Advances in Bioscience and Biotechnology
Vol.4 No.3A(2013), Article ID:29282,5 pages DOI:10.4236/abb.2013.43A058
PCR-DGGE analysis of earthworm gut bacteria diversity in stress of Escherichia coli O157:H7*
![]()
1College of Resources and Environmental Sciences, China Agricultural University, Beijing, China
2Ohio Agricultural Research and Development Center, Department of Veterinary Preventive Medicine, The Ohio State University, Wooster, USA
Email: #sun108@cau.edu.cn
Received 13 January 2013; revised 14 February 2013; accepted 24 February 2013
Keywords: PCR-DGGE; Earthworm Intestinal Bacteria Diversity; Escherichia coli O157:H7 Stress
ABSTRACT
In order to test if the intestinal bacteria play an important role in antibacterial ability of earthworm, we chose Escherichia coli O157:H7, an anthropozoonosis pathogen, as a biological stressor and studied the change of intestinal bacteria community of earthworm by PCR-DGGE analysis. Results showed that the pathogen merely existed 1 - 3 days, then almost disappeared after through the earthworm’s gut. In this period, the diversity and abundance index of intestinal bacteria increased first, then decreased, and finally kept stably after 7 days. The result demonstrated that the intestinal bacteria of earthworm had ability of adjust community structure to eliminate the pathogen E. coli O157:H7, and the amount of bacteria Bacillus increased significantly, which might be the positive antagonism to E. coli O157:H7.
1. INTRODUCTION
The antibacterial ability of earthworm is not only depend on natural immunity, the intestinal microbes also play a important role on cooperative defense [1]. Escherichia coli O157:H7 is an anthropozoonosis pathogen with characters of acid-resisting, low temperature resistance, low infectious dose, strong pathogenicity, no specific medicine for clinical treatment etc. Therefore it can cause gastrointestinal disease with potentially fatal consequences as a result of systemic Shiga toxin activity [2]. The Escherichia coli O157:H7 could threaten human health through the food chain as the natural host of Escherichia coli O157: H7 (like cattle, sheep, dogs and chicken and other animals) is often animal origin food pollution source.
Because of the strong ability of digest, earthworm was used to dispose the wastes and livestock feces for a long time. However, we had hardly found they died of pathogens, even they only existed natural immunity (body wall, cellular immunity and humoral immunity), which is not enough to defend all pathogen, as the lower level in animals [3]. Kumar found the amount of Escherichia coli O157:H7 reduced through digestion of the earthworm [4]. Liu isolated 6 Lactobacillus strains from vermicompost of Eisenia fetida, which was stressed by Escherichia coli O157:H7 and each strain could restricted the pathogen obviously [5]. Base on these studies, we reasonably infer the microbe in gut may help earthworm to defense the pathogen Escherichia coli O157:H7.
In this study, we use PCR-DGGE technique to analysis the dynamics of Eisenia fetida intestinal bacterial community structure and diversity under the stress of pathogen Escherichia coli O157:H7. Furthermore, we try to explain the relationship between the disappearance of pathogen and the change of intestinal bacterial community by which to evaluate if the intestinal bacteria of earthworm play an important role to against pathogen in surrounding environments.
2. MATERIALS AND METHODS
2.1. The Experimental Organisms and Preparation of an Artificial Soil
The earthworm Eisenia fetida, which was bred in our lab, was used as the experimental organism. Adult earthworms were kept in test substrates for one week before experiment. Artificial soil was used as test substrate in the experiments and was prepared according to the improved OECD method, which comprised 10% dried cowdung instead of sphagnum peat, 20% kaolin, 70% sand, and Ca carbonate was used to adjust pH to 6.0 ± 0.05 [6]. Two days before experiments, the earthworms were transferred onto the filter paper, which was soaked with isotonic Lumbricus balanced salt solution, to cleaned the feces in gut [7].
Escherichia coli O157:H7 (the international reference strain EDL933) was purchased from Institute of Epidemiology and Microbiology, Chinese Academy of Preventive Medicine.
2.2. Experimental Procedures
Escherichia coli O157:H7 was maintained in stock culture at 4˚C, and was shock cultivated in Luria-Bertani medium (pH 7.0, Tryptone 1%, Yeast extract 0.5%, NaCl 1%) at 37˚C, 200 rpm for 18 h. The cells of bacteria in the culture were collected by centrifugation at 3000 rpm for 30 min, washed, and resuspended in water. They were added into the artificial soil at final concentrations of approximately 107 CFU g−1 soil [8]. Then, the mixture was thoroughly mixed with water at least 30 min to achieve a homogenous distribution which was adjusted to 35% of the final water content. The soil samples were stored in the plastic containers at room temperature overnight. The control soil without bacterial addition was prepared similarly. Subsequently, the earthworms were transferred to the plastic containers. 300 earthworms were added into each container with 30 kg bacteria-soil mix as prepared in the above steps. The containers were placed in a room which maintained a constant environmental temperature under 23˚C; water was supplemented to maintain the moisture level of the soil [9]. The earthworms were fed with dried cow-dung, which was added onto the soil surface once a week after the dung was thoroughly wetted. The experiments lasted for 21 days.
2.3. The Extraction of the Total DNA of Bacteria in Earthworm Gut
On Day 0, 1, 3, 7, 14, and 21 of the feeding period, ten earthworms were random collected, and the intestinal content was gathered by anatomy. The total DNA of intestinal bacteria was extracted from 0.5 g intestinal content by E.Z.N.A. Soil DNA Kit (obtained from American OMEGA).
2.4. The Conditions of PCR and DGGE
The total DNA of bacteria in earthworm gut was used as a template (0.5 ng) in PCR reactions (2 × Easy taq Super mix were obtained from Beijing TransGen Biotech Co., Ltd.) with primers: GC-968F (5’-CgCCCgGggCgCgCCCCgggCggggCgggggCACggggggAACgCgAAgAACCTTAC-3’) and 1401R (5’-gCgTgTgTACAAgACCC-3’). PCR cycles were as follows: one cycle of 94˚C for 5 min, and 30 cycles of 94˚C, 30 s, 54˚C, 30 s and 72˚C, 30 s, followed by a final extension at 72˚C for 5 min.
DGGE condition was controlled as: 25 μL sample (DNA concentration more than 50 ng/μL at least) has been purified (PCR Cycle Pure Kit were obtained from American OMEGA Co.), then it was add into the gel with 40% - 65% denaturing gradient, ran 7 hour under 120 V.
2.5. Gel Extraction and Sequencing
Gel Extraction Mini kit (obtained from American OMEGA) was used to extract DNA fragments. After that, the products were linked with PMD 18-T vector to ensure the correct sequencing (the primers without GC clamp were used for sequencing).
3. RESULTS
3.1. Behavior of Earthworms under Escherichia coli O157:H7 Stress
At the beginning, few earthworms (about 70 - 80) stayed on the surface of artificial soil, without escaping. Then, all of them dip into the earth during 3 hours. The difference of weight per 100 earthworms between the control and stress groups was not significant (Figure 1).
3.2. The Change of Intestinal Bacteria Diversity
According to the cluster analysis of DGGE fingerprinting, the sample 1st and 3rd were classified to a group; the sample 7th and 14th were classified to the second; the last group contained sample 21st (Figure 2).
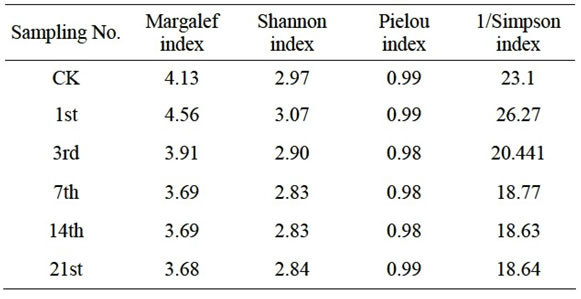
The diversity, abundance and dominance index of Eisenia fetida intestinal bacteria had a short increase in 24 h under Escherichia coli O157:H7 stress, but it reduced rapidly after 3 days, and finally kept stable during the last days (Table 1).
3.3. The Change of Intestinal Bacteria Community Structure
Band L, which means Escherichia coli O157:H7, merely existed in earthworm gut in the first day, and could not be found in the rest of days. With the pathogen disappeared, bond a, d and k, which existed in earthworm
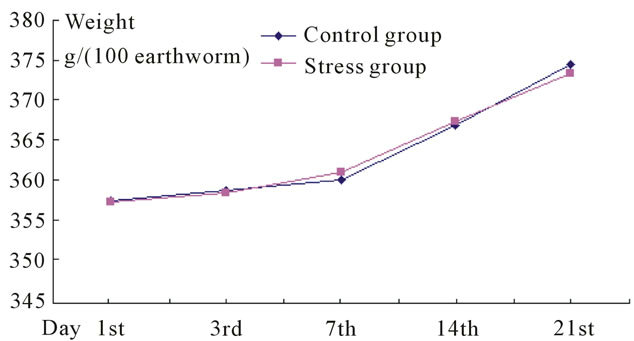
Figure 1. The increasing weight of two groups of earthworm. The growth rate of control group is 4.784%; and the number of stress group is 4.535%.
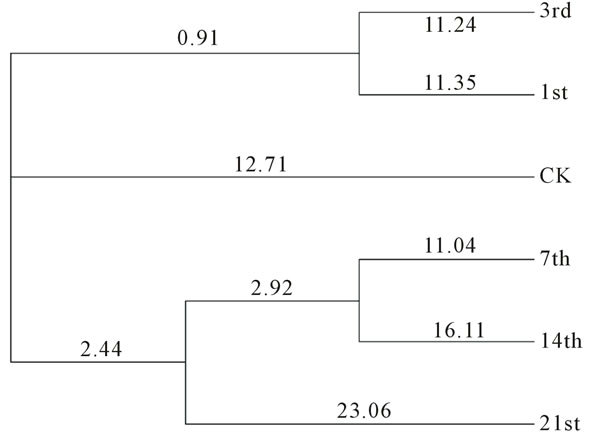
Figure 2. Cluster analysis of PCR-DGGE fingerprinting.
Table 1. Diversity of the intestinal bacteria in Eisenia fetida.
gut originally, also could not be found after the 7th day. Another bond, which need to be noticed is c. It occurred with Escherichia coli O157:H7, and had a high abundance during the culture period (the percentage in bacteria community of each sample was 3.24%, 3.87%, 5.31%, 5.28%, 5.29%) (Figure 3).
The phylotype, which band a, d, c and k represented, belong to Anaerosporobacter, Aeromonas, Bacillus, and Rubritalea, respectively. The band a (Anaerosporobacter), c (Bacillus), k (Rubritalea) had higher genetic relationship than band d (Aeromonas) with Escherichia coli O157:H7. The rest bacteria mainly belong to Clostridiales, Aeromonadales, Chromatiales et al., which belong to phylum Proteobacteria and Firmicutes (Figure 4).
4. DISCUSSION
4.1. The Concentration of Escherichia coli O157:H7 in Artificial Soil
Hutchison found the concentration of Escherichia coli O157:H7 in livestock feces was 103 - 105 CFU/g, and Fukushima found the amount of the pathogens could increase to 108 CFU/g [8]. To ensure the stress effect, the concentration of Escherichia coli O157:H7 was maintained at 107 CFU/g. However, the growth of Eisenia fetida was not affected under these concentration (Figure 1).
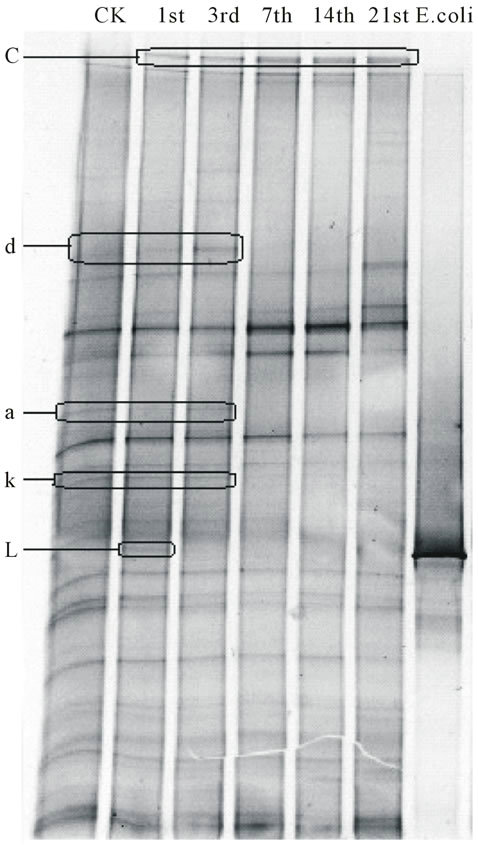
Figure 3. PCR-DGGE fingerprinting of intestinal bacteria in Eisenia fetida gut with Escherichia coli O157:H7.
4.2. The Diversity and Community Structure Change of Eisenia fetida Intestinal Bacteria
The result of cluster analysis show that the dynamics of earthworm intestinal bacteria community have experienced 3 difference periods under the stress of Escherichia coli O157:H7, and it was in accordance with the results of diversity, abundance and dominance index (Table 1). Combined the results of PCR-DGGE fingerprinting and ecology index, we speculated that the appearance of band c lead a short increasing of diversity and abundance index in the first day. In second period, the disappearance of Escherichia coli O157:H7 result in the reducing of diversity index and abundance index, and might have some relationship to the disappearance of band a, d, k. The third period performed relative stable, the indexes and structure basically did not change, and the rebalance might be build in this period.
4.3. The Defense Effection to Escherichia coli O157:H7 of Bacillus
Bacillus is a kind of aerobic microbe, through take biological oxygen to reduce the REDOX, and further reduce the aerobium, so as to keep the intestinal microecological balance [10]. Bacillus could produce a variety of diges-
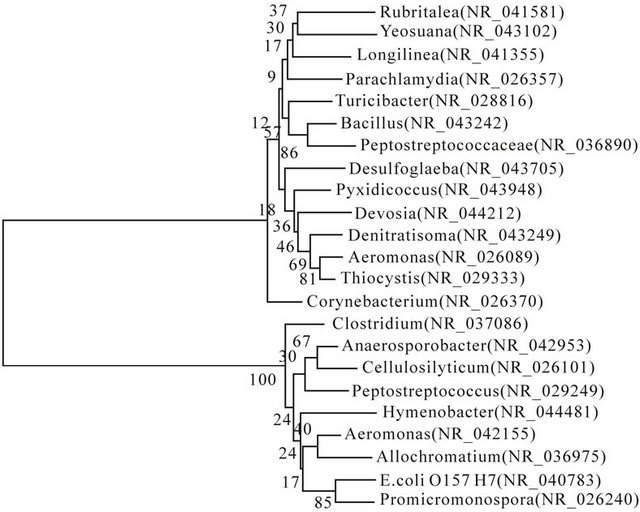
Figure 4. A neighbor-joining phylogenetic tree contain main intestinal bacteria of Eisenia fetida Bootstrap analyses were made with 1000 cycles.
tive enzyme and antibiotics (peptide antibiotics, lipopeptide antibiotics, phosphatide antibiotic, polyketide etc.) in intestinal [11]. Therefore, Bacillus is the source of many probiotic like some bacteria helpful for digestion and antimicrobial.
The bacteria Bacillus, which appeared with Escherichia coli O157:H7, might be a strain with antagonism to this pathogen. The Bacillus usually has a low level in earthworm gut. Therefore DGGE could not detect it. When the intestinal microbes were stressed by Escherichia coli O157:H7, the proliferation of Bacilllus made it to be found. The ability to defense Escherichia coli O157:H7 might depend on the secretion of antimicrobial compound [12]. The isolation of this strain and separation of antimicrobial compounds need to be promoted.
Other interesting bacteria are belong to Anaerosporobacter sp. and Aeromonas sp., which also could lead diarrhea like Escherichia coli O157:H7. They were also found disappeared in 7 - 14 days combined with Bacillus appeared. Whether this Bacillus strain could influence these two pathogens, it needs to be confirmed and complemented with further studies.
REFERENCES
- Pedersen, J.C. and Hendriksen, N.B. (1993) Effect of passage through the intestinal tract of detritivore earthworms (Lumbricus spp.) on the number of selected gram-negative and total bacteria. Biology and Fertility of Soils, 16, 227-232. doi:10.1007/BF00361413
- McNeilly, T.N., Mitchell, M.C., Rosser, T., McAteer, S., Low, J.C., Smith, D.G.E., Huntley, J.F., Mahajan, A. and Gally, D.L. (2009) Immunization of cattle with a combination of purified intimin-531, EspA and Tir significantly reduces shedding of Escherichia coli O157:H7 following oral challenge. Vaccine, 28, 1422-1428. doi:10.1016/j.vaccine.2009.10.076
- Wang, C., Sun, Z.J. and Zheng, D.M. (2006) Research advance in antibacterial immunity ecology of earthworm. Chinese Journal of Applied Ecology, 17, 525-529
- Kumar, A.G. and Sekaran, G. (2005) Enteric pathogen modification by anaecic earthworm Lampito mauritii. Journal of Applied Sciences and Environmental Management, 9, 15-17.
- Liu, X.L., Zhang, N., Cui, D.L., Wang, C. and Sun, Z.J. (2011) Lactic acid bacteria isolated from vermicompost and its antimicrobial activity in E. coli O157:H7. GAB, 1, 1-6
- Organisation for Economic Cooperation and Development (1984) Guideline for testing of chemicals No. 207: Earthworm acute toxicity tests. OECD Publishing.
- Stein, E.A. and Cooper, E.L. (1981) The role of opsonins in phagocytosis by celomocytes of the earthworm, Lumbricus terrestris. Developmental & Comparative Immunology, 5, 415-425.
- Hutchison, M.L., Walters, L.D., Avery, S.M., Synge, B.A. and Moore, A. (2004) Levels of zoonotic agents in British livestock manures. Letters in Applied Microbiology, 39, 207-214. doi:10.1111/j.1472-765X.2004.01564.x
- Jensen, J., Krogh, P.H. and Sverdrup, L.E. (2003) Effects of the antibacterial agents tiamulin, olanquindox and metronidazole and the anthelmintic ivermectin on the soil invertebrate species Folsomia fimetaria (Collembola) and Enchytraeus crypticus (Enchytraeidae). Chemosphere, 50, 437-443. doi:10.1016/S0045-6535(02)00336-3
- Li, H.B., Song, X.L. and Li, Y. (1995) Progress on probiotic bacteria in aquiculture. Progress in Veterinary Medicine, 29, 94-99.
- Liu, C.H., Chiu, C.S. and Ho, P.L. (2009). Improvement in the growth performance of white shrimp. Litopenaeus vannamei, by a protease-producing probiotic, Bacillus subtilis E20, from natto. Journal of Applied Microbiology, 107, 1031-l041. doi:10.1111/j.1365-2672.2009.04284.x
- Kajimura, Y., Sugiyama, M. and Kaneda, M. (1995) Bacillopeptins, new cyclic lipopeptide antibiotics from Bacillus subtilis FR-2. The Journal of Antibiotics, 48, 1095- 1110.
NOTES
*This work was financially supported by National Natural Science Foundation of China (No: 31172091).
#Corresponding author.

