell variability concluded: “Whatever the mechanism, the origin of haploid from diploid or lower degrees of ploidy from higher ones is an established source of somatic cell variation”. Probably the most telling examples of gonomery expressed as two chromosomal rings (rosette figures), and its conservation in higher organisms is from polyploid, parthenogenetic weevils undergoing reduction division [71] . Thus, various unicellular organisms demonstrate early, evolutionary established gonomery, which today are conserved in genomes of multicellular plants [72] and animals [73] , including human and other mammalian species [14] [74] . The base for gonomeric segregation is a separation of mat and pat genomes into two territories of normal diploid metaphase cells, which was also demonstrated for an endotetraploid cell, but note, with larger-sized individual territories from cytoplasmic increase, naturally associated with polyploid transcriptions. In general gonomery has been associated with embryonic conditions of the cells [69] [72] and present cells are of that origin.
The conservation of gonomery in multicellular organisms can become de-repressed in human cells with the consequence that the genome reductive mechanisms (reductive endopolyploidy and diploid-haploidization) could cycle with gonomery-based chromosomal distribution, to transformed progeny cells. However, one drawback for recognition of these transformed in vitro progeny cells is that they are not readily recognizable by mitotic errors in early beginning proliferation. The best markers are prophase divisions (Figure 2(L), Figure 2(M)) directly into anaphase [12] , also an occurrence for budding yeast [75] , and that endo-derived diploid cells can be perpendicularly oriented to the cytoskeleton axis [14] . Clearer notice of transformed cell-growth was apparent in extended proliferation (Figure 3(A), Figure 3(B)), when the patterns of growth were hyperplastic-like and multi-layered focal areas with cell polarity and nuclear morphology changes were present [76] . Nuclear-shape changes (i.e., aniso-nucleosis) are powerful biomarker in cancer-diagnostic pathology, and are often present in dysplastic lesions, noted to contain endoreplicated cells before the transformation to full blown cancer [23] . All in all, the diversity in surviving mitotic errors from haplo-endo-derived inherited special traits merged into the innate, mitotic machineries appeared to promote a neoplastic-like progression, linked to increased fitness (GPA) for these in vitro originally normal cells. Similar suggestions have been expressed for immortalized transformed cells, also showing a phase with diplochromosomes, and origin from (age-related) normal, human cells [10] [11] [30] . However, the route to the immor-talized cells is strictly associated with cells expressing age-related dysfunctional telomeres, senescence and a crisis period (chaotic genome-destructive divisions). The present routes to transformed cells can occur for young cells (normal telomeres), and has as origin mechanistic reductive, orderly, cell-division-systems, which lead to GPA. Whether these transformed cells have gained immortality or proceed to senescence as pre-cancerous cells do, is an ongoing study.
6) Endopolyploid and diploid reductive divisions with links to a cancerous process The link to a cancerous process especially, for reductive endopolyploid divisions, was recently shown for ovarian cancer, which presented giant-cell pathology from endo-replication, not intervened by mitosis [77] . Previously, cancer-cell behavior was compared to life cycles of primitive organisms [78] , which is also mentioned by the authors of the ovarian giant-cell study. They stressed the similarity in the cellular processes to offspring cells from giant-cells and budding yeast, and to amitotic cleavage of polyploid “simple organisms” (see Eudorina, ameba—above). They concluded in unison with earlier scientist’s views and pledges [26] [79] -[81] that giant cells “--- have not attracted much attention in the cancer research community and their roles in tumorigenesis have been largely untested.” This is a sentiment, that should include lower endo-polyploid levels (4n/8C & 8n/16C), which occur in gynecological specimens and in Barrett’ esophagus, recognized to be: “--- beginning/pre-cancerous tissue ---” [22] . Sometimes it is necessary to look back to move forward, considering that diplo-chromosomal-divisions from endo-replication (endomitosis) were demonstrated over a half century ago for mouse ascites-tumor cells [33] .
The above clear-cut connection of endopolyploidy to cancer is not yet available for gonomeric haploidization, but it is almost “there”, because a certain rarer, type of childhood acute lymphoblastic leukemia (ALL) and also rarer chondrosarcomas, proliferate with near-haploid genomes [82] -[85] . The total chromosomal number for one case of ALL-L1 was as low as 26, and strangely, the karyotype showed loss of almost a complete complement (i.e., 26,XX,-1,-2,-3,-4,-5,-6,-7,-8,-9,-10,-11,-12,-13,-15,-16,-17,-18,-19,-20,-22), only X,14 and 21 were in normal disomic condition in these cells. Recent molecular studies determined that this peculiar, non-random loss, (similarly for chondrosarcoma) was a primary event from diploid cells [86] and that this process could be followed by complete or partial genomic duplications to UPDs, resulting in hyperdiploidy with extensive LOH. The most often cited explanations for such loss are a rarer multipolar division or a series of mitoses with chromosome loss from non-disjunction, [or assumed “unstable” regular tetraploidy (92) with multiple chromosomal losses [38] [39] . Neither suggested mechanisms are viable alternatives, to a gonomey-based haploidization process, because they would result in random chromosomal losses. Furthermore, loss by multiple non-disjunctions to haploidy was calculated to be virtually zero for cells with chromosome numbers above 5 - 7 [16] [17] . A rarer multipolar division with random chromosomal distribution can neither be “explained-away” nor can it be proven. Other facts linking the near-haploid leukemia-L1 type to gonomery is that L1 denotes small leukemic cells as compared to the larger L2 type, and also that these cells appear to originate from immature lymphocytes (no Tor B-cell antigens) [83] . These facts also hint to undifferentiated tissue/cells being especially susceptible to gonomery [69] . Although the lymphocyte, leukemias are very rarer, the haploidization process to mat and pat genomes may itself have a higher rate, but hidden in aneuploid-UPD-type hyperdiploidy from genomic duplication [86]
No-where in the literature on cancer-biology has gonomery-based functional divisions with cell volume reduction to 1/2-size compared to normal cells of origin, been considered to play roles in cancer initiation and/or progression. The 1/2-size reduction is in agreement with origin from single territories [13] [74] . Such segregations are not restricted to diploid cells, because preliminary observations indicate that it also occurs for endotetraploid cells (see above). The question is whether endotetraploid gonomery-based segregations back to diploidy would end up as large cells due to tetraploid-associated enlarged cytoplasmic content compared to diploidy? For example, smalland large-cell lung carcinomas are well known, but strangely this cell-size difference is not reflected in different numbers of chromosomes, which often ranges from 43 - 44 to 48 for both cell-types [82] . These cancer-types can occur mixed in the lung which is also a feature in non-Hodgkin’s lymphoma associated with follicular center cells [82] . Thus far, there are no other very likely, explanations for these occurrences except a possible origin from gonomery, which can only be verified by (affordable) SNP-array haplotyping [87] .
7) An in vitro model system for molecular studies of cancer-like traits induced in normal, human cells The inducing system in young cells was carcinogen-free being a nutritional deprivation that has high probability of a natural occurrence in normal somatic cells undergoing replacement growth (e.g., gut and skin). From discussions of Barrett’s esophageal disease, “pathological mitosis” [22] [23] and recent demonstration of ovarian giant cells with genome reductive behavior [77] there is now good evidential material for the conclusion that endopolyploidy can act as initiator of tumorigenesis. The carcinogen-free inducing system is experimentally simple, repeatable, and well proven to change normal, diploid human cells to pre-neoplasia-like GPA [12] -[14] , which next hopefully will be molecularly analyzed for genetic determinants of this GPA. Such knowledge of involved genes, never before available without “contamination” from genomic changes induced by carcinogens, has high probability of significant contributions to a cancer-preventive vaccine-program. The difference from ongoing vaccine-research is that the immunological response would be from healthy immune systems as compared to cancer-victims with poor/destroyed immunological responses. A daring prediction considering very possible gonomery in childhood leukemia, is that the future (soon) will have available a vaccine that will reduce cancer-incidence rate.
The question is now why deficiency of amino acid glutamine with only a day or two exposures to normal cells, caused such dramatic, (undesirable) cellular effects? Earlier several other amino acids in deprivation were also tried on cells with a general response of induced endo-polyploidization to giant cells and chromosomal breakage [88] . But it appears that among all amino acids glutamine is special, because it is a cell-preferred added source for nitrogen and carbon. It fuels glycolysis to pyruvate-kinase, which catalyze the step to energy source, ATP and carbon dioxide, in both normal and cancerous growth [89] . Macro-molecular syntheses to for example, nucleotides for DNA synthesis are dependent on glutamine availability, which has high presence in serum, and is an added growth factor for cell cultures (3 mmol/100). Several studies in pursuit of cellular happenings during poor nutrition, grew normal fibroblast cells under reduced serum content (0.1%) for weeks, followed by revival to mitotic activity in complete medium (10%). Presence-absence of polyploidy was not mentioned, but in one study the recuperating cells showed increased growth-rate, focal dysplastic cell-growth areas, which when introduced into mice produced tumors [90] . Likewise in the “glutamine” studies focal areas with change in cell polarity produced multicellular heaps of cells, and even 3-D tumor-like spheres in extended growth of the endotetraploid primary “transformed” progeny cells [13] [14] [76] . The serum deficient and the glutamine studies complement each other in demonstrating that normal human cells (fibroblasts) can change directly into a cancerous-pathway given the “right” initiating growth-conditions. This judgment is contrary to expectation/prediction of the highly acknowledged “mutation theory” which for cancer development assumes accumulation of mutations in genes leading to GPA [91] . However, at the time of its formulation, it was not known that the human genome shelter evolutionary important, relic genetics for two mechanistic division-systems that directly produce transformed progeny cells.
4. Conclusion
Young, normal, human cells in a carcinogen-free induction system were shown to harbor in their genomes—two different mechanistic reductive division systems, evolutionarily conserved from their establishment in unicellular eukaryotes during primeval evolutionary times. These divisions gave rise to cancer-like cellular characteristics directly to progeny cells such as increased fitness and transformed cell phenotypes. Thus man as a genetic carrier of his cancer fate has the capacity to adopt life-style choices not connected to cancer induction. For instance, life-styles to be avoided are obesity, use of tobacco and “risky” behaviors. However, today there is another rather pressing problem: breast cancer is increasing at alarming rates (1 in 3) with 25% of all such cancers occurring in “young” people, 50 years old and under. This switch from “old to young” has no explanation, except for being caused by a wrong life-style. One untested theory is that present young workers seldom raise their arms much above the computer-level, whereas this was not a couple of generations ago, when the average households were less mechanistic—no washer/dryer and arms did the work. The breast is a secretory organ with the flow of waste products (garbage) from metabolic autophagy (dead cell debris). Arm-stretching (above head) also stretches the breast, which can give push to proper flow for normal discard. Reduced flow could lead to accumulation of the garbage in small, focal areas of the breast, and with likely toxic effect could lead to genomic damage in surrounding normal cells. Importantly, a damaged genome (DNA-breakage) is the basic route for activation of the undesirable, mechanistic, genome reducing systems. “Wellness letters” are stressing “undesirable” consequences from tight braziers, which also could lead to reduce garbage flow-rate. There are no statistics on breast cancer incidence between those that regularly exercise with arm movements and those that do not. Other disturbing facts regarding healthy life-style choices is how to avoid 10 to 20 chemicals in foods, primarily added for increased shelf-life. It is up to the consumer to read labels and be knowledgeable in chemistry, because too often presence of “danger” chemicals is brushed off with “they are within safe limits”. This is a real issue for drinking water, so “plastic” water has taken over, but what does soft plastic give off to the water?—potential cancer-causing petroleum products. But note that glass drinking containers are a choice. Since the present study underscores the importance of diets having adequate amounts of amino acids, the big question is whether today’s different diet craze is neglectful in this regard. Amino acids are “building blocks” for all types of proteins, including nucleotides for normal DNA replication, which is necessary for normal cell divisions. These few pointers for likely reduction of cancer incidence are probably only “the tip of the iceberg”, but would be a big step in the right direction. Further meaningful reduction is speculated to be from a vaccine development against gene products that determine increased fitness of normal human cells. The present simple AAD methodology, free from carcinogen contamination, producing “fast-growing” genome changed cells with experimental ease, which is subjected to molecular technology, could reveal genetic determinants for GPA.
Competing Interest
The author declares that she has no competing interest.
References
- Loewenstein, W.R. (2000) The Touchstone of Life. The Oxford University Press, New York.
- Lynch, M. (2006) The Origin of Eukaryotic Gene Structure. Molecular Biology and Evolution, 23, 450-468. http://dx.doi.org/10.1093/molbev/msj050
- Zimmer, C. (2008) Now: The Rest of the Genome. The New York Times.
- Solari, A.J. (2002) Primitive Forms of Meiosis: The Possible Evolution of Meiosis. Biocell, 26, 1-13.
- Wilkins, A.S. and Holliday, R. (2009) The Evolution of Meiosis from Mitosis. Genetics, 181, 3-12. http://dx.doi.org/10.1534/genetics.108.099762
- Egel, R. and Penny, D. (2008) On the Origin of Meiosis in Eukaryotes: Coevolution of Meiosis and Meiosis from Feeble Beginnings. Genome Dynamics and Stability, 3, 249-288. http://dx.doi.org/10.1007/7050_2007_036
- Bernstein, H. and Bernstein, C. (2010) Evolutionary Origin of Recombination during Meiosis. BioScience, 60, 498-505. http://dx.doi.org/10.1525/bio.2010.60.7.5
- Raikov, I.B. (1982) The Protozoan Nucleus: Morphology and Evolution. Springer Verlag, Vienna and New York.
- Raikov, I.B. (1994) The Diversity of Forms of Mitosis in Protozoa: A Comparative Review. European Journal of Protistology, 30, 253-269. http://dx.doi.org/10.1016/S0932-4739(11)80072-6
- Walen, K.H. (2007) Bipolar Genome Reduction Division of Human Near-Senescent, Polyploid Fibroblast Cells. Cancer Genetics and Cytogenetics, 173, 43-50. http://dx.doi.org/10.1016/j.cancergencyto.2006.09.013
- Walen, K.H. (2007) Origin of Diplochromosomal Polyploidy in Near-Senescent Fibroblast Cultures: Telomeres and Chromosomal Stability (CIN). Cell Biology International, 31, 1447-1455. http://dx.doi.org/10.1016/j.cellbi.2007.06.015
- Walen, K.H. (2012) Genome Reversion Process of Endopolyploidy Confers Chromosome Instability on the Descendent Diploid Cells. Cell Biology International, 36, 137-145. http://dx.doi.org/10.1042/CBI20110052
- Walen, K.H. (2013) Normal Human Cells Acquiring Proliferative Advantage to Hyperplasia-Like Growth-Morphology: Aberrant Progeny Cells Associated with Endopolyploid and Haploid Divisions. Cancer and Clinical Oncology, 2, 1-15.
- Walen, K.H. (2014) Haploidization of Human Diploid Metaphase Cells: Is This Genome Reductive Mechanism Operational in Near-Haploid Leukemia? Journal of Cancer Therapy, 5, 101-114. http://dx.doi.org/10.4236/jct.2014.51013
- Hurst, L.D. and Nurse, P. (1991) A Note on the Evolution of Meiosis. Journal of Theoretical Biology, 150, 561-563. http://dx.doi.org/10.1016/S0022-5193(05)80447-3
- Kondrashov, A.S. (1994) Gradual Origin of Amphimixis by Natural Selection. In: Kirkpatrick, M., Ed., The Evolution of Haploid-Diploid Life Cycles, Vol. 25, 27-51.
- Haig, D. (1993) Alternatives to Meiosis: The Unusual Genetics of Red Algae, Mirosporidia and Others. Journal of Theoretical Biology, 163, 15-31. http://dx.doi.org/10.1006/jtbi.1993.1104
- Walen, K.H. (1965) Spatial Relationships in the Replication of Chromosomal DNA. Genetics, 51, 915-929.
- Kuhn, E.M. and Therman, E. (1986) Cytogenetics of Bloom’s Syndrome. Cancer Genetics and Cytogenetics, 22, 1-18. http://dx.doi.org/10.1016/0165-4608(86)90132-9
- Ohno, S. (1970) Evolution by Gene Duplication. Georg Allen and Unwin, London.
- Wolfe, K.H. (2001) Yesterday’s Polyploids and the Mystery of Diploidization. Nature Reviews Genetics, 2, 333-341. http://dx.doi.org/10.1038/35072009
- Barrett, M.T., Pritchard, D., Palanca-Wessels, C., Anderson, J., Reid, B.J. and Rabinovitch, P.S. (2003) Molecular Phenotype of Spontaneously Arising 4N (G2-tetraploid) Intermediates of Neoplastic Progression in Barrett’s Esophagus. Cancer Research, 63, 4211-4217.
- Steinbeck, R.G. (2004) Dysplasia in View of the Cell Cycle. European Journal of Histochemistry, 48, 203-211.
- Walen, K.H. (2009) Spindle Apparatus Uncoupling in Endo-Tetraploid Asymmetric Division of Stem and Non-Stem Cells. Cell Cycle, 8, 3234-3237. http://dx.doi.org/10.4161/cc.8.19.9570
- Walen, K.H. (2013) Senescence Arrest of Endopolyploid Cells Renders Senescence into One Mechanism for Positive Tumorigenesis. In: Hayat, M.A., Ed., Tumor Dormancy and Cellular Quiescence and Senescence, Vol. 1, Springer, Berlin, 215-226.
- Erenpreisa, J., Salmina, K., Huna, A., Kosmacek, E.A., Cragg, M.S., Ianzini, F. and Anisimov, A. (2011) Polyploid Tumor Cells Elicit Paradiploid Progeny through Depolyploidizing Divisions and Regulated Autophagic Degradation. Cell Biology International, 35, 687-695. http://dx.doi.org/10.1042/CBI20100762
- Brito, D. and Rieder, C.L. (2006) Mitotic Checkpoint Slippage in Humans Occurs via Cyclin B Destruction in the Presence of an Active Checkpoint. Current Biology, 16, 1194-1200. http://dx.doi.org/10.1016/j.cub.2006.04.043
- D’Amato, F. (1989) Polyploidy in Cell Differentiation. Caryologia, 42, 183-211. http://dx.doi.org/10.1080/00087114.1989.10796966
- Lee, H.O., Davidson, J.M. and Duronio, R.J. (2009) Endoreplication: Polyploidy with a Purpose. Genes & Development, 23, 2461-2477. http://dx.doi.org/10.1101/gad.1829209
- Davoli, T. and De Lange, T. (2012) Telomere-Driven Tetraploidization Occurs in Human Cells Undergoing Crisis and Promotes Transformation of Mouse Cells. Cancer Cell, 21, 765-776. http://dx.doi.org/10.1016/j.ccr.2012.03.044
- Fox, D.T. and Duronio, R.J. (2013) Endoreplication and Polyploidy: Insight into Development and Disease. Development, 140, 3-12. http://dx.doi.org/10.1242/dev.080531
- Becak, M.L., Becak, W. and Pereira, A. (2003) Somatic Pairing, Endomitosis and Chromosome Aberration in Snakes (Viperida and Colubridae). Anais da Academia Brasileira de Ciências, 75, 285-300. http://dx.doi.org/10.1590/S0001-37652003000300004
- Levan, A. and Hauschka, T.S. (1953) Endomitotic Reduplication Mechanisms in Ascites Tumors of the Mouse. Journal of the National Cancer Institute, 14, 1-43.
- Nawata, H., Kashino, G., Tano, K., Daino, K., Shimada, Y., Kugoh, H., Oshimura, M. and Watanabe, M. (2011) Dysregulation of Gene Expression in Artificial Human Trisomy Cells of Chromosome 8 Associated with Transformed Cell Phenotypes. PLoS One, 6, Article ID: e25319. http://dx.doi.org/10.1371/journal.pone.0025319
- Davoli, T., Denchi, E.L. and De Lange, T. (2010) Persistent Telomere Damage Induces Bypass of Mitosis and Tetraploidy. Cell, 141, 81-93. http://dx.doi.org/10.1016/j.cell.2010.01.031
- Cleveland, L.R. (1947) The Origin and Evolution of Meiosis. Science, 105, 287-289. http://dx.doi.org/10.1126/science.105.2724.287
- Cleveland, L.R. (1956) Brief Accounts of the Sexual Cycles of the Flagellates of Cryptoserus. Journal of Protozoology, 3, 161-180. http://dx.doi.org/10.1111/j.1550-7408.1956.tb02452.x
- Storckova, Z. and Pellman, D. (2004) From Polyploidy to Aneuploidy, Genomic Instability and Cancer. Nature Reviews Molecular Cell Biology, 5, 45-54. http://dx.doi.org/10.1038/nrm1276
- Storchova, Z. and Kuffer, C. (2008) The Consequences of Tetraploidy. Journal of Cell Science, 121, 3859-3866. http://dx.doi.org/10.1242/jcs.039537
- Schvartzman, J.M., Sotillo, R. and Benezra, R. (2010) Mitotic Chromosomal Instability and Cancer: Mouse Modeling of the Human Disease. Nature Reviews Cancer, 10, 102-115. http://dx.doi.org/10.1038/nrc2781
- Ravid, K., Lu, J., Zimmet, J.M. and Jones, M.R. (2002) Roads to Polyploidy: The Megakaryocyte Example. Journal of Cellular Physiology, 190, 7-20. http://dx.doi.org/10.1002/jcp.10035
- Margulis, L., Enzien, M. and McKhann, H.I. (1990) Revival of Dobell’s “Chromidia” Hypothesis: Chromatin Bodies in Amoebomastigote Paratetramitus jugosus. Biological Bulletin, 178, 300-304. http://dx.doi.org/10.2307/1541832
- Edgar, B.A. and Orr-Weaver, T.I. (2001) Endoreplication Cell Cycles More for Less. Cell, 105, 297-306. http://dx.doi.org/10.1016/S0092-8674(01)00334-8
- Ganem, N.J. and Pellman, D. (2012) Linking Abnormal Mitosis to the Acquisition of DNA Damage. Journal of Cell Biology, 199, 871-881. http://dx.doi.org/10.1083/jcb.201210040
- Gondek, L.P., Tiu, R., O’Keefe, L., Sekeres, M.A., Theil, K.S. and Maciejewski, J.P. (2008) Chromosomal Lesions and Uniparental Disomy Detected by SNP Arrays in MDS, MDS/MPD and MDS Derived AML. Blood, 111, 1534-1542. http://dx.doi.org/10.1182/blood-2007-05-092304
- Nielaender, I., Martin-Subero, J.I., Wagner, F., Martinez-Climent, J.A. and Siebert, R. (2006) Partial Uniparental Disomy: A Recurrent Genetic Mechanism Alternative to Chromosomal Deletion in Malignant Lymphoma. Leukemia, 20, 904-905. http://dx.doi.org/10.1038/sj.leu.2404173
- Mollinedo, F. and Gajate, C. (2003) Microtubules, Microtubule-Interfering Agents and Apoptosis. Apoptosis, 8, 413-450. http://dx.doi.org/10.1023/A:1025513106330
- Tautvydas, K.J. (1976) Evidence for Chromosome Endoreduplication in Eudorina californica, a Colonic Alga. Differentiation, 5, 35-42. http://dx.doi.org/10.1111/j.1432-0436.1976.tb00889.x
- Enzien, M., McKhann, H.I. and Margulis, L. (1989) Ecology and Life History of an Amoebomastigote, Paratetramitus jugosus, from a Microbial Mat: New Evidence for Multiple Fission. Biological Bulletin, 177, 110-129. http://dx.doi.org/10.2307/1541839
- Walen, K.H. (2002) The Origin of Transformed Cells: Studies of Spontaneous and Induced Cell Transformation in Cell Cultures from Marsupials, a Snail and Human Amniocytes. Cancer Genetics and Cytogenetics, 133, 45-54. http://dx.doi.org/10.1016/S0165-4608(01)00572-6
- Walen, K.H. (2010) Mitosis Is Not the Only Distributor of Mutated Cells: Non-Mitotic Endopolyploid Cells Produce Reproductive Genome Reduced Cells. Cell Biology International, 34, 867-872. http://dx.doi.org/10.1042/CBI20090502
- Grell, K.G. and Ruthmann, A. (1964) Uber die Karyologie des Radiolars Aulachanta scolymantha und Feinstruktur seiner Chromosomen. Chromosoma, 15, 185-211. http://dx.doi.org/10.1007/BF00285729
- Saunders, W.S., Shuster, M., Huang, X., Gharaibe, B., Enyenihi, A.H., Petersen, J. and Gollin, S.M. (2000) Chromosomal Instability and Cytoskeleton Defects in Oral Cancer. Proceedings of the National Academy of Sciences of the United States of America, 97, 303-308. http://dx.doi.org/10.1073/pnas.97.1.303
- Gonzalez-Robles, A., Cristobal-Ramos, A.R., Gonzalez-Lazaro, M., Omana-Molina, M. and Martinez-Palomo, A. (2009) Naegleria fowleri: Light and Electron Microscopy Study of Mitosis. Experimental Parasitology, 122, 212-217. http://dx.doi.org/10.1016/j.exppara.2009.03.016
- Brenner, S., Branch, A., Meredith, S. and Berns, M.W. (1977) The Absence of Centrioles from Spindle Poles of Rat Kangaroo PtK1 Cells Undergoing Meiotic-Like Reduction Division in Vitro. Journal of Cell Biology, 72, 368-379. http://dx.doi.org/10.1083/jcb.72.2.368
- Sherratt, D.J. (2003) Bacterial Chromosome Dynamics. Science, 301, 780-785. http://dx.doi.org/10.1126/science.1084780
- Castagnetti, S., Oliferenko, S. and Nurse, P. (2010) Fission Yeast Cells Undergo Nuclear Division in the Absence of Spindle Microtubules. PLoS Biology, 8, Article ID: e1000512. http://dx.doi.org/10.1371/journal.pbio.1000512
- Travis, J. (2007) Return of the Matrix. Science, 318, 1400-1401. http://dx.doi.org/10.1126/science.318.5855.1400
- Johansen, K.M., Forer, A., Yao, C., Girton, J. and Johansen, J. (2011) Do Nuclear Envelope and Intranuclear Proteins Reorganize during Mitosis to form an Elastic, Hydrogel-Like Spindle Matrix? Chromosome Research, 19, 345-365. http://dx.doi.org/10.1007/s10577-011-9187-6
- Swanson, C.P. (1957) Cytology and Cytogenetics. Prentice-Hall, Inc., Englewood Cliffs, NJ, 526-532.
- Rach, E.M. and Wyngaard, G.A. (2008) Gonomery and Chromatin Diminution in Mesocyclops longisetus (Copepoda). Journal of Crustacean Biology, 28, 180-184. http://dx.doi.org/10.1651/07-2847R.1
- Leeb, M., Walker, R., Mansfield, B., Nichols, J., Smith, A. and Wutz, A. (2012) Germline Potential of Parthenogenic Haploid Mouse Embryonic Stem Cells. Development, 139, 3301-3305. http://dx.doi.org/10.1242/dev.083675
- Sarto, G.E., Stubblefield, P.A., Lurain, J. and Therman, E. (1984) Mechanisms of Growth in Hydatidiform Moles. American Journal of Obstetrics & Gynecology, 148, 1014-1023. http://dx.doi.org/10.1016/0002-9378(84)90545-3
- Kukita, Y., Miyatake, K., Stokowski, R., Hinds, D., Higasa, N., Wake, N., et al. (2013) Genome-Wide Definitive Haplotypes Determined Using a Collection of Complete Hydatidiform Moles. Genome Research, 15, 1511-1518. http://dx.doi.org/10.1101/gr.4371105
- Gerstein, A.C., Chun, H.J.E., Grant, A. and Otto, S.P. (2006) Genomic Convergence toward Diploidy in Saccharomyces cerevisiae. PLoS Genetics, 2, 1396-1401.
- Alabrudzinska, M., Skoneczny, M. and Skoneczna, A. (2011) Diploid-Specific Genome Stability Genes of S. cerevisiae: Genomic Screen Reveals Haploidization as an Escape from Persisting DNA Rearrangement Stress. PLoS One, 6, Article ID: e21124. http://dx.doi.org/10.1371/annotation/77daccf9-9976-4d0e-b666-35a900cb2d17
- Nagele, R.G., Freeman, T., McMorrow, L. and Lee, H.Y. (1995) Precise Spatial Positioning of Chromosomes during Prometaphase: Evidence for Chromosomal Order. Science, 270, 1831-1835. http://dx.doi.org/10.1126/science.270.5243.1831
- Bolzer, A., Kreth, G., Solovei, I., Koehler, D., Saracoglu, K., Fauth, C., et al. (2005) Three-Dimensional Maps of All Chromosomes in Human Male Fibroblast Nuclei and Prometaphase Rosettes. PLoS Biology, 3, 826-842.
- Mayer, W., Smith, A., Fundele, R. and Haaf, T. (2000) Spatial Separation of Parental Genomes in Preimplantation Mouse Embryos. Journal of Cell Biology, 148, 629-634. http://dx.doi.org/10.1083/jcb.148.4.629
- Costello, D.P. (1970) Identical Linear Order of Chromosomes in both Gametes of the Acoel Tubularian Polychoerus carmelensis: A Preliminary Note. Proceedings of the National Academy of Sciences of the United States of America, 67, 1951-1958. http://dx.doi.org/10.1073/pnas.67.4.1951
- Stern, C. (1958) The Nucleus and Somatic Cell Variation. Journal of Cellular and Comparative Physiology, 52, 1-34. http://dx.doi.org/10.1002/jcp.1030520404
- Huskins, C.L. and Cheng, K.C. (1950) Segregation and Reduction in Somatic Tissues. IV. Reductional Grouping Induced in Alliun cepa by Low Temperature. Journal of Heredity, 14, 13-18.
- Glass, E. (1957) Das Problem der Genomsonderung in den Mitosen unbehandelter Rattenlebern. Chromosoma, 8, 468-492. http://dx.doi.org/10.1007/BF01259515
- Glazko, T.T. (2004) Chromosome Subdividing to Haploid Sets in Diploid Metaphase Plates of Some Mammalian Species. In: Proceedings of 15th International Chromosome Conference, London, 5-10 September 2004, 63.
- Straight, A.F., Marshall, W.F., Sedat, J.W. and Murray, A.W. (1997) Mitosis in Living Budding Yeast: Anaphase A but No Metaphase. Science, 277, 574-578. http://dx.doi.org/10.1126/science.277.5325.574
- Walen KH. (2011) Normal Human Cell Conversion to 3-D Cancer-Like Growth: Genome Damage, Endopolyploidy, Senescence Escape, and Cell Polarity Change/Loss. Journal of Cancer Therapy, 2, 181-189. http://dx.doi.org/10.4236/jct.2011.22023
- Zhang, S., Mercado-Uribe, I., Xing, Z., Sun, B., Kuang, J. and Liu, J. (2013) Generation of Cancer-Stem-Like Cells through the Formation of Polyploid Giant Cells. Oncogene, 33, 116-128.
- Renpreisa, J. and Cragg, M.S. (2007) Cancer: A Matter of Life Cycles. Cell Biology International, 31, 1507-1510. http://dx.doi.org/10.1016/j.cellbi.2007.08.013
- Wheatley, D.N. (2008) Growing Evidence of Repopulation of Regressed Tumors by the Division of Giant Cells. Cell Biology International, 32, 1029-1030. http://dx.doi.org/10.1016/j.cellbi.2008.06.001
- Shackney, S.E. and Shanky, T.V. (1995) Genetic and Phenotypic Heterogeneity of Human Malignancies: Finding Order in Chaos. Cytometry, 21, 2-5. http://dx.doi.org/10.1002/cyto.990210103
- Puig, P.E., Guilly, M.N., Bouchot, A., Droin, N., Cathelin, D., Bouyer, F., et al. (2008) Tumor Cell Can Escape DNA-Damaging Sisplatin through DNA Endoreduplication and Reversible Polyploidy. Cell Biology International, 32, 1031-1043. http://dx.doi.org/10.1016/j.cellbi.2008.04.021
- Mitelman, F. (1988) Catalog of Chromosome Aberrations in Cancer. Alan Liss, Inc., New York.
- Heim, S. and Mitelman, F. (1995) Cancer Cytogenetics: Chromosomal and Molecular Genetic Aberrations of Tumor Cells. 2nd Edition, Wiley-Liss, Inc., New York.
- Mandahl, N., Johansson, B., Mertens, F. and Mitelman, F. (2012) Disease-Associated Patterns of Disomic Chromosomes in Hyperhaploid Neoplasms. Genes, Chromosomes and Cancer, 51, 536-544. http://dx.doi.org/10.1002/gcc.21947
- Olsson, L., Paulsson, K., Bovee, J.V. and Nord, K.H. (2011) Correction: Clonal Evolution through Loss of Chromosomes and Subsequent Polyploidization in Chondrosarcoma. PLoS ONE, 6, 1-7. http://dx.doi.org/10.1371/annotation/8f845569-8244-416b-b15e-89562177ce32
- Safavi, S., Forestier, E., Golovleva, I., Barbany, G., Nord, K.H., Moorman, A.V., Harrison, C.J., Johansson, B. and Paulsson, K. (2013) Loss of Chromosomes Is the Primary Event in Near-Haploid and Low-Hypodiploid Acute Lymphoblastic Leukemia. Leukemia, 27, 248-250. http://dx.doi.org/10.1038/leu.2012.227
- Peters, B.A., Kermani, B.G., Sparks, A.B., Alferov, O., Alexeev, A., Jiang, Y., et al. (2012) Accurate Whole-Genomic Sequencing and Haplotyping from 10 to 20 Human Cells. Nature, 487, 190-195.
- Freed, J.J. and Schatz, S.A. (1969) Chromosome Aberrations in Cultured Cells Deprived of Single Essential Amino Acids. Experimental Cell Research, 55, 393-409. http://dx.doi.org/10.1016/0014-4827(69)90574-6
- Deberardinis, R.J. and Cheng, T. (2010) Q’s Next: The Diverse Function of Glutamine in Metabolism, Cell Biology and Cancer. Oncogene, 29, 313-324. http://dx.doi.org/10.1038/onc.2009.358
- Zhang, J., Wang, X., Zhao, Y., Chen, B., Suo, G. and Dai, J. (2006) Neoplastic Transformation of Human Diploid Fibroblasts after Long-Term Serum Starvation. Cancer Letters, 243, 101-108. http://dx.doi.org/10.1016/j.canlet.2005.11.022
- Lengauer, C., Kinzler, K.W. and Vogelstein, B. (1998) Genetic Instability in Human Cancers. Nature, 396, 643-649. http://dx.doi.org/10.1038/25292
List of Abbreviations
ML-1,2: meiotic-like first and second divisions GPA: gain of proliferative advantage LOH: loss of heterozygosity UPD: uniparental disomy 3D: three dimensional growth SNP: single nucleotide polymorphism

