Journal of Biophysical Chemistry
Vol.2 No.4(2011), Article ID:8430,7 pages DOI:10.4236/jbpc.2011.24046
Glutathione transferase-P1-1 binding with naturallyoccurring ligands: assessment by docking simulations
1School of Biomedical Sciences, University of Ulster, Coleraine, UK;
2Department of Bimolecular and Sport Sciences, Coventry University, Coventry, UK;
3Department of Pharmacology and Therapeutics, University of Liverpool, Liverpool, UK. *Corresponding Author: r.owusu-apenten@ulster.ac.uk
Received 16 July 2011; revised 16 September 2011; accepted 25 September 2011.
Keywords: Glutathione Transferase; Docking; Vitamins; Phytochemicals
ABSTRACT
Glutathione transferase-P1-1 (hGSTP1-1), which is associated with acquired drug resistance in some tumour cells, requires two identical subunits for full activity. Naturally occurring inhibitors for GSTP1-1 quaternary structure could be interesting therapeutic agents. The aim of this study was to investigate potential binding sites for hGSTP1-1 interaction with ligands many of which occur naturally. Simulations were performed with commercial docking software and with GST monomer or dimer as template. Docking results using hGSTP1-1 dimer showed one binding site for most of the ligands tested. Lycopene, glutathione, ellagic acid, ethacrynic acid, quercetin, caffeic acid, ferulic acid, porphyrin, curcumin, cinnamic acid, and also α-tocopherol bound at the enzyme dimer subunit-subunit interface. In contrast, investigations using hGSTP1-1 monomer revealed three additional sites for ligand binding. In conclusion, the docking simulations suggest that the enzyme subunit interface may be important for hGSTP1-1 interactions with ligands. These findings may provide valuable insights for further research to identify naturally occurring therapeutic agents.
1. INTRODUCTION
Glutathione transferases (glutathione-S transferase; GST, EC 2.5.1.18) are phase-II enzymes associated with acquired drugs resistance in tumour cells [1]. The GSTs are involved also with intracellular signalling and other functions unrelated to catalysis; see [2] for review. The three major families of GST are, mitochondrial GST, microsomal GST or MAPEG (membrane associated proteins involved in eicosanoid and glutathione metabolism), and cytosolic GST. Seven classes of cytosolic GST are differentiated (zeta, theta, omega, sigma, alpha, mu, pi/P1-1) by their sequence homology, kinetic properties, immunological characteristics or subunit structure [1,2]. Human GST isoform P1-1 (hGSTP1-1) is over-expressed in several tumours, suggesting that its selective targeting may have chemotherapeutical potential [1,2].
Each hGSTP1-1 enzyme molecule contains two identical subunits (A and B) with an accessible surface area (ASA) of 8975 Å2 per monomer. Dimerization of the enzyme subunits reduces the total ASA by 13% owing to the burial of some residues at the subunit-subunit interface [3,4]. The dimerization of hGSTP1-1 seems to be essential for activity [5,6]. The globular structure of each GST subunit includes one N-terminal α/β domain (domain-1, residues 1-74) and an all-helix C-terminal domain (residues 81-207). Each subunit of hGSTP1-1 has two binding sites for substrate molecules. There is a glutathione binding site (G-site) located within domain-1 (except for residues Glu97 and Asp98) with Tyr8 and Asp99 identified as essential for activity [3,4]. A second binding site for electrophilic substrates (the H-site) is composed of residues from domain-1 and 2 notably Tyr8, Phe9, Pro10, Val11, Val36, Tyr104, Tyr109 and Gly206 [7]. Finally, hydrophobic non-substrate molecules bind with the so-called ligandin site (L-site) located within the H-site [8,9] thought to be near the interface between the two enzyme subunits [3-5,7]. Currently, the effect of non -substrate ligands on GST activity is not fully understood [5].
Although the complexes with hGSTP1-1 have yet to be studied, there is growing interest in naturally occurring compounds such as tocopherol [10,11], quercetin [12], ellagic acid and cucumin [13] which apparently inhibit GST. Docking simulations have been successfully applied to model the interaction of GST from filaria [14-16] and insects [17,18]. The aims of this study were to examine hGSTP1-1 interaction with some naturally occurring ligands using docking simulations. To allow for possible receptor-site steric hindrance in the design of this study, we selected both the native enzyme dimer and a single GST subunit for docking simulations. The results are considered in terms of binding site motifs and their possible interrelationships with GST activity and structure. This research may provide valuable insights into the effect of naturally occurring compounds on GST and the role of such compounds as chemotherapeutic agents.
2. MATERIALS AND METHODS
The crystal structures for hGSTP1-1 (pdb id: 3HJM) used were from the Protein Data Bank (www.rcsb.org; [19]. The quality of structures was evaluated using a global QMEAN scoring function (http://swissmodel.expasy.org/ qmean/cgi/index.cgi). PROCHECK (http://www.ebi.ac. uk/thornton-srv/software/PROCHECK/) was used to assess the stereo-chemical properties of the polypeptide structure. The structure for ligands were downloaded from the ChEBI database (www.ebi.ac.uk/chebi/ init.do) using CHEBI id, CAS numbers (www.chemicalbook.com) or their PDB id code (www.ligand-expo.rcsb.org). Ligand structures were cleaned using gradient optimisation after adding explicit H-atoms using Marvin Sketch 5.0.7 software. Molecular dynamics was performed for 1000 cycles using Dreiding force fields at 300 K. Ligand structures were selected by energy minimization and geometry, and saved using a *pdb file format.
Docking simulations employed Hex version 5.1 (from http://www.loria.fr/~ritchied /hex/) which uses a special polar Fourier correlation to attain precision and speed of operation. Modelling operations were performed using a 4GB-64-bit computer to facilitate large 5D rotational grids. The structure for either hGSTP1-1 dimer or single monomer was loaded to Hex 5.1 keeping default parameters. Shape, electrostatics and potential distributions were used for docking control. Simulations were conducted in the full rotation mode where the ligand is flexible but the receptor is rigid. Docked poses were analysed based on the minimum values for the interaction energy (ΔEmin); ΔEmin = ΔEvdw + ΔEele where, ΔEvdw refers to non-bonded van der Waals interactions and ΔEele refers to electrostatic interactions. Structures with low values for ΔEmin were selected and amino acid residues within 4 Å of the ligand were noted as possible binding sites. Graphical displays were generated using a Discovery Studio Visualizer (http://accelrys.com/products/discovery-studio/visualization.html).
3. RESULTS AND DISCUSSION
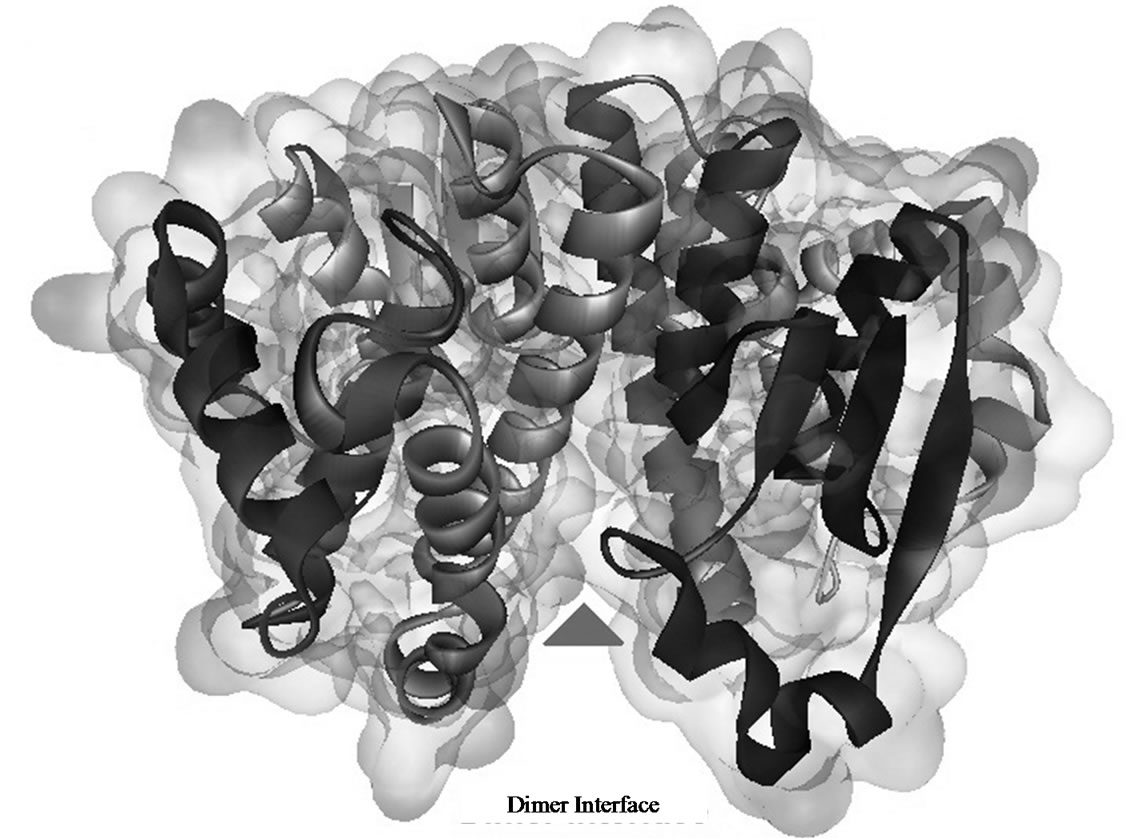
Glutathione transferases are phase-II enzymes associated with detoxification and resistance to drugs and xenobiotics. The GSTs are also implicated in intracellular signalling [1,2]. The active form of hGSTP-P1 consists of two subunits [5,6]. However, the role of enzyme subunit-subunit interactions on GST structure and activity is not fully understood. Claims that hGSTP-P1 single subunit was involved in the inhibition of c-JUN kinase [20] were discounted recently in favour of the dimer [21]. In the present work, docking investigation using hGSTP1-1 dimer (Figure 1(a)) showed that the majority ligands tested bound at sites bordering the enzyme subunit-subunit interface (site D2/D3; Table 1). Amino acid residues from sites D2/ D3 (Table 3) were partly from the 310- helix (residues 45 - 46), strand b4 (residues 51 - 64) or αC helix (residues 63 - 74) from subunit A. Other residues from subunit-B helices αD (residues 81 - 85) and αE (residues 109 - 132) were also involved in ligand binding. According to crystallographic data, some 27 residues form the inter-subunit contact [4]. However, only a sub-set of amino acid residues stabilize interactions between hGSTP1-1 subunits, notably Tyr50 which functions as a key whilst Met92, Gly96, Pro129, Phe130 and Leu133 functions as a lock [22,23]. Mutation of Tyr50 had a profound effect on the stability and kinetic properties of hGSHP1-1 [22]; Glu65, Ser66, Arg67 and Phe104 were also reported as important for subunit-subunit contact [23]. The residues important for the stabilization of hGSTP1-1 dimer are highlighted in Table 3.
The list of residues for site D2 (Tables 1 and 3) agrees closely with the only other study to our knowledge that employed hGSTP1-1 dimer as the template for docking. It was shown that 89% of docking sites for four green tea catechins occurred at the hGSTP1-1 subunit-subunit interface [24]. According to the literature, residues within 5 Å of the binding site for the catechins included (from subunit A) Gln52, Pro54, Asn67, Glu97, Asp94 and also residues that overlap with the G-site (Tyr8, Phe9, Arg14, Trp39, Gln66, Ser67, Glu98) or the H-site (Tyr104, Tyr109, Gly208). Other residues for catechin binding were (Thr68, Arg71, Arg75, Asp91, Asp95 and Asp99) located at the subunit-subunit interface [24]. Not all amino acid residues mentioned above are important for enzyme subunit-subunit association [4,19,21,22].
In the current study, glutathione and ethacrynic acid bound with the interface site D2 from hGSTP1-1 dimer rather than G-site or H-site as expected from X-ray data [7,8]. A possible explanation for this result is that using the enzyme dimer leads to steric hindrance and docking poses with unreliable energy minima [25]. As support of this view, when we used hGSTP1-1 monomer as template, glutathione bound with the G-site (Table 2 and Ta

Table 1. Energy values (ΔEmin) of ligand binding with glutathione transferases dimer*.
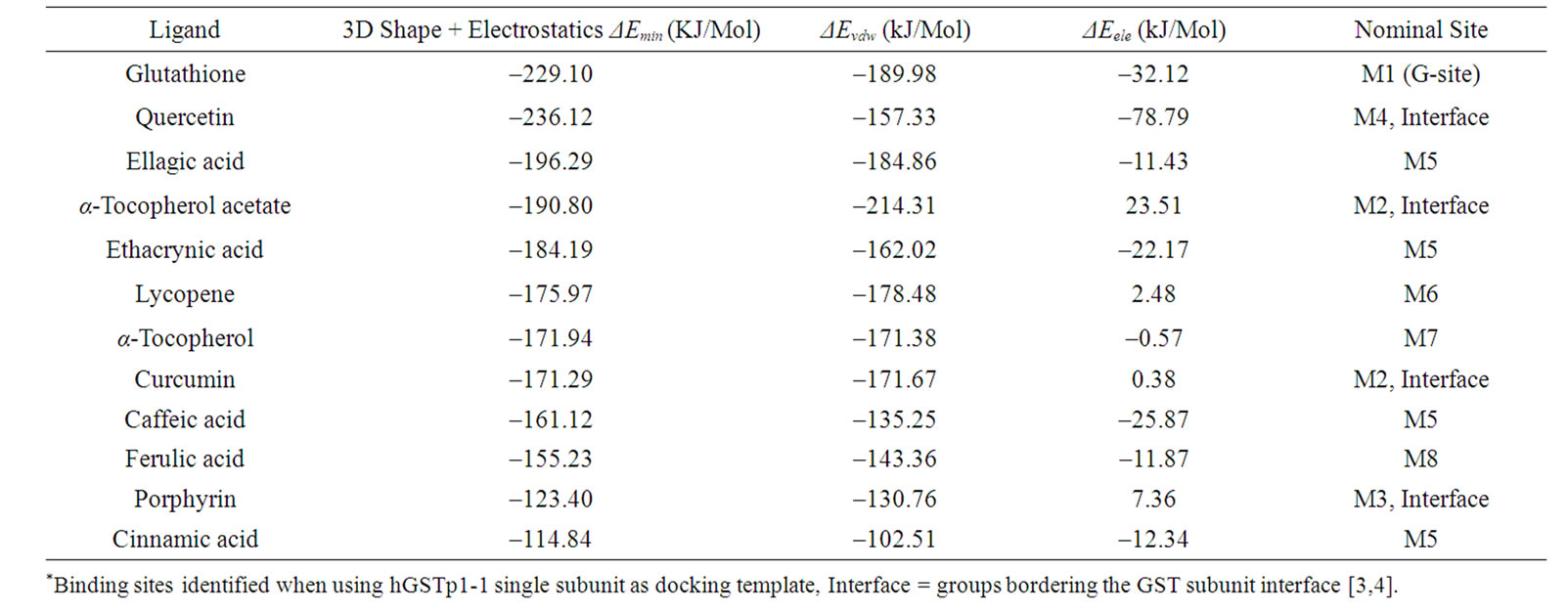
Table 2. Energy values (ΔEmin) of ligand binding with glutathione transferases monomer*.
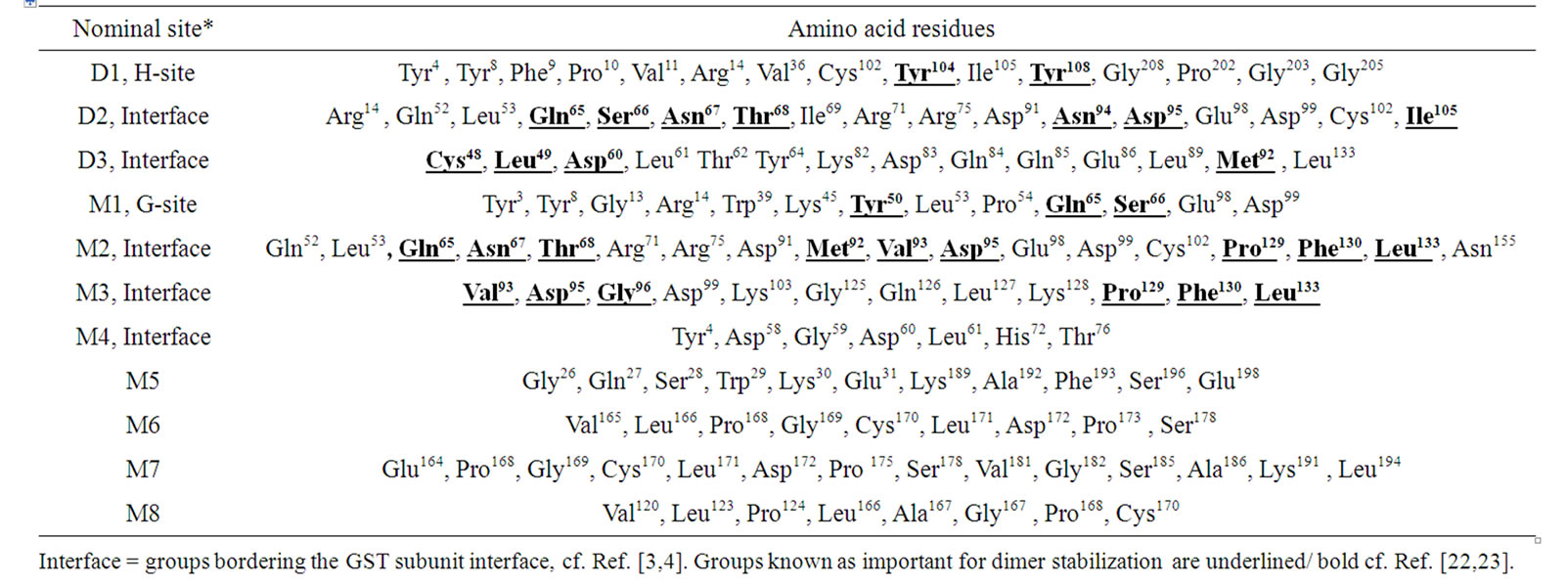
Table 3. The ligand binding sites identified using glutathione transferases dimer (D) or monomer (M) as docking template.
ble 3) near Tyr 8 (Figure 1(b)). The G-site residues identified using the hGSTP1-1 monomer as the docking template [3,7,8] was apparently inaccessible when hGSTP1-1 dimer was used as template. Clearly, the docking simulations may not accommodate conformational changes important for substrate binding to hGSTP1-1 [4,19,22, 23]. These and other limitations of docking simulations have been discussed extensively [25; and reference cited therein].
We now turn to docking simulations using a single hGSTP1-1subunit. The rational for including the GST monomer in the study was to consider possible steric restrictions to binding sites arising from the presence of two subunits. Another possible justification for including GST monomer in this study comes from the results of a recent investigation using variety of indices which showed that, the dissociation constant (Kd) for hGSTP1-1 dimer was 1.0 nanomolar [21]. Apparently, the GST will not dissociate to from the monomer at the concentration of enzyme employed in most previous studies. Current evidence also suggests that GST monomer is catalytically inactive [21]. It has also been found that substrate binding to the active sites of GST dimer may affect interface groups and vice versa [5]. Docking simulations comparing either one or two GST subunits offers a prospect for identifying ligands which bind according to the state of enzyme dissociation equilibrium and which might alter Kd and GST activity (see below).
1.0 nanomolar [21]. Apparently, the GST will not dissociate to from the monomer at the concentration of enzyme employed in most previous studies. Current evidence also suggests that GST monomer is catalytically inactive [21]. It has also been found that substrate binding to the active sites of GST dimer may affect interface groups and vice versa [5]. Docking simulations comparing either one or two GST subunits offers a prospect for identifying ligands which bind according to the state of enzyme dissociation equilibrium and which might alter Kd and GST activity (see below).
The docking studies using a single hGSTP1-1 subunit (M) as template showed a variety of binding sites (numbered M2-M8 in Table 2). Three binding sites from the single GST subunit (M2-M4; Tables 2 and 3) contained amino acid residues from the enzyme subunit-subunit interface. Site M2 (for α-tocopherol acetate) included many residues previously noted for site D2 from hGSTP1-1 dimer (Table 1). The binding site for ellagic acid, ethacrynic acid, caffeic acid, and cinnamic acid (M5; Tables 2 and 3) contained amino acid residues from helix αA of GST domain 1, and also helices αH and αI from GST domain 2 [4]. Lycopene and α-tocopherol bound with site M6 and M7 (respectively) which shared overlapping amino acid residues (Table 2). Interestingly, α-tocopherol acetate and α-tocopherol did not bind to a similar site. Binding site M6, M7 and also M8 occurred within domain-2, involving helixes αF and αG [4].
We now turn to the docking interaction energy (ΔEmin) estimates for hGSTP1-1 (Tables 1 and 2). Results using the single GST monomer with ethacryinic acid showed ΔEmin was –181.9 kJ/mol which compares with ΔEmin of –162 kJ/mol for filarial GST binding with ethacrynic acid [14]. Since two entirely different enzyme isoforms were used in these studies, some disparity between ΔEmin estimates is to be expected. Overall, the interaction energy values (and association constants, Ka) obtained from docking simulations were substantially different from values determined from wet-lab experiments. Practical measurements using isothermal titration calorimetry showed that the Gibbs free energy change (DG) for hGSTP1-1 binding with ethacrynic acid was –28 kJ/mol (Ka = 7.64 × 10–4 M–1 ) [23]. The differences between virtual and actual binding energies are probably due to inherent limitations of the docking approach [25].
In the following section, we consider the significance of docking results in terms the reported activity of selected ligands. Previous work has shown that α-tocopherol inhibits hGSTP1-1 non-competitively with the concentration for 50% inhibition (IC50) equal to (0.5 - 0.8) × 10–6 M [10,11]. Interestingly also, curcumin or ellagic acid inhibits hGSTP1-1 with an IC50 value of 1 × 10–6 M and 5 × 10–6 M and the Ka value of 1.1 × 105 M–1 (DG = –34 kJ/mol) or 8.8 × 103 M–1 (DG = –11 kJ/mol), respectively [13]. Quercetin also inhibits hGSTP1-1 but the process is irreversible, probably leading to covalent modification of Cys47 followed by disulfide bond formation. The net reaction involved quercetin oxidation of GST [12]. There was some evidence from this study that lycopene may be an important ligand for hGSP1-1 (Table 1; Figure 1(c)). However, there is no literature support for an inhibitory action of lycopene towards GST. Cell culture based studies indicate that hGSP1-1 expression is increased by the presence of lycopene [26]. It has been proposed that hGSTP1-1 may function as a carotenoidbinding protein in the eye [27]. Clearly, a wide range of interactions appear to be possible between GST and different ligands. Phenolic ligands, such as quercetin and cucumin which bind to GST reversibly at first can then produce enzyme irreversible inhibition following autoxidation to form quinone derivatives [12,13]. Perhaps in view of the solvents effects referred to above, and the wide range of interactions possible with different ligands, current docking interaction energies should not be considered as quantitative indicators of bioactivity.
Finally, it may be useful to compare docking interacttion energies observed from modelling studies using either the single GSTP1 subunit or the enzyme dimer as template. In principle, the relative strength of ligand binding to hGSTP1-1 monomer and dimer was calculated from DEmin values (Tables 1 and 2) and the relation:
Ka1/Ka2 = exp [(DEmin,1 – DEmin,2)/RT where DEmin,1 and DEmin,2 is the interaction binding energy for a particular ligand with hGSTP1-1 dimer or monomer, and Ka1 or Ka2 are the corresponding association constants. For ligands which bind preferentially to the hGSTP1-1 monomer, the ratio Ka1/Ka2 1.0. From the preceding analysis, Ka1/Ka2 ranged from 6.78 × 1014 for quercetin, 1.89 × 1014 for a-tocopherol, and 7.67 ×
1.0. From the preceding analysis, Ka1/Ka2 ranged from 6.78 × 1014 for quercetin, 1.89 × 1014 for a-tocopherol, and 7.67 ×
 (a)
(a)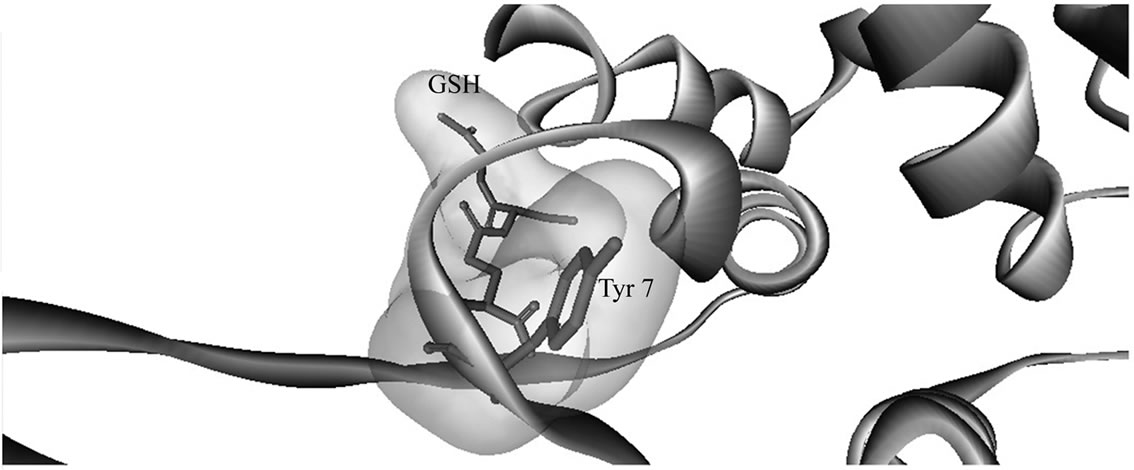 (b)
(b)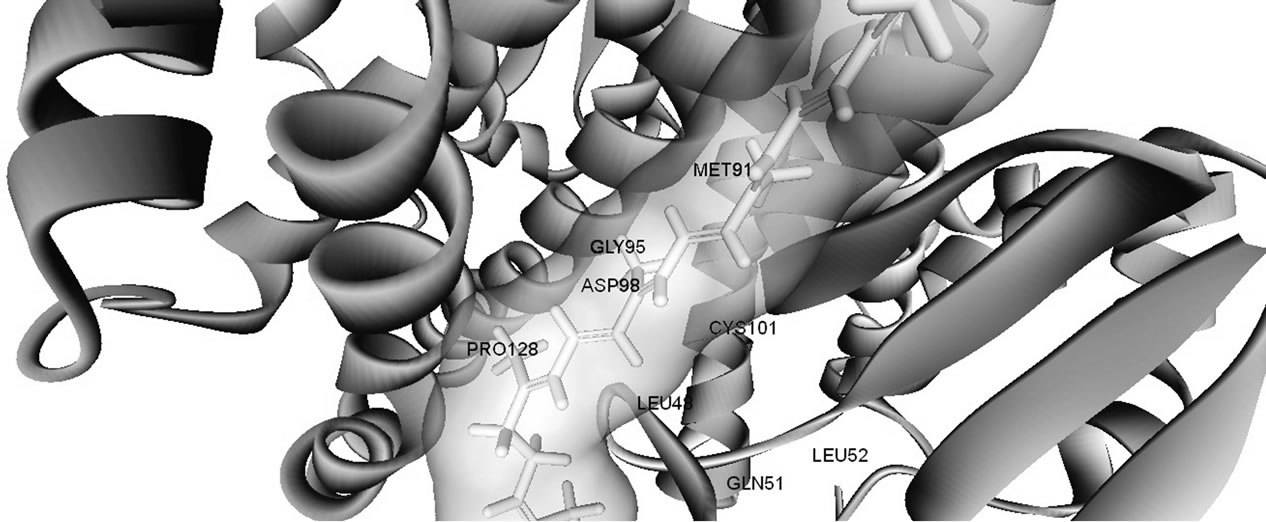 (c)
(c)
Figure 1. Three dimensional structures of human glutathione transferase P1-1 (hGSTp1-1). (a) Enzyme dimer—arrow shows the dimer interface; (b) enzyme complex with glutathione at G-site—docking simulations were with a single GST subunit; and (c) binding of lycopene at hGSTp1-1 dimer subunit interface. Docking posses were rendered with Discovery Studio Visualizer (see text for details).
1011 for curcumin. These ligands were predicted to bind more strongly to the GSTP1-1 monomer and less strongly to the enzyme dimer. By contrast, the values for Ka1/Ka2 were (= (1.0 - 5.42) × 103) moderate for ferulic acid, glutathione, caffeic acid, ellagic acid and cinnamic acid, suggesting that these ligands have only moderately higher affinity for the hGSTP1-1 monomer compared to dimer. A similar analysis showed that α-tocopherol and lycopene (Ka1/Ka2 = (3.7 - 4.87) ×10–7) may bind preferentially to hGSTP1-1 dimer compared to the monomer. These predictions suggest that the presence of different ligands might alter the dimerization state of hGSTP1-1.
In conclusion, docking simulations using the dimer form of GST (corresponding to the native enzyme) showed that most ligands bound to the interface between the two enzyme subunits. By contrast, the single enzyme subunit was found to provide a wider range of binding sites for naturally occurring ligands. Some inherent limitations of docking were considered perhaps explaining the low level of agreement between docking interaction energies and observed experimental values for enzyme-ligand binding. Finally, evidence is presented also for differential ligand binding to the GST monomer or dimer which, if confirmed by direct measurements, could open the intriguing possibility that the single hGSTP1-1 subunit might be formed under some physiological conditions. More research is needed to determine whether naturally occurring ligands can disturb the structure and stability of hGSTP1-1.
4. ACKNOWLEDGEMENTS
We are grateful to the School of Biomedical Sciences for supporting this work.
REFERENCES
- Townsend, D.M. and Tew, K.D. (2003) The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene, 22, 7369-7365. doi:10.1038/sj.onc.1206940
- Tew, K.D. and Townsend D.M. (2011) Regulatory functions of glutathione S-transferase P1-1 unrelated to detoxification. Drug Metabolism Reviews, 43, 179-193. doi:10.3109/03602532.2011.552912
- Reinemer, P., Dirr, H.W., Ladenstein, R., Schäffer, J., Gallay, O. and Huber, R. (1991) The three-dimensional structure of class π glutathione S-transferase in complex with glutathione sulfonate at 2.3 Å resolution. EMBO Journal, 10, 1997-2005. doi:10.1016/0022-2836(92)90692-D
- Dirr, H., Reinemer P. and Huber R. (1994) Refined crystal structure of porcine Pi glutathione S-transferase (pG ST P1-1) at 2.1 Å resolution. Journal of Molecular Biology, 234, 72-92. doi:10.1006/jmbi.1994.1631
- Balchin, D., Fanucchi, S., Achilonu, I., Adamson R.J., Burke, J., Fernandes, M., Gildenhuys, S. and Dirr, H.W. (2010) Stability of the domain interface contributes towards the catalytic function at the H-site of class alpha glutathione transferase A1-1. Biochimica et Biophysica Acta-Proteins and Proteomics, 1804, 2228-2233. doi:10.1016/j.bbapap.2010.09.003
- Fabrini, R., De Luca, A., Stella, L., Mei, G., Orioni, B., Ciccone, S., Federici, G., Bello, M.L. and Ricci, G. (2009) Monomer-dimer equilibrium in glutathione transferases: A critical re-examination. Biochemistry, 48, 10473-10482. doi:10.1021/bi901238t
- Oakley, A.J., Rossjohn, J., Lo Bello, M., Caccuri, A.M., Federici, G. and Parker, M.W. (1997) The three-dimensional structure of the human P1-1 class glutathione transferase P1-1 in complex with the inhibitor ethacrynic acid and its glutathione conjugate. Biochemistry, 36, 576- 585. doi:10.1021/bi962316i
- Oakley, A.J., Bello, M.L., Nuccetelli, M., Mazzetti, A.P. and Parker, M.W. (1999) The ligandin (non-substrate) binding site of human P1-1 class glutathione transferase is located in the electrophile binding site (H-site). Journal of Molecular Biology, 291, 913-926. doi:10.1006/jmbi.1999.3029
- Ralat, L.A. and Colman, R.F. (2004) Glutathione Stransferase Pi has at least three distinguishable xenobiotic substrate sites close to its glutathione-binding site. Journal of Biological Chemistry, 279, 50204-50213. doi:10.1074/jbc.M407445200
- Van Haaten, R.I.M., Evelo, C.T.A., Penders, J., Eijnwachter, M.P., Haenen, G.R. and Bast, A. (2001) Inhibition of human glutathione S-transferase P1-1 by tocopherols and α-tocopherol derivatives. Biochimica Biophysical Acta―Protein Structure and Molecular Enzymology, 1548, 23-28. doi:10.1016/S0167-4838(01)00211-4
- van Haaften, R.I.M., Evelo, C.T.A., Haenen, G.R.M.M. and Bast, A. (2001) α-Tocopherol inhibits human glutathione S-transferase. Biochemical and Biophysical Research Communications, 280, 631-633. doi:10.1006/bbrc.2000.4174
- van Zanden, J.J., Ben Hamman, O., van Lersel, M.L.P.S., Boeren, S., Cnubben, N.H.P., Bellod, M.L., Vervoort J., van Bladerena, P.J. and Rietjensa, I.M.C.M. (2003) Inhibition of human glutathione S-transferase P1-1 by the flavonoid quercetin. Chemico-Biological Interactions, 145, 39-148. doi:10.1016/S0009-2797(02)00250-8
- Hayeshi, R., Mutingwende, I., Mavengere, W., Masiyanise, V. and Mukanganyama, S. (2007) The inhibition of human glutathione S-transferases activity by plant polyphenolic compounds ellagic acid and curcumin. Food and Chemical Toxicology, 45, 286-295. doi:10.1016/j.fct.2006.07.027
- Nathan, S.T., Mathew, N., Kalyanasundaram, M. and Balaraman, K. (2005) Structure of glutathione S-transferase of the filarial parasite Wuchereria bancrofti: A target for drug development against adult worm. Journal of Molecular Modelling, 11, 194-199. doi:10.1007/s00894-005-0234-0
- Yadav, M., Singh, A., Rathaur, S. and Liebau E. (2010) Structural modelling and simulation studies of Brugia malayi glutathione-S-transferase with compounds exhibiting antifilarial activity: Implications in drug targeting and designing. Journal of Molecular Graphics and Modelling, 28, 435-445. doi:10.1016/j.jmgm.2009.10.003
- Srinivasan, L., Mathew, N. and Muthuswamy, K. (2010) In-vitro antifilarial activity of glutathione S-transferase inhibitors. Parasitology Research, 105, 1179-1182. doi: 10.1007/s00436-009-1534-6
- Kapoli, P., Axarli, I.A., Platis, D., Fragoulaki, M., Paine, M., Hemingway, J., Vontas, J. and Labrou, N.E. (2008) Engineering sensitive glutathione transferase for the detection of xenobiotics. Biosensors and Bioelectronics, 24, 498-503.
- Setzer, W. N. (2010) The molecular mechanism for DDT detoxification in Anopheles gambiae: A molecular docking study. Journal of Biophysical Chemistry, 2, 135-136. doi.10.4236/jbpc.2011.22016
- Quesada-Soriano, I., Parker, L.J., Primavera, A., CasasSolvas, J.M., Vargas-Berenguel, A., Barón, C., Morton, C.J., Mazzetti, A.P., Lo Bello, M., Parker, M.W. and García-Fuentes, L. (2009) Influence of the H-site residue 108 on human glutathione transferase P1-1 ligand binding: Structure-thermodynamic relationships and thermal stability. Protein Science, 18, 2454-2470. doi:10.1002/pro.253
- Adler, V., Yin, Z.M., Fuchs, S.Y., Benezra, M., Rosario, L., Tew, K.D., Pincus, M.R., Sardana, M., Henderson, C.J., Wolf, C.R., Davis, R.J. and Ronai, Z. (1999) Regulation of JNK signalling by GSTp. EMBO Journal, 18, 1321-1334. doi:10.1093/emboj/18.5.1321
- Gildenhuys, S., Wallace, L.A., Burke, J.P., Balchin, D., Sayed, Y. and Dirr H.W. (2010) Class Pi glutathione transferase unfolds via a dimeric and not monomeric intermediate: Functional implications for an unstable monomer. Biochemistry, 49, 5074-5081. doi:10.1021/bi100552d
- Stenberg, G., Abdalla, A.M. and Mannervik, B. (2000) Tyrosine 50 at the subunit interface of dimeric human glutathione transferase P1-1 is a structural key residue for modulating protein stability and catalytic function. Biochemical and Biophysical Research Communications, 27, 59-63. doi:10.1006/bbrc.2000.2579
- Wongsantichon, J. and Ketterman, A. J. (2006) An intersubunit lock-and-key ‘Clasp’ motif in the dimer interface of Delta class glutathione transferase. Biochemical Journal, 394, 135-144. doi:10.1042/BJ20050915
- Artali, R., Beretta, G., Morazzoni, P., Bombardelli, E. and Meneghetti, F. (2009) Green tea catechins in chemoprevention of cancer: A molecular docking investigation into their interaction with glutathione-S-transferase (GSTP1-1). Journal of Enzyme Inhibition and Medicinal Chemistry, 24, 287-295.
- Leach, A.R., Shoichet, B.K. and Peishoff, C.E. (2006) Predictions of protein-ligand interactions. Docking and scoring: Successes and gaps. Journal of Medicinal Chemistry, 49, 5851-5855.
- Leal, M., Shimada, A., Ruiz, F. and González de Mejía, E. (1999) Effect of lycopene on lipid peroxidation and glutathione-dependent enzymes induced by T-2 toxin in vivo. Toxicology Letters, 109, 1-10. doi:10.1016/S0378-4274(99)00062-4
- Bhosale, P., Larson, A.J., Frederick, J.M., Southwick, K., Thulin, C.D. and Bernstein, P.S. (2004) Identification and characterization of a Pi isoform of glutathione S-transferase (GSTP1) as a zeaxanthin-binding protein in the macula of the human eye. Journal of Biological Chemistry, 279, 49447-49454.

