World Journal of AIDS
Vol. 2 No. 2 (2012) , Article ID: 19724 , 7 pages DOI:10.4236/wja.2012.22010
Genotypic Distribution of Different Variants of Oncogenic Human Papilloma Virus (HPV) among the Sexually Active HIV-1 Positive Female Population from Manipur, India
![]()
1HIV/AIDS Laboratory, Division of Virology, National Institute of Cholera & Enteric Diseases (Indian Council of Medical Research), Government of India, Kolkata, India; 2Regional Institute of Medical Sciences, Imphal, India.
Email: {ranajoymullick, shanjbati, kamalesh.sarkar}@gmail.com, ronisarkar@zapak.com, #drsekharchakrabarti@yahoo.co.in
Received January 17th, 2012; revised February 21st, 2012; accepted April 1st, 2012
Keywords: HPV-HIV coinfection; oncogenic variants; high risk groups; genotyping
ABSTRACT
A total of 531 sexually active female populations from Manipur (north-eastern India), was chosen for this study. Out of 531 females from Manipur, 111 (20.9%) were HIV positive and 420 were HIV negative (79%). PCR amplification of the MY region of the HPV L1 gene using consensus MY09/11 primer showed 3.7% positivity in Manipur. Interestingly HPV infection among the HIV infected population from Manipur was found to be higher (9%) compared to the non HIV infected population from Manipur (3.7%). Further, single PCR for detecting the 16/18 genotypes and multiplex PCR for the other high risk groups showed equal prevalence of 16 and 18 and other high risk groups (1.3% each) in Manipur. This result was further confirmed by the sequencing and phylogenetic analysis. Thus, our study showed fairly high HPV prevalence rate among the HIV infected population compared to the non HIV cases in Manipur and also equal prevalence of 16/18 genotypes with other high risk groups. According to our knowledge this is the first kind of a genotypic study among the HPV-HIV co-infected population from northeastern states of India.
1. Introduction
The 16 and 18 oncogenic variants of the human papilloma virus are the p revalent genotypes responsible for causing cervical cancer among sexually active female population worldwide and in India. The main objective of our study was to find the HPV prevalence among the HIV-1 infected sexually active female population from Manipur with identification of the number of oncogenic variant types 16/18, circulating among this group of population.
Cervical cancer has become one of the major sexually transmitted diseases worldwide especially in the developing countries like India. Human papilloma virus (HPV) is considered to be the sole etiological agent for the cervical cancer cases worldwide. India has the highest number of annual incident cases and mortality rates for cervical cancer worldwide. The 16 and 18 variants of HPV has been found to be the major cause of cervical cancer cases in India. A recent study conducted in Chennai, South India had shown that HPV 16 had a high prevalence of uterine cancer compared to HPV 18 cases, although the development of invasive cancer from precancerous lesions was more for HPV18 compared to HPV16 [1]. Study conducted in North India had detected prevalence of 16/18 among women attending a local hospital with intraepithelial lesions [2]. Another study among the young married women from North India had identified HPV16 as the commonest high-risk type (3%) followed by HPV52 (1.2%) and HPV51 (0.8%) [3]. Recent study from four different regions across India also showed that other than the prevalent HR (High Risk) variant 16 followed by 18 (79.6%), there has also been prevalence of other high risk types (12.4%) such as 45, 73, 31, 56, 52, 58, 59, 33, 68, 51, 35, 26, and 39 [4].
Additionally, highly oncogenic HPV16 and 18 variants might be expected to have a particularly aggressive natural history in the presence of diminished host immune status, such as in HIV-positive women [5-8]. Women with human immunodeficiency virus (HIV)/ AIDS are at elevated risk of cervical cancer, as shown by studies of population-based US AIDS and cancer registries [9,10]. In this context Manipur has always been in focus of our study due to the prevalence of different wild type and recombinant variants of HIV-1 found in this region as shown by our earlier studies [11,12]. To our knowledge there haven’t been any study from this part of India which dealt with the molecular epidemiology point of view of the prevalence rate of HPV infection with HIV-1 infection. The main objective of our study was to compare the HPV prevalence rate among the HIV infected and noninfected sexually active high risk and low risk population of Manipur, India and also to identify the oncogenic variant types circulating among this group of population.
2. Materials & Methods
2.1. Collection of Cervical Swab Samples
Cervical swab samples were collected with the help of cytobrush from the sexually active female patients in HIViral Transport Kit (HIMEDIA, India) from the clinics of Manipur and transported to laboratory within a few hours. A total of 531 samples from Manipur were collected. Out of 531 samples from Manipur, 420 were HIV negative and 111 was HIV positive. After receiving the samples, collection tubes were vortexed thoroughly and the samples were aliquoted in a micro centrifuge tube and stored at –70˚C until further processing.
2.2. Extraction of Viral DNA
The integrated HPV DNA was extracted from the cervical cells with the QIAamp Blood DNA Mini Kit (Qiagen, Germany) using the manufacturers’ instruction. Carrier RNA (5 - 10 µg) was added directly to the lysis buffer prior to lysis. Elution of the viral genomic DNA was done at 60 µl.
2.3. PCR Amplification of the HPV Concensus M Region of L1 Gene, 16 and 18 Genotypes
The HPV L1 My region PCR amplification was carried out using the concensus primer pairs My09/11 resulting in a 450 bp product. Final concentrations of 1× PCR Buffer, 1.5 mM MgCl2, 0.8 mM of dNTP mix, 20 pmole/µl of My09 and My11 primer pairs, 2.5 U of ampli Taq gold (Applied Biosystems, USA) and appropriate amount of the template DNA. The PCR condition followed was 94˚C for 15 min, 35 cycles of 94˚C for 30 sec, 57˚C for 30 sec and 72˚C for 1 min with final extension of 72˚C for 10 min. An aliquot was analyzed in 1% agarose gel (Figure 1).
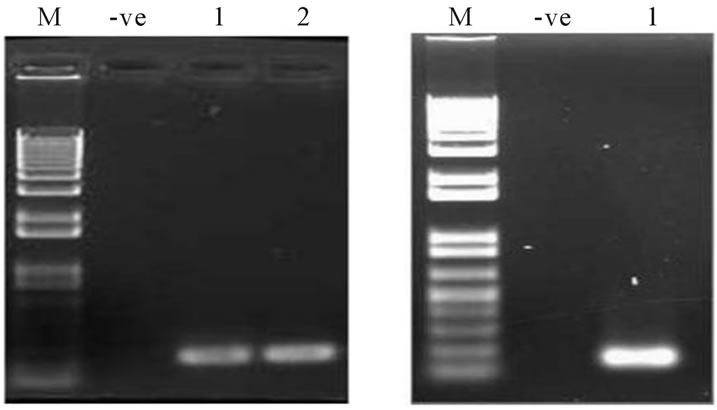
This was followed by type specific PCR of the high risk oncogenic groups 16 and 18 for all the samples from both the population. The protocol and condition followed for both 16 and 18 genotypes were same. Final concentrations of 1× PCR Buffer, 1.5 mM MgCl2, 0.8 mM of dNTP mix, 20 pmole/µl of type specific primer pairs 16F/16R and 18F/18R for 16 & 18 respectively, 2.5 U of ampli Taq gold (Applied Biosystems) and appropriate amount of the template DNA. The PCR condition followed for both was 94˚C for 15 min, 30 cycles of 94˚C for 30 sec, 53˚C for 30 sec and 72˚C for 1 min with final extension of 72˚C for 10 min. An aliquot was analyzed in 1% agarose gel (Figures 2(a) and (b)). PCR amplification for the 16 and 18 genotype resulted in a 250 bp and 145 bp fragments respectively. Samples were also tested for HPV genotyping using the Linear Array HPV genotyping test kit (Roche) following the manufacturer’s instruction.
2.4. PCR Amplification of the Other High Risk Groups
After identification of the circulating 16 and 18 genotypes, multiplex PCR was done for other positive strains in order to identify any possible circulating other high

Figure 1. PCR amplification of the My region of HPV positive samples. Lane Nos. 1 - 8 corresponds to the My amplicons. M corresponds to 500 bp DNA ladder. -ve corresponds to negative control.
 (a) (b)
(a) (b)
Figure 2. PCR amplification of HPV 16 & 18 genotypes. (a) Lane nos. 1 - 2 denotes amplicons corresponds to HPV16 genotype; (b) Lane1 denotes to amplicon corresponding to HPV18 amplicon. M corresponds to 1Kb plus DNA ladder. -ve corresponds to negative control.
risk groups among the two groups of population. The reaction mixture included final concentrations of 1× PCR Buffer, 1.5 mM MgCl2, 0.8 mM of dNTP mix, 20 pmole/µl of cocktails of set of four pairs of primers for major high risk groups other than 16 and 18 (Table 1), 2.5 U of ampli Taq gold (Applied Biosystems) and appropriate amount of the template DNA. The PCR condition followed was 94˚C for 15 min, 30 cycles of 94˚C for 30 sec, 55˚C for 30sec and 72˚C for 1 min with final extension of 72˚C for 10 min. An aliquot was analyzed in 1% agarose gel. The sequences of the 18 type specific primers used in the study [13] and their corresponding position in the viral genome is shown in Table 1. For HPV6 and 11 only one specific primer pair was used because of their sequence homologies and biological similarity.
2.5. Sequencing & Phylogenetic Analysis
In order to identify the genotypes of the other circulating variants of HPV among the sexually active female populations of Manipur, sequencing and phylogenetic analysis of all the positive samples (My positive) other than those positive for 16 & 18 was done. Amplicons corresponding to the 450 bp of My region were purified by a QIAquick PCR purification kit (QIAGEN, Germany) and subjected to cycle sequencing reactions using fluorescent dye-labeled dideoxy nucleotides in an ABI PRISM 3100 automated sequencer following the manufacturer’s protocol.


Table 1. Sequences of type-specific PCR primers used in this study (genotypes in bold are the detected high risk groups among the study population).
All the sequences were edited using BIOEDIT program (version 5.0.6; Department of Microbiology, North Carolina State University) [http://www.mbio.ncsu.edu/BioEdit/ BioDoc.pdf]. Nucleotide sequences were analyzed with the respective sequences of HPV strains isolated from different geographic regions available in the HPV sequence database (http://www.stdgen.lanl.gov/). All the nucleotide sequences obtained in this present study were screened using the BASIC BLAST program to search for sequence similarities for previously reported sequences in the databases, and to rule out potential laboratory errors. Sequences were aligned with all the reference strains, using CLUSTAL W (version 1.4) multiple sequence alignment program. Evolutionary distances were measured by a Kimura two-parameter distance matrix method and a phylogenetic tree was constructed by Neighbor-joining (NJ) method using the Interior Branch Test of Phylogeny with 1000 bootstrap replicates, in MEGA version 4.
3. Result
3.1. PCR Amplification of the HPV Concensus My Region of L1 Gene, 16 and 18 Genotypes from Manipur
PCR amplification of the HPV concensus My region of L1 gene from 531 samples from Manipur showed 20 samples as HPV positive (3.7%). Again the prevalence rate among the HIV infected population from Manipur was higher 10/111 (9%) compared to the noninfected population 15/420 (3.5%) of the same (Figure 3). Statistical analysis among the two group of population showed an unbiased standard deviation of 3.889, a mean value of 6.250 and the corrected standard error of the mean of 2.747. The results were found to be statistically significant.

Figure 3. Graph showing percentage prevalence of HPV infection among HIV-1 infected (1) and non HIV infected (2) sexually active female population from Manipur. The error bar represents the values for the Standard Errors. The percentage values were found to be statistically significant.
PCR amplification of the 16 and 18 genotypes from 20 positive samples from Manipur showed 7 samples as 16 positive (RIMS209, RIMS229, RIMS189, RIMS73, SASO10, SASO29, SASO34) and none as 18 positive i.e. a total of 7 positive cases for oncogenic 16/18 variants out of 531 (1.3%). The results were consistent with the results obtained from the Roche kit (data not shown).
3.2. Detection of Other High Risk Groups by Multiplex PCR
PCR amplification of the other high risk groups from the positive samples by multiplex PCR other than 16 and 18, revealed presence of quite a good number of high risk groups both in Manipur. Out of 20 positive samples 7 were found to be positive for high risk groups other than 16 and 18. Analysis showed samples were positive for the high risk groups HPV58, HPV33 and HPV39 (Figure 4). The population therefore has a prevalence rate of 1.3% (7/531) for the high risk groups of other types.
3.3. Sequencing and Phylogenetic Analysis
Sequencing and phylogenetic analysis results were consistent with the results obtained from the multiplex PCR. Analysis of the HPV My region of the Manipur samples showed sample no. SASO34, RIMS30; SASO83, SASO90; RIMS110 were positive for the high risk groups HPV58, HPV33 and HPV39 respectively (Figure 5). On the other hand SASO52, SASO88; RIMS68, RIMS150 were positive for the probable high risk group HPV66 and HPV53. Sample number RIMS314 and SASO01 were positive for the low risk group HPV61. The GenBank accession numbers for the Manipur sequences are available as JN383596- JN383608.
4. Discussion
Establishment of a persistent infection by oncogenic
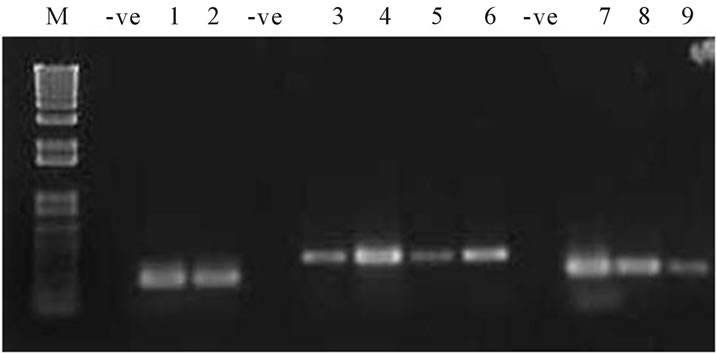
Figure 4. PCR amplification of the HPV high risk and probable high risk groups. Lane nos. 1 - 2 denotes amplicons corresponding to HPV 66 genotype. Lane nos. 3 - 6 corresponds to HPV 33, Lane nos. 7, 8 corresponds to HPV 58 and Lane 9 corresponds to HPV 39. M corresponds to 1Kb plus DNA ladder. -ve corresponds to negative control.

Figure 5. Phylogenetic analysis of the HPV My region of the 13 samples from Manipur which were positive other than those positive for 16 and 18 genotypes, showed prevalence of different high risk groups along with the probable high risk and low risk groups.
human papillomavirus (HPV) is thought to be central to the development of most cases of cervical cancer. Over 95% of cervical tumour specimens harbour HPV DNA. Two HPV types, HPV 16 and 18, alone account for approximately 50% and 14% of cervical cancer cases worldwide respectively [14,15].
Studies have further confirmed that HPV persistence is moderately increased by HIV coinfection. In this study we have investigated the incidence or prevalence of Human Papilloma Virus infection among the sexually active female population from Manipur (north-eastern India), a certain percentage (20.9%) of which were HIV positive as well as HIV negative (79%). PCR analysis of the My region of the HPV L1 gene among these population showed 3.7% of positive cases among the female populations from Manipur. Further the prevalence level of HPV infection among the HIV infected population from Manipur was also found to be higher (9%) compared to the non HIV infected population from Manipur (3.5%).
Interestingly genotyping of the positive samples for the high risk groups other than 16/18 by multiplex PCR showed equal prevalence of other high risk groups in Manipur compared to 16/18. For Manipur, the percentage was same (1.3%) in both the cases. The primers used for type specific amplification for other high risk groups were very specific and did not give any nonspecific amplification. The result was further confirmed by the sequencing and phylogenetic analysis for the My region for respective populations.
A recent study from Eastern India [16] has also shown prevalence of HPV 16, 18 among HIV positive females from Kolkata (32.2%; n = 30) was higher than HIV negative females (9.1%; n = 101). About 53% (23/43) of cases with oncogenic HPV were infected with genotypes other than 16, 18 either as single/multiple infections.
Further observing the increasing trend towards prevalence of different high risk groups other than 16/18 in both Kolkata and Manipur, we also agree according to Pillai et al. 2010 that considering the prevalence of other high risk groups circulating among these two groups of population, other than the prophylactic HPV-16/18-L1 vaccine a broad-based vaccine approach would be appropriate for India. At present there are two HPV vaccines under trial in India: Gardasil (Donated by Merck & Co., Inc.) and Cervarix (GlaxoSmithKline). Both vaccines have demonstrated high efficacy against precancerous lesions caused by 16/18 types of HPV. Gardasil, a quadrivalent human papillomavirus vaccine is designed based on HPV types 6, 11, 16, 18. Cervarix is based on HPV types 16 and 18. But considering the prevalence of high risk groups other than 16 and 18 among Indian population as mentioned above, there is a need to think about a suitable candidate vaccine for the Indian population. According to our knowledge this is a first kind of study among the HPV-HIV coinfected population from north-eastern states of India. Further indepth study is necessary to understand the dynamics of HPV in presence of the HIV infection among the study population.
5. Acknowledgements
We thank Department of Biotechnology, Govt. of India for financial assistance and all the staff members of OB/Gyn dept of RIMS, Imphal Manipur for their support.
REFERENCES
- V. M. B. Grace, “HPV Type 18 Is More Oncopotent than HPV16 in Uterine Cervical Carcinogenesis Although HPV16 is the Prevalent Type in Chennai, India,” Indian Journal of Cancer, Vol. 46, No. 3, 2009, pp. 203-207.
- R. Arora, A. Kumar, B. K. Prusty, U. Kailash, S. Batra and B. C. Das, “Prevalence of High-Risk Human Papillomavirus (HR-HPV) Types 16 and 18 in Healthy Women with Cytologically Negative Pap Smear,” European Journal of Obstetrics & Gynecology and Reproductive Biology, Vol. 121, No. 1, 2005, pp. 104-109.
- P. Datta, N. Bhatla, L. Dar, A. R. Patro, A. Gulati, A. Kriplani and N. Singh, “Prevalence of Human Papillomavirus Infection among Young Women in North India,” Cancer Epidemiology, Vol. 34, No. 2, 2010, pp. 157-161.
- R. M. Pillai, J. M. Babu, V. T. Jissa, S. Lakshmi, S. V. Chiplunkar, M. Patkar, et al., “Region-Wise Distribution of High-Risk Human Papillomavirus Types in Squamous Cell Carcinomas of the Cervix in India,” International Journal of Gynecological Cancer, Vol. 20, No. 6, 2010, pp. 1046-1051.
- A. K. Chaturvedi, J. A. Brinkman, A. M. Gaffga, J. Dumestre, R. A. Clark, P. S. Braly, K. Dunlap, P. J. Kissinger and M. E. Hagensee, “Distribution of Human Papillomavirus Type 16 Variants in Human Immunodeficiency Virus Type 1-Positive and -Negative Women,” Journal of General Virology, Vol. 85, No. 5, 2004, pp. 1237-1241.
- M. H. Mayrand, F. Coutlee, C. Hankins, N. La pointe, P. Forest, M. De Ladurantaye and M. Roger, “Detection of Human Papillomavirus Type 16 DNA in Consecutive Genital Samples Does Not Always Represent Persistent Infection as Determined by Molecular Variant Analysis,” Journal of Clinical Microbiology, Vol. 38, No. 9, 2000, pp. 3388-3393.
- L. Perez-Gallego, G. Moreno-Bueno, D. Sarrio, A. Suarez, C. Gamallo and J. Palacios, “Human Papillomavirus-16 E6 Variants in Cervical Squamous Intraepithelial Lesions from Hiv-Negative and HIV-Positive women,” American Journal of Clinical Pathology, Vol. 116, No. 1, 2001, pp.143-148. doi:10.1309/42U5-Q2EV-RCBM-WPGJ
- L. F. Xi, C. W. Critchlow, C. M. Wheeler, et al.,” Risk of Anal Carcinoma in Situ in Relation to Human Papillomavirus Type 16 Variants,” Cancer Research, Vol. 58, No. 17, 1998, pp. 3839-3844.
- J. J. Goedert, T. R. Cote, P. Virgo, S. M. Scoppa, D. W. Kingma, M. H. Gail, et al., “Spectrum of AIDS Associated Malignant Disorders,” The Lancet, Vol. 351, No. 9119, 1998, pp. 1833-1839. doi:10.1016/S0140-6736(97)09028-4
- S. M. Mbulaiteye, R. J. Biggar, J. J. Goedert and E. A. Engels, “Immunodeficiency and Risk for Malignancy among Persons with AIDS,” Journal of Acquired Immune Deficiency Syndromes, Vol. 32, No. 5, 2003, pp. 527-533. doi:10.1097/00126334-200304150-00010
- P. Bhanja S. Sengupta, N. Y. Singh, K. Sarkar, S. K. Bhattacharya and S. Chakrabarti, “Determination of Gag and Env Subtypes of HIV-1 Detected among Injecting Drug Users (IDUs) in Manipur, India: Evidence for Intersubtype Recombination,” Virus Research, Vol. 114, No. 1-2, 2005, pp. 149-153.
- R. Mullick, S. Sengupta, K. Sarkar and S. Chakrabarti, “Molecular Characterization of Tat Gene and Long Terminal Repeat Region of Human Immunodeficiency Virus Type-1 Detected among the Injecting Drug Users (IDUs) of Manipur, India: Identification of BC Recombinants,” Virus Research, Vol. 147, No. 2, 2010, pp. 195-201. doi:10.1016/j.virusres.2009.10.024
- K. Sotlar, D. Diemer, A. Dethleffs, Y. Hack, A. Stubner, N. Vollmer, S. Menton, M. Menton, K. Dietz, D. Wallwiener, R. Kandolf and B. Bültmann, “Detection and Typing of Human Papillomavirus by e6 Nested Multiplex PCR,” Journal of Clinical Microbiology, Vol. 42, No. 7, 2004, pp. 3176-3184.
- F. X. Bosch, M. M. Manos, N. Muñoz, M. Sherman, A. M. Jansen, J. Peto, M. H. Schiffman, V. Moreno, R. Kurman and K. V. Shah, “Prevalence of Human Papillomavirus in Cervical Cancer: A Worldwide Perspective. International Biological Study on Cervical Cancer (IBSCC) Study Group,” Journal of the National Cancer Institute, Vol. 87, No. 11, 1995, pp. 796-802. doi:10.1093/jnci/87.11.796
- J. M. Walboomers, M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer and N. Muñoz, “Human Papillomavirus is a Necessary Cause of Invasive Cervical Cancer Worldwide,” The Journal of Pathology, Vol. 189, No. 1, 1999, pp. 12-19.
- K. Sarkar, R. Pal, B. Bal, B. Saha, S. Bhattacharya, S. Sengupta, P. P. Mazumdar and S. Chakrabarti, “Oncogenic HPV among HIV Infected Female Population in West Bengal, India,” BMC Infectious Diseases, Vol. 11, No. 22, 2011, pp. 11-72.
NOTES
*Both the authors contributed equally.
#Corresponding author.

