World Journal of AIDS
Vol.4 No.2(2014), Article
ID:46781,8
pages
DOI:10.4236/wja.2014.42019
Two Highly Variable Vpr84 and Vpr85 Residues within the HIV-1-Vpr C-Terminal Protein Transduction Domain Control Transductionnal Activity and Define a Clade Specific Polymorphism
Jean-Hervé Colle1*, Thiery Rose2, Christine Rouzioux3, Alphonse Garcia4
1Unité de Biologie des Populations Lymphocytaires, Institut Pasteur, Paris, France
2Unité d’Immunogénétique Cellulaire, Institut Pasteur, Paris, France
3EA 3620 Université Paris Descartes, AP-HP, Laboratoire de Virologie, Hôpital Necker, Paris, France
4Laboratoire E3 Phosphatases, Unité Signalisation Moléculaire et Activation Cellulaire, Institut Pasteur, Paris, France
Email: *jhcolle@pasteur.fr, *jean-herve.colle@pasteur.fr, thierry.rose@pasteur.fr, christine.rouzioux@nck.aphp.fr, agarcia@pasteur.fr
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 19 February 2014; revised 19 March 2014; accepted 30 March 2014
ABSTRACT
The virally encoded HIV-1 viral protein R (VPR) is a multifunctional factor that is required for induced HIV-1 pathogenesis. VPR is also a cell-penetrating protein found in biological fluids from HIV-1 infected individuals. In this regard, we previously published that the C-terminal VPR77-92 sequence from HIV-1 89.6, but not from pNL4.3 strain, is a new pro-apoptotic and protein transduction domain (PTD). Here we report on a sequence analysis of VPR77-92 domain using the Los Alamos HIV-1 sequence database. The analysis showed that the two residues of the domain VPR84 and VPR85 are highly variable and differently biased in HIV-1 clade B and HIV-1 clade C. Furthermore, when Jurkat lymphoblastoid cells or PBMC were incubated with chemically synthesized peptides containing distinct VPR77-92 C-terminal sequences from clades B or C, we found that a clade-dependent polymorphism in VPR84 and VPR85 residues controlled the transducing activity of the C-terminal HIV-1 VPR77-92 domain. Together our data indicate that clade-dependent polymorphism in the VPR84 and VPR85 residues defines the transducing properties mediated by the C-terminal domain of HIV-1 VPR. Identification of this VPR polymorphism suggests new approaches to understand the HIV-1 biology and/or pathogenesis.
Keywords:VPR, HIV-1 Subtypes, Polymorphism, Transduction

1. Introduction
HIV-1 VPR is a small 14 kDa accessory virion-associated protein encoded by HIV-1. VPR supports multiple functions in vitro and is required for viral pathogenesis. Structural and mutagenesis studies have characterized various functional domains and their relationships with structural domains within VPR protein (see Ref. [1] for review). Nuclear magnetic resonance studies have revealed a flexible N-terminal region (residues 1 - 16), three a-helical domains (residues 17 - 33, 38 - 50, 54 - 77) and a flexible C-terminal region (residues 78 - 96), which is rich in basic amino acids. The first N-terminal a-helix is required for incorporation of VPR into virion and for nuclear import of the pre-integration complex (PIC) [2] . The second a-helical domain is involved in the oligomerization of VPR and the translocation of PIC in the perinuclear region [2] [3] . The third a-helix is required for the binding of VPR to uracil-DNA-glycosylase (UNG2) and for UNG2 targeting to proteosomal degradation and also for VPR dimerization [4] . This region with the flexible C-terminal sequence is involved in cell cycle arrest and apoptosis. Although the non-structured C-terminal domain appears to be essential for G2 arrest [5] , the link between G2 arrest and apoptosis and the mechanism of induction of apoptosis by VPR is not definitively established yet. The C-terminal domain is also required for the long terminal repeat-directed transcription [6] . Both R80A mutation of residue VPR80 and deletion of 18 residues of Vpr C-terminal domain unable VPR-mediated induction of G2 arrest [7] . Concerning apoptosis is a key apoptotic mechanism that requires the physical interaction of VPR with the adenine nucleotide translocator (ANT), and a component of the permeability transition pore of mitochondria has been firstly described [8] [9] . Mutational analyses showed that the region spanning amino acid residues 71 - 82, which encompass 2 tightly conserved H(F/S)RIG polypeptide motifs, are a ANT binding site providing mitochondriotoxic properties to the complex [9] . This ANT domain contains the VPR71-82 sequence that is mostly located at the end of the third α-helix of VPR (VPR55-77) but also partially overlapping with the flexible C-terminal domain (VPR77-96). In HIV-1 VPR subtype B, this ANT binding site contains three highly basic arginine residues (R73, R77, and R80) accessible for interaction with the first cytoplasmic loop of ANT. Furthermore, VPR point mutations (R77Q, R77A, and R80A), that inhibit ANT-dependent apoptosis, enhance HIV-1 replication in NL4.3 isolate suggesting a link between VPR-mediated apoptosis and HIV-1 infection [10] . However, Andersen and coworkers [11] have reported that VPR-induced apoptosis is unaffected following knockdown of ANT but requires the mitochondrial pore protein Bax. Recently we have showed that the penetrating C-terminal VPR77-92 sequence could bind to PP2A1 and induced apoptosis in human cells [12] suggesting that several issues remain to be solved to fully understand the role of VPR-mediated apoptosis induced by the virus.
In addition, it has been reported that HIV-1 VPR is a penetrating protein possessing cell transduction activities when it is added to cell culture [13] [14] . It was found that free VPR as well as VPR cleaved products are present in the culture medium of HIV-1-infected cells [15] . Consistently entire proteins or partial fragments of HIV-1 VPR have been found in serum and cerebrospinal fluid from HIV-1 infected individuals [16] [17] suggesting that both full length VPR proteins and C-terminal processed fragments could also penetrate into uninfected cells and be responsible of bystander effect.
In this study, following our previous finding that the 89.6-VPR77-92, but not the NL4.3-VPR77-92 C-terminal VPR sequence displays cell transducing capacities [12] , we reported on a genetic variability of the C-terminus domain of VPR proteins that influence its cell transducing efficiency. This variability appears to be clade-dependent. Consequence for biological significance of this newly identified HIV-1 VPR polymorphism is discussed.
2. Materials and Methods
2.1. Peptides
Carboxy-end-biotinylated (Eurogentec) or amino-end-biotinylated VPR peptides and Tat peptide (Neo system) were prepared by solid-phase peptide synthesis and purified by high-performance liquid chromatography. Peptides were dissolved in EtOH 50% and 150 mM NaCl and stored at −20˚C until use.
2.2. Cell Culture and Reagents
Jurkat lymphoblastoide T cells (clone E6, ATCC: TIB-152) were cultured in RPMI 1640 Glutamax Medium (Gibco; Invitrogen) supplemented with 10% fetal calf serum (Invitrogen).
2.3. Sequence Analysis
Based on the consensus RHSRIGIX1X2QRRaRNG sequence that recovers 85% of total VPR sequences in the Los Alamos HIV-1 sequence database, we performed statistical analysis of residue occurrence at positions 84 - 85 in two major HIV-1subtypes (clade B and C).
2.4. Transducing Studies Based on Quantification of Peptides Internalization
The peptides were pre-incubated at room temperature for 1 h with streptavidin-HRP conjugate in a 4:1 ratio prior to be incubated at 37˚C with 2.5 × 105 exponentially proliferating Jurkat cells. After 6 h, cells were washed twice in PBS, and counted prior to lysed in 300 ml of 0.1 M Tris pH 7.4, and 0.5% Nonidet P-40 buffer for 10 min on ice. A total of 50 ml of cell lysate was mixed with 50 ml of OPD buffer (51.4 mM Na2HPO4 and 24.3 mM citric acid) then mixed with 100 ml of OPD solution according to manufacturer’s instructions (Sigma). After 10 to 20 min, reaction was stopped by adding 100 ml of 1 M HCl, and optical density read at 490 nm. Cells incubated with streptavidin-HRP conjugate were used as negative controls.
3. Results
3.1. VPR84 and VPR85: Two Variable Residues Localized in the C-Terminus of HIV-1 VPR Proteins
The two homologous VPR proteins from LAI or NL4.3 strains have been commonly used in numerous studies. Previously we published that A89T and IIQ83-85VTR substitution in NL4.3 suppress the biological effects of the 89.6-VPR77-92 sequence [12] . We explore further the conservation of this domain among different HIV-1 subtypes, 727 VPR sequences from different isolates belonging to either clade B or clade C were retrieved from the Los Alamos HIV-1 sequence data base and aligned using Clustal [18] . Multi-sequence alignment provided a consensus sequence encompassing the VPR77-92 domain. This consensus is shown Figure 1 (upper panel) and compared to the homologous sequences from HIV-1LAI, NL4.3 and 89.6 strains. Occurrence of each amino acid in VPR77-92 domain was analyzed. We found that all positions in VPR77-92 appeared highly conserved (>95%), excepted the positions 84 and 85, which are variable and frequently occupied (>70%) by one of the following height residues: [VTMIL] and [RPQ] at the positions 84 and 85, respectively (Figure 1 lower panel). Interestingly we noted that the representation of the residues 84 and 85 diverged between the clade B and C. Around 70% of VPR84-85 variable residues correspond to [IT] and [RPQ] in clade B and to [IL] and [RQ] in clade C. This difference was largely increased when the 84 - 85 pair residue frequencies were considered in each clade (Figure 1 lower panel).
3.2. VPR84-85 Variability Influences the Cell Transduction Activity of VPR77-92 Sequences
HIV-1 89.6 VPR77-92 sequence both acts as transducing domain and induces apoptosis in human cells. To further study the functional consequences of the variability found at the level of 84 and 85 residues in clade B/clade C, we carried out experiments with several biotinylated VPR77-92 sequences to evaluate their internalization efficiencies. Nine similar biotinylated peptides were contained different side chain at the position 84 and 85. This panel of peptides represents 80% of VPR84-85 sequences that had been retrieved from the database as shown above. Experiments were carried out to evaluate the peptides ability to deliver a protein marker (Streptavidin-HRP) into Jurkat and PBL cells. Since VPR is detectable in nM concentration range in plasma from HIV-1 patients [13] , each peptide was tested at 25 nM. As shown in Figure 2 (upper panel), when compared to the well-characterized Tat penetrating domain, VPR84-85 peptides imported cargos in Jurkat cells in PBMC as well as (data not shown). And the cargo efficiencies varied according the nature of the two residues at the position 84 and 85 (Figure 2(a), Figure 2(b)). The presence of I84 or R85 resulted in elevated internalization potency, and
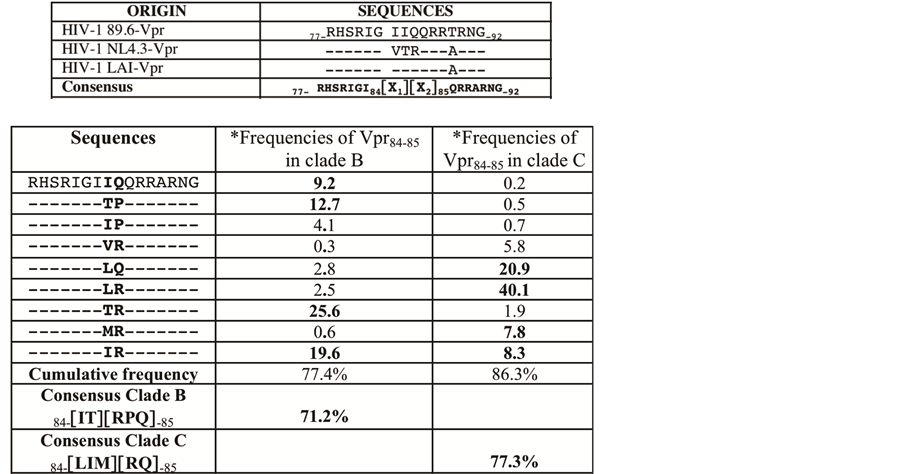
Figure 1. Residue occurrence at the two positions 84 and 85 of HIV-1 VPR is highly variable. Upper panel: Consensus sequence of VPR, VPR77-92, is presented and compared with three different 89.6, NL4.3 and LAI isolate sequences. The consensus sequence was deduced by multi sequence alignements using Clustal IX [18] . 727 VPR sequences, clade B or clade C, from the Los Alamos HIV-1 sequence database were analyzed. The residues VPR84 [X1] and VPR85 [X2] were found variable contrasting with the other flanking residues, well conserved. Single letter code for amino acids (aa) is used. Lower panel: We present the frequencies (percentage) of the most frequent associations of VPR 84 and 85 residues calculated in clade B (316 sequences) and clade C (411 sequences). The most frequent residues at the positions 84 and 85, found in each clade, are indicated as their respective cumulating frequencies.
the pairing I84/R85 provided the higher cargo capability and reached a level similar to that observed with the Tat penetrating peptide used as positive reference. Conversely the presence of V84 or Q85 residues lowered strongly the transducing efficiency. The different pairs of amino acids tested at 84 - 85 provided the following ordering deceasing efficiencies IR > TR > MR > LR > LQ > IP > VR > TP > IQ. All together our results show that the variability at the positions 84 and 85 influences critically the internalization efficiency of the C-terminal VPR77-92 sequences in the two HIV-1 clades B and C (Figure 2(b)).
4. Discussion
The HIV-1 VPR protein possesses numerous biological activities and it has been reported to be a highly conserved protein. Recently we reported that VPR77-92 C-terminal sequence of HIV-1 VPR could bind the phosphatase PP2A1 protein and possess both proapoptotic and transducing properties [12] . In this work, we aligned 727 VPR sequences deposited in the Los Alamos HIV-1 sequence database deduced from virus belonging to either clade B or C isolated from different patients. We found that within the carboxy-end the positions 84 and 85 corresponded to a spot of side chain variability contrasting with the very well conservation of the other flanking residues in the VPR77-92 sequences. The variability of residues 84 and 85 is mainly supported by three amino acids [LIT] and [RPQ] respectively. Height combinations of these six residues account roughly for 80% of the sequences of VPR retrieved from the database suggesting that the amino acid usage remains under the control of a selective pressure. Interestingly the frequency differences of the two variable residues between the clade B and clade C lead to distinct consensus sequences: 84 [IT] and 85 [RPQ] in clade B, mostly present in Northern countries [L] and [RQ] in clade C, responsible of the major epidemics in southern Africa. In addition we found that the internalization efficiencies of the VPR77-92 sequences was dictated by the nature of both residues 84 and 85.
 (a)
(a)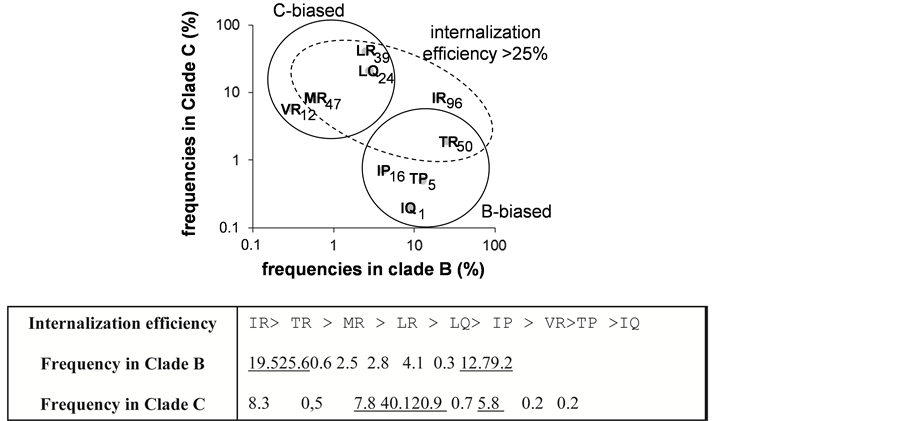 (b)
(b)
Figure 2. Transduction effect of VPR77-92 peptide and influence of residues 84 and 85. (a) Transduction activity of the most frequent peptides: Nine biotinylated peptides accounting largely for the variability of the two residues 84 and 85, found in the C-terminal domain of VPR, were used to test the delivery of streptavidin-peroxidase, when coupled to the peptides into Jurkat cells. TAT peptide signal was used to normalize the results and was expressed as 100 units = TAT signal. SD is shown for n = 3. The sequence of invariant residues was: RHSRIGI-{X1-X2}-RRARNG, and the pair of X1-X2 residues is indicated in the figure with their corresponding efficiencies. (b) Relationship between the pair residues (VPR84-85) frequencies in clade B and C and peptide internalization efficiencies: Panel: X1-X2 residue pair frequencies in clade B versus C. The internalization efficiencies are indicated. Pairs are grouped by clade bias (plain circle) or internalization frequency > 25% (dashed ellipse). The table summarizes the peptide transducing efficiencies order with their frequency in clade B or clade C.
This is illustrated on one hand by the exchange of residues I or V at position 84 and R or Q at 85 resulting in a dramatic decrease of peptide internalization efficiency and on the other hand the presence of I84 or R85 giving rise to elevated transducing effects, the pair I84/R85 providing the higher cargo effect.
VPR has been classified as an auxiliary protein since it was found dispensable for viral replication. It has been also recognized that VPR expression is required for the occurrence of HIV-1-induced pathology. Since more than twenty years, VPR has elicited a huge number of studies that have led to deciphering the variety of VPR biological functions and their relationship with its structure. The two homologous VPR proteins from LAI or NL4.3 strains were almost exclusively used in the studies. To our knowledge the question of the natural variability and function of VPR remains incompletely documented in the literature.
Our results raise the question about the biological relevance of this polymorphism in the control of VPR protein transduction domain efficiency. Lysine or arginine-rich motifs are the hallmark of cell penetrating proteins and they enable the protein transduction domains (PTDs) to cross biological membranes and to enter the cytoplasm. However, in contrast to HIV-1 TAT protein, a sequence VPR accounting for a PTD activity was not clearly established. First Sherman et al. [19] have previously reported that entire soluble VPR from NL4.3 isolate, but neither amino-end (residues 1 - 47) or carboxy-end (residues 52 - 96) can enter cells. In addition these authors showed that the entire VPR protein was also able to enter cells when fused with b-galactosidase and this effect is energy-independent and does not require any cellular receptor. As a possible mechanism to explain the absence of PTD in VPR NL4.3 sequences may be connected to previous reports indicating that this protein can form a channel in lipid bilayers and cellular membranes [20] and the amino-terminal region of 40 amino acids, including flexible amino-terminal and first a-helix VPR, which is required for the ion channel formation [21] . More recently Taguchi et al. [22] identified a shorter sequence DTWPGVEALIRILQQLLFIH FRIGCQH corresponding to the residues 52 - 78 of the third a-helix domain of a HIV-1 VPR protein (strain 98IN022) that supported transducing activity. Surprisingly, the homologous region from NL4.3 contains R77Q substitution when compared with the strain 98IN022, suggesting that this residue could be critically involved in the PTD function supported by the third helical domain of HIV-1 VPR. Interestingly we found in our study that Q in position 77 is very frequent in clade C (80%). All together these data suggest that several mechanisms might be involved in VPR transducing activity.
The role of our newly identified PTD variable sequence VPR7-92 in HIV-1 biology remains to be established. Interestingly, the well characterized and frequently used YGKKRRQRRR penetrating sequence, derived from HIV-1 TAT47-57 PTD, promotes the delivery of proteins into cells [23] [24] . It is noteworthy that this arginine-rich domain strongly resembles the motif R85QRRAR found in efficient transducing VPR7-92 sequences. Consistent with a key role of this motif, R85Q substitution strongly decreases the transducing efficiency of VPR77-92 PTD.
In addition, a specific proteolytic processing of VPR occurring at a very well conserved protein convertase cleavage site, and 85RQRR88¯ located within carboxy-end domain of VPR pNL4.3 was described [25] . It is noteworthy that such cleavage involves the R85 residue, which critically modulates the transducing efficiency of VPR77-92 PTD suggesting that protein convertases may specifically regulate the biological effects of VPR77-92 domain in some viral subtypes.
Acknowledgments
This work was supported by Institut Pasteur. The authors thank Xavier Cayla INRA Tours for helpful discussion.
References
- Romani, B. and Engelbrecht, S. (2009) Human Immunodeficiency virus Type 1 Vpr Functions and Molecular Interactions. Journal of General Virology, 90, 1795-1805.
http://dx.doi.org/10.1099/vir.0.011726-0 - Kamata, M., Nitahara-Kasahara, Y., Miyamoto, Y., Yoneda, Y. and Aida, Y. (2005) Importin-Alpha Promotes Passage through the Nuclear Pore Complex of Human Immunodeficiency Virus Type 1 Vpr. Journal of Virology, 79, 3557-3564. http://dx.doi.org/10.1128/JVI.79.6.3557-3564.2005
- Singh, S.P., Tomkowicz, B., Lai, D., Cartas, M., Mahalingam, S., Kalyanaraman, V.S., Murali, R. and Srinivasan, A. (2000) Functional Role of Residues Corresponding to Helical Domain II (Amino Acids 35 to 46) of Human Immuno-deficiency Virus Type 1 Vpr. Journal of Virology, 74, 10650-10657. http://dx.doi.org/10.1128/JVI.74.22.10650-10657.2000
- Schrofelbauer, B., Yu, Q., Zeitlin, S.G. and Landau, N.R. (2005) Human Immunodeficiency Virus Type 1 Vpr Induces the Degradation of the UNG and SMUG Uracil-DNA Glycosylases. Journal of Virology, 79, 10978-10987. http://dx.doi.org/10.1128/JVI.79.17.10978-10987.2005
- Mahalingam, S., Ayyavoo, V., Patel, M., Kieber-Emmons, T. and Weiner, D.B. (1997) Nuclear Import, Virion Incorporation and Cell Cycle Arrest/Differentiation Are Mediated by Distinct Functional Domains of Human Immunodeficiency virus Type 1 Vpr. Journal of Virology, 71, 6339-6347.
- Forget, J., Yao, X.J., Mercier, J. and Cohen, E.A. (1998) Human Immunodeficiency Virus Type 1 Vpr Protein Transactivation Function: Mechanism and Identification of Domains Involved. Journal of Molecular Biology, 284, 915-923. http://dx.doi.org/10.1006/jmbi.1998.2206
- DeHart, J.L., Zimmerman, E.S., Ardon, O., Monteiro-Filho, C.M., Arganaraz, E.R. and Planelles, V. (2007) HIV-1 Vpr Activates the G2 Checkpoint through Manipulation of the Ubiquitin Proteasome System. Virology Journal, 4, 57. http://dx.doi.org/10.1186/1743-422X-4-57
- Jacotot, E., Ravagnan, L., Loeffler, M., Ferri, K.F., Vieira, H.L., Zamzami, N., Costantini, P., Druillenec, S., Hoebeke, J., Briand, J.P., Irinopoulou, T., Daugas, E., Susin, S.A., Cointe, D., Xie, Z.H., Reed, J.C., Roques, B.P. and Kroemer, G. (2000) The HIV-1 Viral Protein R Induces Apoptosis via a Direct Effect on the Mitochondrial Permeability Transition Pore. Journal of Experimental Medicine, 191, 33-46.
- Jacotot, E., Ferri, K.F., El Hamel, C., Brenner, C., Druillennec, S., Joebeke, J., Rustin, P., Métivier, D., Lenoir, C., Geuskens, M., Vieira, H.L., Loeffler, M., Belzacq, A.S., Briand, J.P., Zamzami, N., Edelman, L., Xie, Z.H., Reed, J.C., Roques, B.P. and Kroemer, G. (2001) Control of Mitochondrial Membrane Permeabilization by Adenine Nucleotide Translocator Interacting with HIV-1 Viral Protein R and Bcl-2. Journal of Experimental Medicine, 193, 509-519.
- Rajan, D., Wildum, S., Rucker, E., Schindler, M. and Kirchhoff, F. (2006) Effect of R77Q, R77A and R80A Changes in Vpr on HIV-1 Replication and CD4 T Cell Depletion in Human Lymphoid Tissue Ex Vivo. AIDS, 20, 831-836. http://dx.doi.org/10.1097/01.aids.0000218546.31716.7f
- Andersen, J.L., De Hart, J.L., Zimmerman, E.S., Ardon, O., Kim, B. and Jacquot, G. (2006) HIV-1 Vpr-Induced Apoptosis Is Cell Cycle Dependent and Requires Bax But Not ANT. PLoS Pathogens, 2, e127. http://dx.doi.org/10.1371/journal.ppat.0020127
- Godet, A.N., Guergnon, J., Croset, A., Cayla, X., Falanga, P.B., Colle, J-H. and Garcia, A. (2010) PP2A1 Binding, Cell Transducing and Apoptotic Properties of Vpr77-92 a New Functional Domain of HIV-1 Vpr Proteins. PloS ONE, 5 e13760. http://dx.doi.org/10.1371/journal.pone.0013760
- Henklein, P., Bruns, K., Sherman, M.P., Tessmer, U., Licha, K., Kopp, J., de Noronha, C.M.C., Greene, W.C., Wray, V. and Schubert, U. (2000) Functional and Structural Characterization of Synthetic HIV-1 Vpr That Transduces Cells, Localizes to the Nucleus and Induces G2 cell Cycle Arrest. Journal of Biological Chemistry, 275, 32016-32026.
- Huang, M.B., Weeks, O., Zhao, L.J., Saltarelli, M. and Bond, V.C. (2000) Effects of Extracellular Human Immunode-ficiency virus Type 1 Vpr Protein in Primary Rat Cortical Cell Cultures. Journal of Neurovirology, 6, 202-220.
- Hoshino, S., Sun, B., Konishi, M., Shimura, M., Segawa, T., Hagiwara, Y., Koyanagi, Y., Iwamoto, A., Mimaya, J., Terunuma, H., Kano, S. and Ishizaka, Y. (2007) Vpr in Plasma of HIV Type 1-Positive Patients Is Correlated with the HIV Type 1 RNA Titers. AIDS Research and Human Retroviruses, 23, 391-397. http://dx.doi.org/10.1089/aid.2006.0124
- Levy, D.N., Refaeli, Y., MacGregor, R.R. and Weiner, D.B. (1994) Serum Vpr Regulates Productive Infection and Latency of Human Immunodeficiency Virus Type 1, 3. Proceedings of the National Academy of Sciences USA, 91, 10873-10877. http://dx.doi.org/10.1073/pnas.91.23.10873
- Levy, D.N., Refaeli, Y. and Weiner, D.B. (1995) Extracellular Vpr Protein Increases Cellular Permissiveness to Human Immunodeficiency Virus Replication and Reactivates Virus from Latency. Journal of Virology, 69, 1243-1252.
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F. and Higgins, D.G. (1997) The CLUSTAL_X Windows Interface: Flexible Strategies for Multiple Sequence Alignment Aided by Quality Analysis Tools. Nucleic Acids Research, 25, 4876-4882. http://dx.doi.org/10.1093/nar/25.24.4876
- Sherman, M.P., Schubert, U., Williams, S.A., de Noronha, C.M.C., Kreisberg, J.F., Henklein, P. and Greene, W.C. (2002) HIV-1 Vpr Displays Natural Protein-Transducing Properties: Implications for Viral Pathogenesis. Virology, 302, 95-105. http://dx.doi.org/10.1006/viro.2002.1576
- Piller, S.C., Ewart, G.D., Premkumar, A., Cox, G.B. and Gage, P.W. (1996) Vpr Protein of Human Immunodeficiency Virus Type 1 form Cation-Selective Channels in Planar Lipid Bilayers. Proceedings of the National Academy of Sciences USA, 93, 111-115. http://dx.doi.org/10.1073/pnas.93.1.111
- Piller, S.C., Ewart, G.D., Jans, D.A., Gage, P.W. and Cox, G.B. (1999) The Amino-Terminal Region of Vpr from Human Immunodeficiency Virus Type 1 Forms Ion Channels and Kills Neurons. Journal of Virology, 73, 4230-4238.
- Taguchi, T., Shimura, M., Osawa, Y., Suzuki, Y., Mizoguchi, I., Niino, K., Takabu, F. and Ishizaka, Y. (2004) Nuclear Trafficking of Macromolecules by an Oligopeptide Derived from Vpr of Human Immunodeficiency Virus Type-1. Biochemical and Biophysical Research Communications, 320, 18-26. http://dx.doi.org/10.1016/j.bbrc.2004.05.126
- Schwarze, S.R., Ho, A., Vocero-Akbani, A. and Dowdy, S.F. (1999) In Vivo Protein Transduction: Delivery of a Biologically Active Protein into the Mouse. Science, 285 1569-1572.
http://dx.doi.org/10.1126/science.285.5433.1569 - Caron, N.J., Torrente, Y., Camirand, G., Bujold, M., Chapdelaine, P., Leriche, K., Bresolin, N. and Tremblay, J.P. (2001) Intracellular Delivery of a Tat-eGFP Fusion Protein into Muscle Cells. Molecular Therapy, 3, 310-318. http://dx.doi.org/10.1006/mthe.2001.0279
- Xiao, Y., Chen, G., Richard, J., Rougeau, N., Li, H., Seidah, N.G. and Cohen, E.A. (2008) Cell-Surface Processing of Extracellular Human Immunodeficiency Virus Type 1 Vpr by Proprotein Convertases. Virology, 372, 384-397. http://dx.doi.org/10.1016/j.virol.2007.10.036
NOTES

*Corresponding author.

