American Journal of Molecular Biology
Vol.3 No.2(2013), Article ID:30720,4 pages DOI:10.4236/ajmb.2013.32013
Research of total levels on DNA methylation in plant based on HPLC analysis
![]()
1Experimental Center, Shenyang Normal University, Shenyang, China
2Department of Horticulture, Shenyang Agricultural University, Shenyang, China
3College of Chemistry and Life Science, Shenyang Normal University, Shenyang, China
Email: *chq2012lunwen@126.com
Copyright © 2013 Qiang Chen et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 20 February 2013; revised 10 April 2013; accepted 20 April 2013
Keywords: DNA Methylation; Cytosine; 5-Methylcytosine; High Performance Liquid Chromatography; Mobilephase
ABSTRACT
HPLC analysis is important for determination of total level on DNA methylation in plants. It can be used to help characterise epigenetic changes during growth, development and stress. HPLC methods have been optimised for mammalian and microbial DNA, but not for plants. This article examines several important factors in the HPLC analysis of plant DNA methylation including extraction and purification of DNA and HPLC conditions choice by using leaves of rice seedling. The experimental results showed that RNA of nucleic acid was removed by using RNase A. This study also identified critical components of HPLC analysis. With the optimized method of HPLC conditions, the better result was achieved in the chromatogram of cytosine and 5-methylcytosine in genomic DNA acid hydrolysis. The study would offer a comprehensive guide for the stringent analysis of DNA methylation in plants.
1. INTRODUCTION
DNA methylation is important regulation mechanism of gene regulation and expression in biological process. DNA methylation plays a key role in plant such as morphogenesis, development, stress, acclimation and adaptation [1,2]. In addition, DNA methylation is concerned with controlling the morphogenetic responses in plant tissue cultures and maintaining their genetic stability, which is of particular interest to biotechnologists. At present, there are a wide variety of methods in determination of DNA methylation such as high performance liquid chromatography (HPLC) [3,4], bisulphite sequencing [5], methylation-sensitive amplified fragment length polymorphism (MSAP) [6,7]. Random primers PCR, denaturing gradient gel electrophoresis (DGGE) and so on. HPLC is an appropriate technique for assessing the larger-scale epigenetic effects of stress and acclimation responses in plants, while this approach does not differentiate between coding and non-coding sequences of DNA, nor detect subtle differences in the methylation status of individual genes [8]. HPLC is considered the most reliable and sensitive technique to determine total DNA methylation [9,10]. HPLC methods have been optimised for mammalian and microbial DNA, but not for plants [11,12]. Consequently, this study identifies critical components of HPLC analysis and offers a reference for the stringent analysis of DNA methylation in plants.
2. MATERIALS AND METHODS
2.1. Chemicals
Chemicals for DNA extraction were supplied by Sinopharm Chemical Reagent Co., Ltd. (Shenyang, China). Cytosine and 5-methylcytosine for HPLC analysis were supplied by Sigma; HPLC-grade methanol was supplied by Fisher Scientific. Deionised H2O (18 MX·cm–1) was used for all experiments.
2.2. Plant Material, Nucleic Acids Extractions
Rice (Oryza sativa L) were used for nucleic acid extractions.Nucleic acids were extracted from 2 to 3-week-old leaf tissue according to the CTAB based on Miao [6]. Deoxyribonucleic acid extractions purified were resuspended in TE buffer (0.2 m-MTris-HCl, 1 mM EDTA, pH 7.5), Nucleic acid concentration and purity was estimated by measuring the absorbance at 260 nm (assuming 1 OD equates to 50 µg·ml–1) and samples were stored at −40˚C.
2.3. Gel Electrophoresis
Nucleic acid samples (3 µl) of Oryza sativa L treated with RNases were mixed with 1 µl of loading buffer (0.25% w/v bromophenol blue). The agarose gel (1.2% w/v) of 100 ml, adding ethidiumbromide 4 µl(0.5 µg·ml–1) before solidification, were loaded onto a 65 mm × 65 mm. Then it was prepared in TBE buffer (0.089 MTris, 0.089 M boric acid, 0.01 M EDTA, pH 8.0) and performed at 90 V, 15 and 30 min .Then observed and photographed electrophoresis band by using a Digital gel image analysis system of Jeda (Jiangsu, China).
2.4. Acid Hydrolysis of DNA
Acid hydrolysis of DNA refered to [13]. DNA solution 100 μl (containing 30 μg DNA) was added 50 μl perchloric acid (70%), and then was hydrolysed for 60 minutes in boiled water. Acid hydrolyzates PH was adjusted to 3 - 5 and centrifuged with 12,000 rpm, 10 min. Then take supernate fluid filtered to perform HPLC program.
2.5. Preparation of C and 5-mC Standard Stock Solutions
The C and 5-mC standard stock solutions refer to [14] weighing C 0.4444 mg and 5-mC 0.55145 g, and dissolving 0.1% perchloric acid, then concentrations of C and 5-mC mother liquor were 4 × 102 μmol/L and 20 μmol/L.
2.6. HPLC-UV Detection
HPLC analysis was performed using Agilent LC 1200 with Quat pump (Germany), a manual injection valve fitted with a 20 µl loop, a Tcc UV absorbance detector (Germany) set to 285 nm. A column for Diamonsil C18 (250 × 4.6 mm, 5 μm) was used in this detection. The HPLC system was flushed with deionised H2O (0.22 µm filtered) at 0.5 ml·min–1 for 15 min to remove all traces of phosphate at the end of each day, followed by 15 min at the same flow-rate with 90% v/v methanol (0.22 µm filtered) to clean the column.
2.7. Calculation of the Percentage 5-mC in DNA
The level of 5-MedC present in the DNA samples was expressed as a percentage of the level of dC which was calculated using the following equation:
% 5-mC = [5-mC/dC + 5-MedC] × 100.
3. RESULTS AND DISCUSSION
3.1. Centrifugation Again after Extracting Liquid Supernatant
There are a small number of impurities in liquid supernatant of DNA regardless of sucking liquid supernatant carefully. The result could be observed on Centrifugal tube bottom. Consequently, another centrifugation is essential after extracting liquid supernatant of DNA (data not shown).
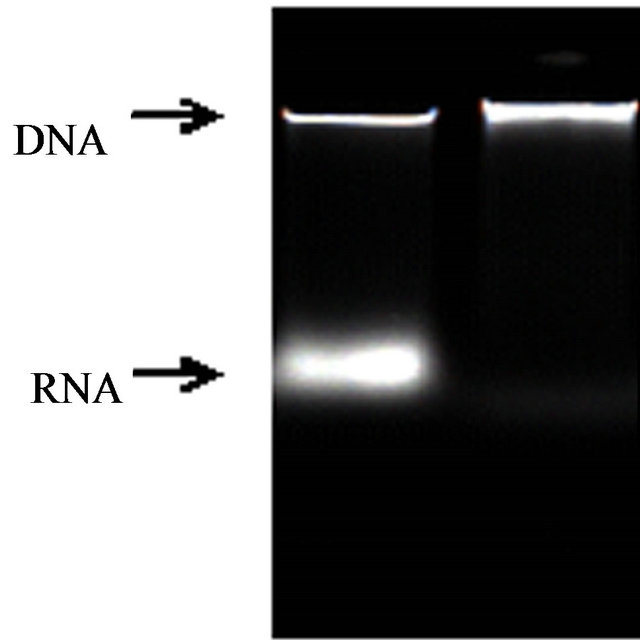
3.2. Adding RNase A to Remove Residual RNA in DNA
RNA in genome DNA should be cleansed as far as possible because the determination of total level on DNA methylation is effected by RNA of DNA. The most common method to remove RNA from DNA is to treat extractions with RNase A [15]. An agarosegel showed RNase A had effectively removed residual RNA from rice DNA. (Figures 1 and 2) The results of ultraviolet spectrophotometry showed that the number of DNA optical density (OD) treated with RNase A approachhed between 1.80 and 2.0 (1.80 < OD < 2.0) much more, but the number without RNase A is almost over 2.0 (OD > 2.0).
3.3. Separation Characteristics 5-Methylcytosine and Cytosine under Different Length of Column
C18 column is currently used for HPLC analysis of DNA
 (a) (b)
(a) (b)
Figure 1. Ethidiumbromide stained agarose (1.2% w/v) gel containing (a) no-treatment; (b) RNase A.
 (a) (b)
(a) (b)
Figure 2. Ethidiumbromide stained agarose (1.2% w/v) gel containing (a) no-treatment; (b) RNase A.
methylation. The study showed that relative retention times and separation degree of cytosine and 5-methylcytosine had changed obviously (Figures 3 and 4). In this detection, C18 column improved the separation degree much better.
3.4. Separation Characteristics of 5-Methylcytosine and Cytosine under Different the pH of the Mobile Phase
The study showed the pH of the mobile phase affected cytosine and 5-methylcytosine peak separation and shape (Figures 5 and 6). A lowering in pH from 5.8 to 4.5 reduced the retention time of cytosine and 5-methylcytosine (Figures 5 and 6). Gehrke et al. [16] found similar results when studying a narrower pH range of 4.0 - 5.0. In addition, the strongest effects of pH for improvement of peak separation occurred at pH values below 4.0 [8]. In conclusion, the PH of the mobile phase effected greatly in the retention time and peak shape of C and 5-mC.
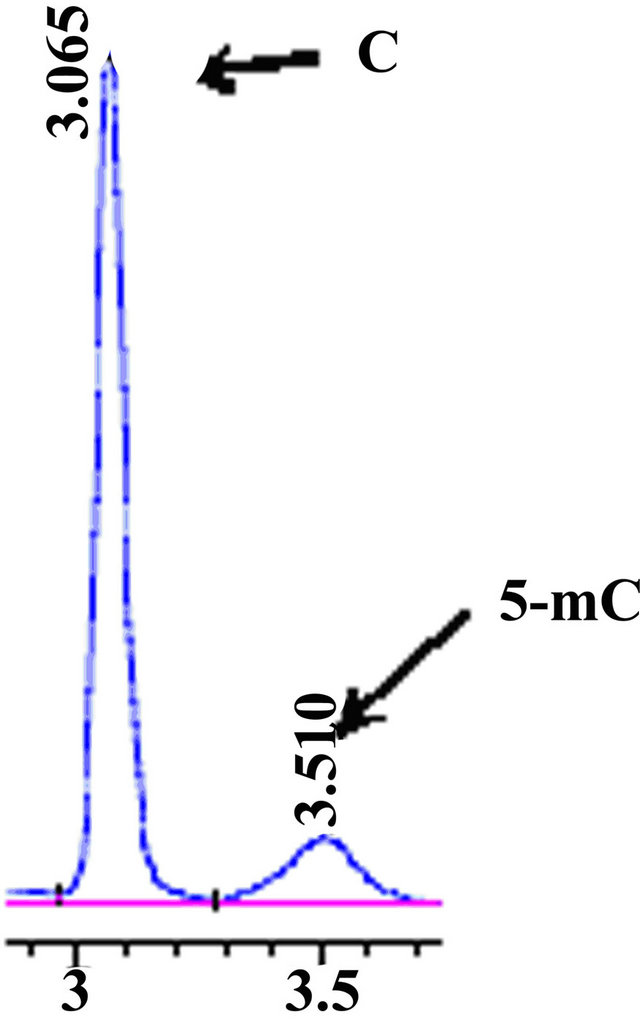
Figure 3. HPLC chromatograms of accustandard cytosine and 5- ethylcytosine. A 150 mm C18 column utilised for this.

Figure 4. HPLC, chromatograms of accustandard C and 5-mC, A 250 mm C18 column utilised for this experiment.

Figure 5. HPLC chromatograms of accustandard C and 5-mC, the mobile phase pH 4.5, flow rate 0.4 ml·min−1.
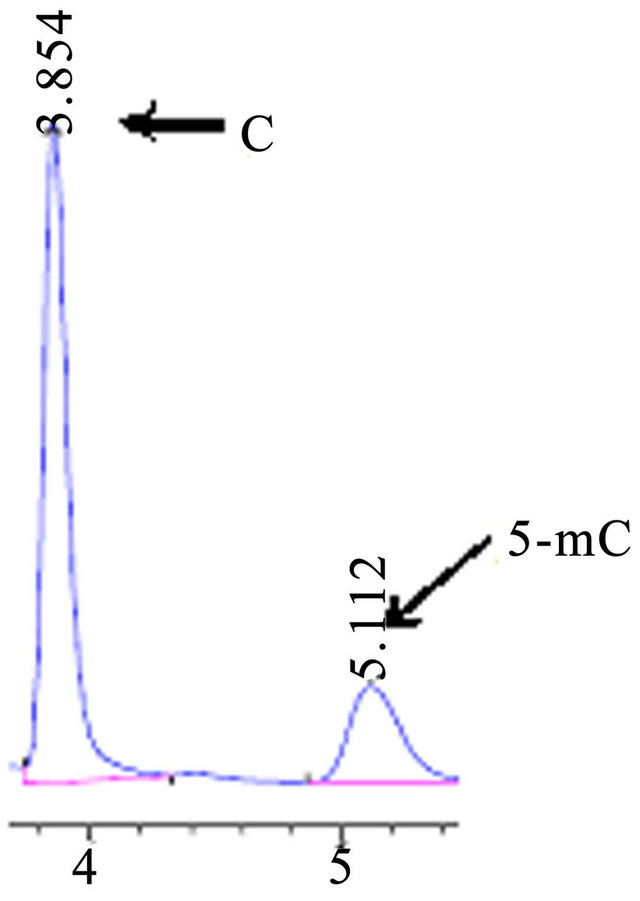
Figure 6. HPLC chromatograms of accustandard C and 5-mC, the mobile phase pH 5.8, flow rate 0.4 ml·min−1.
3.5. Calibration Lines for C and 5-mC, Actual Sample Chromatogram
The calibration lines were constructed by the dilution of the stock standard solutions for C and 5-mC, which were used to determine level of 5-MedC and dC in the hydrolysed DNA samples. The results show that C and 5-mC had the better linear relation between the peak area and concentration of the standard solutions (data not shown). In addition, C and 5-mC were separated in Actual sample chromatogram with the condition for 50 mmol·L−1 KH2PO4, PH 5.8, flow rate 0.5 ml·min−1 (Figure 7).
4. CONCLUSION
This study has demonstrated that several factors need consideration before undertaking HPLC analysis of DNA methylation, including choice of column and mobile phase pH, RNA removal and so on. This study has optimised the procedure for HPLC analysis of DNA methy-
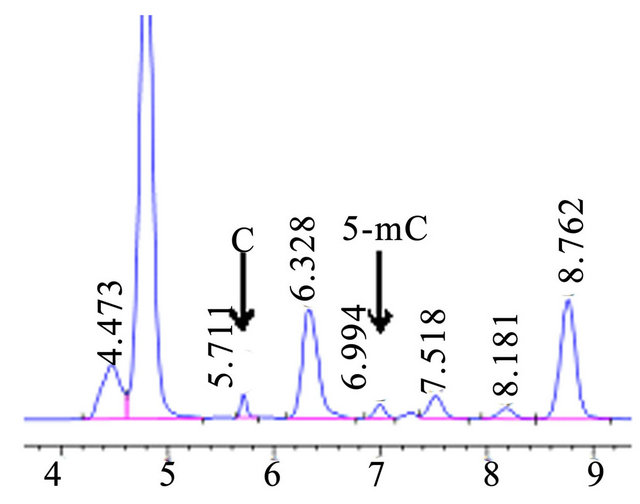
Figure 7. HPLC chromatograms of actual sample in Oryza sativa L.
lation, and identified the critical issues for accurately determining total methylation changes in plant DNA.
5. ACKNOWLEDGEMENTS
The authors acknowledge financial support from National natural science foundation of China (No. 30870205) and Shen yang Normal University Experimental center director foundation (No. SY201104 and No. SY201004).
REFERENCES
- Finnegan, E.J. and Kovac, K.A. (2000) Plant DNA methyltransferases. Plant Molecular Biology, 43, 189-201. doi:10.1023/A:1006427226972
- Chan, S.W., Henderson, I.R. and Jacobsen, S.E. (2005) Gardening the genome: DNA methylation in arabidopsis thaliana. Nature, 6, 351-360.
- Wagner, I. and Capesius, I. (1981) Determination of 5- methylcytosine from plant DNA high-performance liquid chromatography. Biochimica et Biophysica Acta, 654, 52- 56. doi:10.1016/0005-2787(81)90135-0
- Pan, Y.J., Fu, B.Y., Wang, D., Zhu, L.H. and Li, Z.K. (2009) Spatial and temporal profiling of DNA methylation induced by drought stress in rice. Scientia Agricultura Sinica, 9, 3009-3018.
- Hajkova, P., et al. (2002) DNA-methylation analysis by the bisulfite-assisted genomic sequencing method. Methods in Molecular Biology, 200, 143-154.
- Robertson, K.D. and Wolffe. A.P. (2000) DNA methylation in health and disease. Nature Reviews Genetics, 1, 11-19.
- Lu, Y.L., Rong, T. and Cao, M.J. (2008) Analysis of DNA methylation in different maize tissues. Journal of Genetics and Genomics, 35, 41-48.
- Johnston, J.W., Harding, K., Bremner, D.H., et al. (2005) HPLC analysis of plant DNA methylation: A study of critical methodological factors. Plant Physiology, 43, 844-853.
- Fraga, M.F. and Esteller, M. (2002) DNA methylation: A profile of methods and applications. Bio-Technique, 33, 632-649.
- Wagner, I. and Capesius, I. (1981) Determination of 5- methylcytosine from plant DNA high-perfmance liquid “chromatography”. Biochimica et Biophysica Acta, 654, 52-56.
- Perrett, D. (1986) Nucleotides, nucleosides and bases. In: Lim, C.K., Ed., HPLC of Small Molecules, IRL Press Limited, Oxford, 221-259.
- Ramsahoye, B.H. (2002) Measurement of genomewide DNA methylation by reversed-phase high-performance chromatography. Methods, 27, 156-161.
- Demeulemeester, M.A.C., Stallen, N.V. and Droft, P.M.P. (1999) Degree of DNA methylation to chicory (Cichorium intybus L.): Influence of plant age and vernalization. Plant Science, 142, 101-108.
- Dong, Y.J., Cheng, Z., Li, S., et al. (2009) Determination based-on HPLC of DNA methylation levels for different ages in ginseng. Chinese Traditional and Herbal Drugs, 9, 1416-1418.
- Sambrook, J., Fritsch, E.F. and Maniatis, T. (1989) Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, New York.
- Gehrke, C.W., McCune, R.A., Gamasosa, M.A., Ehrlich, M. and Kuo, K.C. (1984) Quantitative reversed-phase high performance liquid chromatography of major and modified nucleoside in DNA. Journal of Chromatography, 301, 199-219.
NOTES
*Corresponding author.

