Journal of Cancer Therapy
Vol.4 No.8A(2013), Article ID:36108,7 pages DOI:10.4236/jct.2013.48A005
Effects of ADH1C, ALDH2, and CYP2A6 Polymorphisms on Individual Risk of Tobacco-Related Lung Cancer in Male Japanese Smokers*
![]()
1Showa Pharmaceutical University, Machida, Tokyo, Japan, 2Mitsui Memorial Hospital, Chiyoda-ku, Tokyo, Japan; 3Hokkaido University, Sapporo, Japan.
Email: #hyamazak@ac.shoyaku.ac.jp
Copyright © 2013 Makiko Shimizu et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received June 25th, 2013; revised July 26th, 2013; accepted August 3rd, 2013
Keywords: Alcohol Dehydrogenase 1C; Aldehyde Dehydrogenase 2; Cytochrome P450 2A6; Lung Cancer; Tobacco
ABSTRACT
Recent genome-wide association studies have identified several lung cancer susceptibility loci. We previously carried out a replication study in male Japanese smokers that focused on chromosome 5p15 (telomerase reverse transcriptase) and 3q28 (tumor protein p63) (Shimizu et al., Journal of Cancer Therapy, Vol. 2, No. 5, 2011, pp. 690-696). The current study was performed to confirm the association of traditional susceptibility loci [i.e., alcohol dehydrogenase 1C (ADH1C) and aldehyde dehydrogenase 2 (ALDH2)] in 1039 male Japanese smokers (573 lung cancer patients and 466 healthy control subjects) who were previously enrolled in a study to investigate the low odds ratio for lung cancer risk associated with functionally impaired and deletion polymorphisms in cytochrome P450 2A6 (CYP2A6). The minor allele frequency of rs671 in ALDH2 (0.304) was significantly higher in lung cancer cases than in controls (0.226), with an odds ratio of 1.42 [95% confidence interval (CI) of 1.12 - 1.80, p = 0.0033]. No significant association of rs698 in ADH1C with lung cancer risk was found in this population of male Japanese smokers. For light smokers categorized according to the 50th percentile Brinkman index value among the control subjects (620 daily cigarettes × years) and for the CYP2A6*1 wild-type non-carrier sub-population, significantly high odds ratios of 1.98 and 1.68 (95% CI of 1.28 - 3.06, p = 0.0022, and 1.07 - 2.66, p = 0.025), respectively, were observed for rs671 in ALDH2. The present results support the association of ALDH2 loci with lung cancers and suggest a specific effect of ALDH2 loci resulting in a higher risk of lung cancer in light smokers. CYP2A6 polymorphisms, including copy number polymorphisms, may lower the risk of heavy tobacco use-related lung cancer.
1. Introduction
Cigarette smoking and alcohol consumption affect susceptibility to etiologically complex diseases, including various tumors [1,2]. Polymorphisms of the alcohol-metabolizing enzymes alcohol dehydrogenase 1C (ADH1C) and aldehyde dehydrogenase 2 (ALDH2) influence the risks of various cancers such as those of the esophagus [3], breast [4], pancreas [5], stomach [6], mouth [7], lung [8], and liver [1]. The International Lung Cancer Consortium was established in 2004 and coordinates genotyping activities and ongoing genome-wide association studies (GWAS) [9]. Recent association findings for smokingrelated diseases implicate genetically derived individual differences [10-14]. Several reports from GWAS with respect to lung cancer risk in different ethnic populations have suggested a variety of susceptibility genes, such as telomerase reverse transcriptase (TERT, rs2736100) [15-17] and tumor protein p63 (TP63, rs4488809, rs9816619, or rs10937405) [17,18]. In one of our previous reports [19], we demonstrated that genetic polymorphisms in cytochrome P450 2A6 (CYP2A6) are determinants of smoking behavior and tobacco-related lung cancer risk, particularly for squamous cell and small cell carcinoma, which are known to be associated with cigarette smoking [13] in male Japanese heavy smokers. In addition, significant associations of rs4488809 (TP63) and rs2736100 (TERT) with the risk of lung adenocarcinoma were identified [20]. Some of the single nucleotide polymorphisms (SNPs) found by GWAS have been investigated extensively; however, there have been few reports of the effects of SNPs in combination in a typical population.
We performed further study of the traditional candidate genes for lung cancer risk (i.e., ADH1C and ALDH2) in a population of male Japanese smokers. Herein, we confirm the increased susceptibility to lung cancers associated with ALDH2 SNPs, especially for light smokers. Also, CYP2A6 polymorphisms, including copy number polymorphisms that were not fully studied in GWAS (except for one computer-based special analytical study [18]), were found to be a major influence reducing the risk of heavy tobacco use-related lung cancer.
2. Materials and Methods
2.1. Subjects
This study was approved by the ethics committees of Hokkaido University and Showa Pharmaceutical University. The sample population comprised 1039 unrelated male Japanese smokers (573 case and 466 control subjects) selected from the 1705 participants of our previous study [19,20]. The other 666 subjects could not be included in this study because of genomic DNA sample limitations; there were apparently no differences in demographic factors such as ages and smoking status between case and control groups after exclusion of these subjects. The patient group consisted of 573 men who had received a pathological diagnosis of lung cancer [squamous cell or small cell carcinoma (n = 285) or adenocarcinoma (n = 288)] with a mean (±SD) age of 63.4 ± 9.3 years (range 29 - 86 years). The control group consisted of 466 male smokers with a mean age of 52.2 ± 12.2 years (range 20 - 92 years) without a history of cancer. The age of the lung cancer patients was defined at the time of pathological diagnosis of lung cancer.
Smokers included current and ex-smokers and were defined as individuals who had ever smoked cigarettes with a minimum smoking history of 10 cigarettes per day for at least 1 year. Light and heavy smokers were categorized by the 50th percentile Brinkman index value (daily cigarettes × years) among control subjects and was found to be less than 620 (n = 233) and more than or equal to 620 (n = 233), respectively. Pathological classification of lung cancers was determined by at least three pathologists according to the criteria described in the literature [19].
2.2. Genotyping
Genomic DNA was prepared from peripheral lymphocytes [19]. Genotyping of ADH1C Ile350Val (rs698) [21] and ALDH2 Glu504Lys (rs671) [3] polymorphisms was carried out according to previously described methods. Genotyping of CYP2A6 was carried out by the methods described previously [19,20] with respect to rs5031016 for CYP2A6*7 and rs28399433 for CYP2A6*9 and the wholegene deletion polymorphism of CYP2A6 (CYP2A6*4). CYP2A6 variants were major alleles in control.
2.3. Statistical Analyses
The associations between the genotype distributions and patient status were assessed by odds ratios and 95% confidence intervals (CIs) that were calculated by unconditional logistic regression adjusting for age and cigarette smoking (Brinkman index value), unless otherwise mentioned. A p value less than 0.05 was considered to be statistically significant. Statistical computations were carried out using the statistical software SAS, version 5.1 (SAS Institute, Inc., Cary, NC) or PLINK 1.07 [22].
3. Results
Table 1 shows the association results of polymorphisms in three genes analyzed in terms of lung cancer risk in a
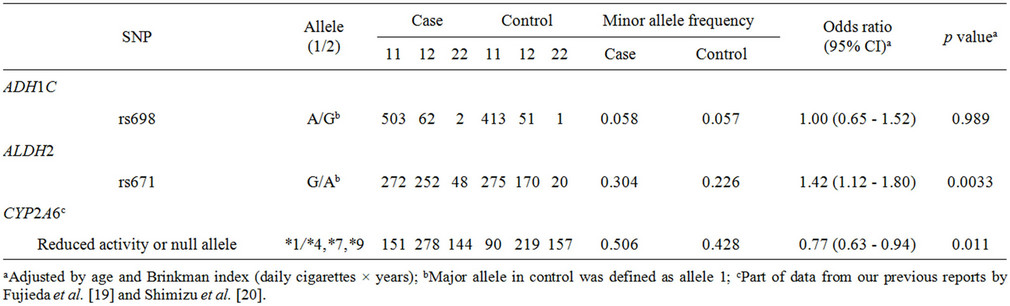
Table 1. Association of polymorphisms of ADH1C, ALDH2, and CYP2A6 with lung cancer risk.
population of 1039 male Japanese smokers. The minor allele frequencies of ADH1C SNPs were not significantly different between case and control groups (p ≥ 0.05). The SNP rs671 in ALDH2 was significantly associated with lung cancer risk (odds ratio of 1.42, 95%CI of 1.12 - 1.80, p = 0.0033). The odds ratio for subjects carrying CYP2A6 variants was calculated to be 0.77 (95%CI of 0.63 - 0.94, p = 0.011), suggesting a decreased risk of tobacco-related lung cancer in subjects with variants of CYP2A6, a finding previously seen in expected phenotype groups estimated from CYP2A6 gene analyses [19,20].
Subgroup analysis according to histological cancer type classified the lung cancer cases into two groups: adenocarcinoma cases and squamous cell or small cell carcinoma cases (Table 2). High odds ratios of 1.40 and 1.41 (95%CI of 1.07 - 1.83, p = 0.014; 95%CI of 1.05 - 1.88, p = 0.021) were observed for the rs671 A allele in ALDH2 in the adenocarcinoma subgroup and the squamous cell or small cell carcinoma subgroup, respectively. In contrast, a lower odds ratio of 0.72 (95% CI of 0.56 - 0.92, p = 0.0099) was obtained for CYP2A6 variants in the squamous cell or small cell carcinoma subgroup than that of 0.79 (95% CI of 0.63 - 0.99, p = 0.040) in the adenocarcinoma group.
The effects of genetic polymorphisms were analyzed in terms of smoking status subgroups (Table 3). Light smokers and heavy smokers were categorized according to the 50th percentile Brinkman index value among the control subjects (620 daily cigarettes × years). For light smokers, a significantly high odds ratio of 1.98 (95%CI of 1.28 - 3.06, p = 0.0022) was observed for rs671 in ALDH2, whereas no significant association was observed for heavy smokers (odds ratio of 1.24, p = 0.130). On the other hand, a significantly lower odds ratio of 0.74 (95%CI of 0.58 - 0.94, p = 0.014) was obtained for CYP2A6 variants in heavy smokers.
Consequently, a further subgroup analysis was performed (Table 4) to understand the different contri- butions of polymorphisms of ADH1C, ALDH2, and CYP2A6 to odds ratios in tobacco use-related lung cancer risk. In patients carrying no CYP2A6*1 wild-type alleles, the highest odds ratio of 1.68 (95%CI of 1.07 - 2.66, p = 0.025) was observed for rs671 in ALDH2. The low but significant odds ratio of 1.32 (95%CI of 1.01 - 1.74, p = 0.049) was obtained in subjects harboring the CYP2A6*1 wild-type allele.
4. Discussion
In the analysis of all subjects included in this study, rs671 in ALDH2 was significantly associated with lung cancer risk (p = 0.0033), as shown in Table 1. A similar influence of ALDH2 genetic polymorphism on the risk of lung cancer has been reported in Japanese [23] and Korean [24] populations. In the cancer type subgroup analysis shown in Table 2, the association of this SNP was detected in both the adenocarcinoma group and the squamous cell and small cell carcinoma group, with similar odds ratios of 1.40 (p = 0.014) and 1.41 (p = 0.021), respectively. These results suggest that ALDH2 polymorphisms are associated with lung cancer risk, irrespective of the histological cancer type. Furthermore, in the subgroup analyses shown in Tables 3 and 4, rs671 in ALDH2 was associated with lung cancer risk for light smokers (but not for heavy smokers) (Table 3) and for non-wild-type CYP2A6 genotypes (associated with a low cigarette consumption phenotype) [19] (Table 4) with similar odds ratios of 1.98 (p = 0.0022) and 1.68 (p = 0.025), respectively.
A recent meta-analysis has suggested that the ADH1C
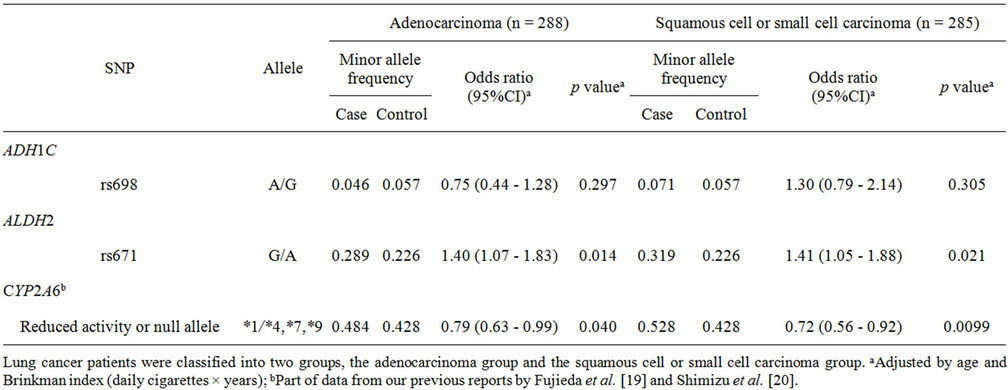
Table 2. Association of polymorphisms of ADH1C, ALDH2, and CYP2A6 with lung cancer risk: subgroup analysis of histological type of lung cancer.
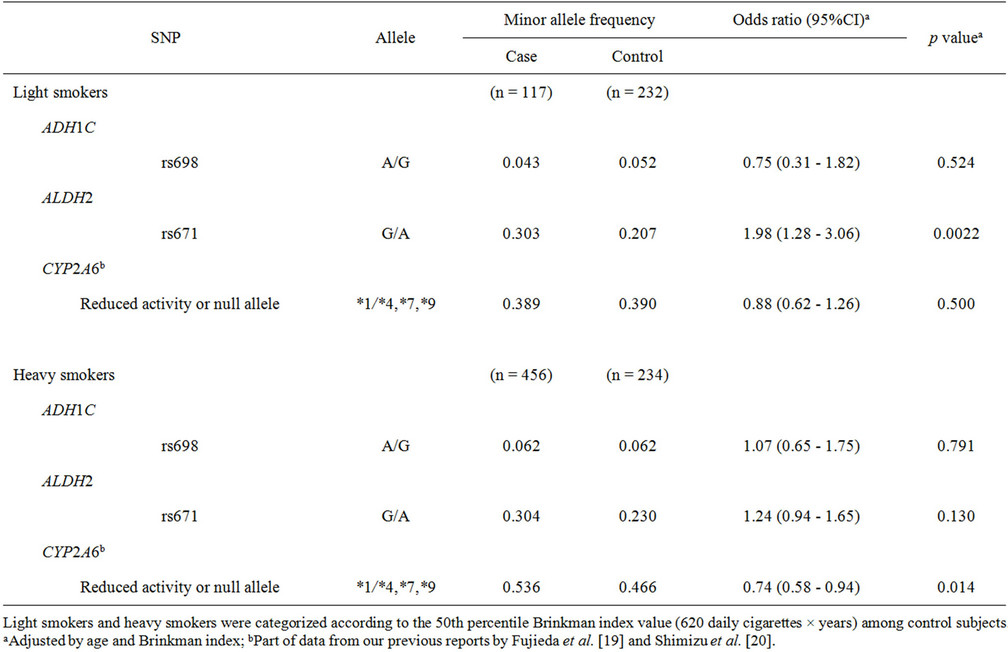
Table 3. Association of polymorphisms of ADH1C, ALDH2, and CYP2A6 with lung cancer risk: subgroup analysis of smoking status.
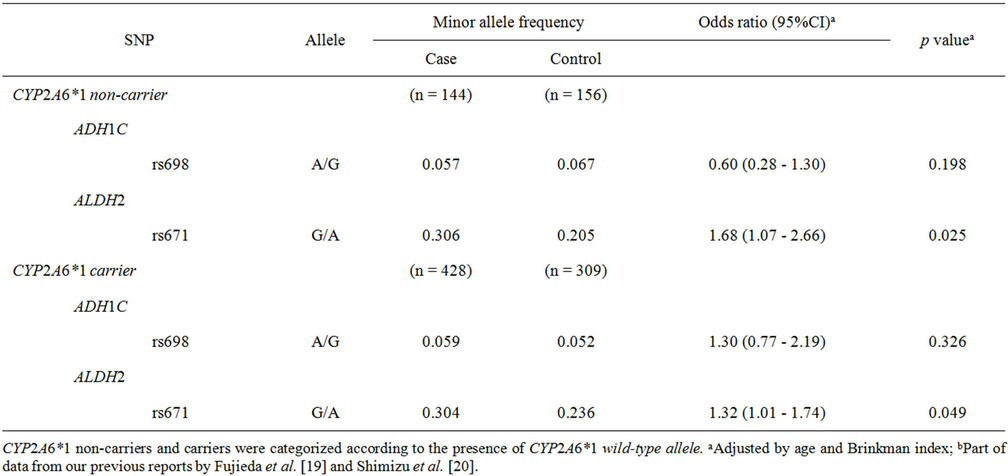
Table 4. Association of polymorphisms of ADH1C, ALDH2, and CYP2A6 with lung cancer risk: subgroup analysis of CYP2A6 genotypes.
rs698 polymorphism may contribute to cancer risk among Africans and Asians [25]. In this study, however, the association of this variant with lung cancer risk was not replicated (Table 1). No association of this SNP with esophageal cancer risk in a Chinese population has also been reported [26]. The cause of these different findings is probably the extremely low frequency of this SNP in the Asian population, as has been reported in a European cohort [6].
Another meta-analysis (which included our previous study [19]) suggested that CYP2A6*4 is associated with susceptibility to lung cancer in Asians [27]. The ethnic differences in CYP2A6 allele frequencies between Caucasians and Japanese should be also considered: lower frequencies of CYP2A6*9, CYP2A6*4 (rare) and CYP2A6*7 (never detected) are evident in Caucasians, whereas these alleles are relatively common in Asians (from about 15% to 20% for each allele) [19]. The whole-gene deletion of CYP2A6 may decrease the risk of lung cancer in Asians [19,27]. In the analysis of all subjects in the current study, CYP2A6 variants were significantly associated with a decrease in lung cancer risk (Table 1). As we previously reported [19,20], CYP2A6 seems to be associated with smoking-dependent lung cancer risk (Tables 3 and 4). We recently reported the different effects of TERT, TP63, and CYP2A6 polymorphisms [20] on associations of individual lung cancer risk in a population of male Japanese smokers who were previously studied for lung cancer risk [19].
Overall, the different roles of ALDH2 and CYP2A6 variants were highlighted in subgroups indicating smoking status in male Japanese smokers. Taken together with the candidate genes identified in GWAS, lung cancer risk in Asian smokers may be accounted for by individual smoking status and multiple gene variations such as those in TERT, TP63, and CYP2A6, as reported previously [20], and those in ALDH2 and CYP2A6 as demonstrated in this study. Individual lung cancer risk and the details of chemical metabolism or disposition will be difficult to elucidate [28], but the following mechanisms are most likely involved. Carcinogenic nicotine-derived N-nitrosamines are probably genotoxically activated by polymorphic CYP2A6 [29], and reactive oxygen species in tobacco smoke may contribute to lung carcinogenesis. Detection of lipid peroxidation-induced DNA adducts have been reported in human autopsy tissues [30]. Severe systemic acetaldehydemia may cause multicentric field cancerization [3], because acetaldehyde has been also detected in mainstream cigarette smoke [31].
In conclusion, we clarified that the previously established association of ALDH2 with lung cancer risk was replicated in the current case–control study of an unrelated population of male Japanese smokers for all participants and for all subgroup analyses, including a high odds ratio for the light-smoker subgroup. In addition, our findings suggest that genetic variation in CYP2A6 may be a major influence on heavy tobacco use-related lung cancer risk in male Japanese smokers.
5. Acknowledgements
This work was supported in part by the Ministry of Education, Culture, Sports, Science and Technology of Japan and by an SRF Grant for Biomedical Research in Japan. The authors thank Drs. Masaki Fujieda, Kazuma Kiyotani, Hirotoshi Dosaka-Akita, Yuichi Sawamura, Jun Yokota, Asaki Kase, Ren Sawaya, and Norie Murayama for their support.
REFERENCES
- M. Munaka, K. Kohshi, T. Kawamoto, S. Takasawa, N. Nagata, H. Itoh, et al., “Genetic Polymorphisms of Tobaccoand Alcohol-Related Metabolizing Enzymes and the Risk of Hepatocellular Carcinoma,” Journal of Cancer Research and Clinical Oncology, Vol. 129, No. 6, 2003, pp. 355-360. doi:10.1007/s00432-003-0439-5
- J. D. McKay, T. Truong, V. Gaborieau, A. Chabrier, S. C. Chuang, G. Byrnes, et al., “A Genome-Wide Association Study of Upper Aerodigestive Tract Cancers Conducted within the INHANCE Consortium,” PLoS Genetics, Vol. 7, No. 3, 2011, Article ID: e1001333. doi:10.1371/journal.pgen.1001333
- A. Yokoyama, T. Muramatsu, T. Ohmori, T. Yokoyama, K. Okuyama, H. Takahashi, et al., “Alcohol-Related Cancers and Aldehyde Dehydrogenase-2 in Japanese Alcoholics,” Carcinogenesis, Vol. 19, No. 8, 1998, pp. 1383- 1387. doi:10.1093/carcin/19.8.1383
- J. Y. Choi, J. Abel, T. Neuhaus, Y. Ko, V. Harth, N. Hamajima, et al., “Role of Alcohol and Genetic Polymorphisms of CYP2E1 and ALDH2 in Breast Cancer Development,” Pharmacogenetics, Vol. 13, No. 2, 2003, pp. 67-72. doi:10.1097/00008571-200302000-00002
- J. Kanda, K. Matsuo, T. Suzuki, T. Kawase, A. Hiraki, M. Watanabe, et al., “Impact of Alcohol Consumption with Polymorphisms in Alcohol-Metabolizing Enzymes on Pancreatic Cancer Risk in Japanese,” Cancer Science, Vol. 100, No. 2, 2009, pp. 296-302. doi:10.1111/j.1349-7006.2008.01044.x
- E. J. Duell, N. Sala, N. Travier, X. Munoz, M. C. Boutron-Ruault, F. Clavel-Chapelon, et al., “Genetic Variation in Alcohol Dehydrogenase (ADH1A, ADH1B, ADH1C, ADH7) and Aldehyde Dehydrogenase (ALDH2), Alcohol Consumption and Gastric Cancer Risk in the European Prospective Investigation into Cancer and Nutrition (EPIC) Cohort,” Carcinogenesis, Vol. 33, No. 2, 2012, pp. 361- 367. doi:10.1093/carcin/bgr285
- T. Asakage, A. Yokoyama, T. Haneda, M. Yamazaki, M. Muto, T. Yokoyama, et al., “Genetic Polymorphisms of Alcohol and Aldehyde Dehydrogenases, and Drinking, Smoking and Diet in Japanese Men with Oral and Pharyngeal Squamous Cell Carcinoma,” Carcinogenesis, Vol. 28, No. 4, 2007, pp. 865-874. doi:10.1093/carcin/bgl206
- S. Zienolddiny, D. Campa, H. Lind, D. Ryberg, V. Skaug, L. B. Stangeland, et al., “A Comprehensive Analysis of Phase I and Phase II Metabolism Gene Polymorphisms and Risk of Non-Small Cell Lung Cancer in Smokers,” Carcinogenesis, Vol. 29, No. 6, 2008, pp. 1164-1169. doi:10.1093/carcin/bgn020
- R. J. Hung, D. C. Christiani, A. Risch, O. Popanda, A. Haugen, S. Zienolddiny, et al., “International Lung Cancer Consortium: Pooled Analysis of Sequence Variants in DNA Repair and Cell Cycle Pathways,” Cancer Epidemiology, Biomarkers & Prevention, Vol. 17, 2008, pp. 3081-3089. doi:10.1158/1055-9965.EPI-08-0411
- The Tobacco and Genetic Consortium, “Genome-Wide Meta-Analyses Identify Multiple Loci Associated with Smoking Behavior,” Nature Genetics, Vol. 42, 2010, pp. 441-447. doi:10.1038/ng.571
- J. Z. Liu, F. Tozzi, D. M. Waterworth, S. G. Pillai, P. Muglia, L. Middleton, et al., “Meta-Analysis and Imputation Refines the Association of 15q25 with Smoking Quantity,” Nature Genetics, Vol. 42, 2010, pp. 436-440. doi:10.1038/ng.572
- N. L. Saccone, R. C. Culverhouse, T. H. Schwantes-An, D. S. Cannon, X. Chen, S. Cichon, et al., “Multiple Independent Loci at Chromosome 15q25.1 Affect Smoking Quantity: A Meta-Analysis and Comparison with Lung Cancer and COPD,” PLoS Genetics, Vol. 6, No. 8, 2010, Article ID: e1001053. doi:10.1371/journal.pgen.1001053
- E. D. Pleasance, P. J. Stephens, S. O’Meara, D. J. McBride, A. Meynert, D. Jones, et al., “A Small-Cell Lung Cancer Genome with Complex Signatures of Tobacco Exposure,” Nature, Vol. 463, 2009, pp. 184-190. doi:10.1038/nature08629
- T. Truong, R. J. Hung, C. I. Amos, X. Wu, H. Bickeboller, A. Rosenberger, et al., “Replication of Lung Cancer Susceptibility Loci at Chromosomes 15q25, 5p15, and 6p21: A Pooled Analysis from the International Lung Cancer Consortium,” Journal of the National Cancer Institute, Vol. 102, No. 13, 2010, pp. 959-971. doi:10.1093/jnci/djq178
- J. D. McKay, R. J. Hung, V. Gaborieau, P. Boffetta, A. Chabrier, G. Byrnes, et al., “Lung Cancer Susceptibility Locus at 5p15.33,” Nature Genetics, Vol. 40, 2008, pp. 1404-1406. doi:10.1038/ng.254
- M. T. Landi, N. Chatterjee, K. Yu, L. R. Goldin, A. M. Goldstein, M. Rotunno, et al., “A Genome-Wide Association Study of Lung Cancer Identifies a Region of Chromosome 5p15 Associated with Risk for Adenocarcinoma,” American Society of Human Genetics, Vol. 85, No. 5, 2009 pp. 679-691. doi:10.1016/j.ajhg.2009.09.012
- D. Miki, M. Kubo, A. Takahashi, K. A. Yoon, J. Kim, G. K. Lee et al., “Variation in TP63 Is Associated with Lung Adenocarcinoma Susceptibility in Japanese and Korean Populations,” Nature Genetics, Vol. 42, 2010, pp. 893- 896. doi:10.1038/ng.667
- N. Kumasaka, M. Aoki, Y. Okada, A. Takahashi, K. Ozaki, T. Mushiroda, et al., “Haplotypes with Copy Number and Single Nucleotide Polymorphisms in CYP2A6 Locus Are Associated with Smoking Quantity in a Japanese Population,” PLoS One, Vol. 7, No. 9, 2012, Article ID: e44507. doi:10.1371/journal.pone.0044507
- M. Fujieda, H. Yamazaki, T. Saito, K. Kiyotani, M. A. Gyamfi, M. Sakurai, et al., “Evaluation of CYP2A6 Genetic Polymorphisms as Determinants of Smoking Behavior and Tobacco-Related Lung Cancer Risk in Male Japanese Smokers,” Carcinogenesis, Vol. 25, No. 12, 2004, pp. 2451-2458. doi:10.1093/carcin/bgh258
- M. Shimizu, K. Kiyotani, H. Kunitoh, T. Kamataki, H. Yamazaki, “Different Effects of TERT, TP63, and CYP2A6 Polymorphism on Individual Risk of TobaccoRelated Lung Cancer in Male Japanese Smokers,” Journal of Cancer Therapy, Vol. 2, No. 5, 2011, pp. 690-696. doi:10.4236/jct.2011.25093
- A. Groppi, J. Begueret and A. Iron, “Improved Methods for Genotype Determination of Human Alcohol Dehydrogenase (ADH) at ADH 2 and ADH 3 Loci by Using Polymerase Chain Reaction-Directed Mutagenesis,” Clinical Chemistry, Vol. 36, No. 10, 1990, pp. 1765-1768.
- J. C. Barrett, B. Fry, J. Maller and M. J. Daly, “Haploview: Analysis and Visualization of LD and Haplotype Maps,” Bioinformatics, Vol. 21, No. 2, 2005, pp. 263-265. doi:10.1093/bioinformatics/bth457
- J. Y. Park, K. Matsuo, T. Suzuki, H. Ito, S. Hosono, T. Kawase, et al., “Impact of Smoking on Lung Cancer Risk Is Stronger in Those with the Homozygous Aldehyde Dehydrogenase 2 Null Allele in a Japanese Population,” Carcinogenesis, Vol. 31, No. 4, 2010, pp. 660-665. doi:10.1093/carcin/bgq021
- S. Y. Eom, Y. W. Zhang, S. H. Kim, K. H. Choe, K. Y. Lee, J. D. Park, et al., “Influence of NQO1, ALDH2, and CYP2E1 Genetic Polymorphisms, Smoking, and Alcohol Drinking on the Risk of Lung Cancer in Koreans,” Cancer Causes Control, Vol. 20, No. 2, 2009, pp. 137-145. doi:10.1007/s10552-008-9225-7
- Y. Xue, M. Wang, D. Zhong, N. Tong, H. Chu, X. Sheng, et al., “ADH1C Ile350Val Polymorphism and Cancer Risk: Evidence from 35 Case-Control Studies,” PLoS One, Vol. 7, No. 5, 2012, Article ID: e37227. doi:10.1371/journal.pone.0037227
- M. Wu, S. C. Chang, E. Kampman, J. Yang, X. S. Wang, et al., “Single Nucleotide Polymorphisms of ADH1B, ADH1C and ALDH2 Genes and Esophageal Cancer: A Population-Based Case-Control Study in China,” International Journal of Cancer, Vol. 132, No. 8, 2013, pp. 1868-1877. doi:10.1002/ijc.27803
- L. Wang, W. Zang, J. Liu, D. Xie, W. Ji, Y. Pan, et al., “Association of CYP2A6*4 with Susceptibility of Lung Cancer: A Meta-Analysis,” PLoS One, Vol. 8, No. 4, 2013, Article ID: e59556. doi:10.1371/journal.pone.0059556
- H. Sugimura, H. Tao, M. Suzuki, H. Mori, M. Tsuboi, S. Matsuura, et al., “Genetic Susceptibility to Lung Cancer,” Frontiers in Bioscience (Scholar Edition), Vol. 3, 2011, pp. 1463-1477.
- H. Yamazaki, Y. Inui, C. H. Yun, F. P. Guengerich and T. Shimada, “Cytochrome P450 2E1 and 2A6 Enzymes as Major Catalysts for Metabolic Activation of N-Nitrosodialkylamines and Tobacco-Related Nitrosamines in Human Liver Microsomes,” Carcinogenesis, Vol. 13, No. 10, 1992, pp. 1789-1794. doi:10.1093/carcin/13.10.1789
- P. H. Chou, S. Kageyama, S. Matsuda, K. Kanemoto, Y. Sasada, M. Oka, et al., “Detection of Lipid PeroxidationInduced DNA Adducts Caused by 4-Oxo-2(E)-Nonenal and 4-Oxo-2(E)-Hexenal in Human Autopsy Tissues,” Chemical Research in Toxicology, Vol. 23, No. 9, 2010, pp. 1442-1448. doi:10.1021/tx100047d
- P. J. Branton, K. G. McAdam, D. B. Winter, C. Liu, M. G. Duke and C. J. Proctor, “Reduction of Aldehydes and Hydrogen Cyanide Yields in Mainstream Cigarette Smoke Using an Amine Functionalised Ion Exchange Resin,” Chemistry Central Journal, Vol. 5, 2011, p. 15. doi:10.1186/1752-153X-5-15
Abbreviations
ADH1C: alcohol dehydrogenase 1C;
ALDH2: aldehyde dehydrogenase 2;
CI: confidence interval;
CYP2A6: cytochrome P450 2A6;
GWAS: genome-wide association studies;
SNP: single nucleotide polymorphism;
TERT: telomerase reverse transcriptase;
TP63: tumor protein p63.
NOTES
*There are no competing financial interests in relation to this study.
#Corresponding author.

