American Journal of Plant Sciences
Vol.4 No.2A(2013), Article ID:28457,8 pages DOI:10.4236/ajps.2013.42A052
Insight to the Mode of Action of Allium sativum Leaf Agglutinin (ASAL) Expressing in T3 Rice Lines on Brown Planthopper
![]()
1Division of Plant Biology, Bose Institute, Kolkata, India; 2The Protein Analysis Facility, Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland.
Email: bala.arpita9@gmail.com, amitroy_81@yahoo.co.in, #sampa@bic.boseinst.ernet.in
Received December 19th, 2012; revised January 20th, 2013; accepted January 27th, 2013
Keywords: Allium sativum Leaf Agglutinin (ASAL); Hemipteran Pest Management; Brown Planthopper; NADH Quinone Oxidoreductase (NQO); Fecundity; Transgenic Rice; Brush Boarder Membrane Vesicle (BBMV) Protein
ABSTRACT
Brown planthopper, the sap sucking hemipteran pest, is one of the major contributors to the yield loss of rice through the world. To combat the situation researchers are interested identifying genes from plant origin having potentiality to develop hemipteran pest resistance. Interestingly, it was observed that rice plants expressing ASAL, a monocot mannose binding lectin, showed significant resistance to brown planthopper and green leafhopper. Additionally, antibiotic resistant marker gene free ASAL expressing rice lines were developed to overcome the biosafety issues. However, the basis behind the resistance against planthoppers is still not clearly understood. Ligand blot assay was performed with total BBMV protein from BPH and a ~56 kDa receptor protein was detected. LC MS/MS analysis revealed that the receptor protein is NADH quinone oxidoreductase (NQO), a key player in electron transport chain, insect defense response and male/female gametogenesis. Presumably interaction of ASAL with NQO may lead to toxicity and loss of fecundity among BPH feeding on ASAL expressing transgenic rice plants. These findings provide a stable scientific basis for considering these transgenic ASAL expressing rice plants as significant product for combating BPH attack associated yield loss of rice.
1. Introduction
Rice (Oryza sativa L.) is the most important food crops in the world and about 90% of the world’s rice is grown and consumed in Asia [1]. Production of this crop is greatly hampered by several biotic and abiotic factors of which sap sucking hemipteran pests namely green leafhopper (GLH, Nephotettix virescens) and brown planthopper (BPH, Nilaparvata lugens) are highly devastating that cause huge amount of yield loss every year [2].
Between the two hemipterans, BPH is the most damaging pest of rice in several Asian countries as reported in the last 10 years which may be avoided if rice ecosystem is better managed [3]. Direct feeding practice of BPH drains out plant fluids and essential nutrients. During high infestation, plants become brown and finally die.
These hoppers not only damage the plants by sucking the plant sap but also act as vectors of plant viruses [4]. To develop BPH resistant lines, several genes such as Bph1 and Bph2 conferring resistance to the pest have been identified and incorporated in different popular rice varieties by conventional breeding method [5]. Unfortunately, the major genes resistant to BPH were not stable [6]. Therefore, introduction of exotic pest-resistant genes into popular rice cultivars through Agrobacterim mediated genetic transformation technique has been preferred.
Among different important insect resistant genes, Bt toxins are one of the most promising proteins but unfortunately Bt toxins remain ineffective against this group of sap-sucking pests [7]. On the other hand plant lectins, specially monocot mannose binding lectins like GNA, ASAL etc. showed significant insecticidal efficacy both in native as well as expressed conditions [8-12]. Interestingly, Allium sativum leaf agglutinin (ASAL) from garlic leaf expressed in transgenic rice exhibited significant anti-metabolic effect towards BPH and GLH [13].
However, scientists have great concern about the public acceptance of these plants due to unwanted incorporation of antibiotic resistance gene, Hygromycin phosphotransferase (hpt II), as plant selection marker. Therefore attempts were made to achieve successful elimination of hpt II marker gene by using the cre/lox recombination system. Bacteriophage P1 specific loxP sites were introduced into the gene construct, flanking the antibiotic resistance gene. Independent ASAL-lox-hpt II-lox plants were generated. Cre recombinase gene was introduced separately in rice line. Both cre/lox were brought together by crossing between ASAL-lox T0 and cre T0 plants. The resultant ASAL expressing hybrid rice plants were obtained without antibiotic resistant marker gene sequences which showed significant degree of resistance against BPH and GLH. The T3 progeny plants of one such line were particularly used in the present study. Among the several strategies used for eliminating antibiotic or herbicide resistant marker gene, cre/lox P sitespecific recombination system has the key advantage where no commercial enzyme mixture is required for recombination. The 38 kDa cre recombinase specifically recognizes & induces precise recombination & DNA excision between two directly repeated asymmetric 34 bp loxP sites flanking the target DNA sequences to be eliminated [14-16].
Although insecticidal activity of transgenic marker gene free ASAL positive rice lines was well documented [17] against N. lugens and other leafhoppers, the mode of action of ASAL against leafhoppers are still unknown and needs to be explored before utilization of the transgenic lines in future IPM programme. Therefore, to achieve a sound scientific basis behind the on field application of transgenic plants, the actual mode of action of ASAL against leaf hoppers needs to be well documented. The aim of this study is to elucidate the mode of action of expressed ASAL in transgenic rice plants against N. lugens and establish the potentiality of transgenic antibiotic resistant marker gene free ASAL expressing rice plants for minimizing planthopper associated yield loss of the crop.
2. Materials and Methods
2.1. Materials Used
Transgenic rice seeds cv IR64 used for this study were developed by the present investigating group [17]. BPH were obtained from Regional Rice Research Station at Chinsurah, West Bengal for all the experiments.
2.2. Seed Germination
Rice seeds (T3) were surface sterilized for 30 minutes in 1.5% (v/v) sodium hypochlorite solution, rinsed thoroughly (3 - 4 times) in sterile water and sown on half strength MS [18] germinating medium with 2% sucrose and 0.7% w/v agar [17]. 30 days old plants were used for molecular analysis.
2.3. PCR Screening and Segregation Analysis of Transgenic Line
Multiplex PCR was carried out with seven T3 plants (randomly chosen) of T2 parental line No. L03C04 (6) using ASAL gene specific primer pairs. Genomic DNA was isolated from young green leaves of T3 progeny plants and control rice plants followed by CTAB extraction method [19]. Multiplex PCR was subjected using Qiagen Multiplex PCR Kit (GmbH, Hilden, Germany). Each 50 µl PCR reaction mixture contained 1x PCR master mix including Hot Star Taq DNA polymerase. Primers (Table 1) were used in 0.2 µM concentration. The reaction started with incubation for 15 minutes at 94˚C followed by 35 cycles of [30 s at 94˚C, 90 s at 58˚C, 90 s at 72˚C] and final extension with 10 minutes at 72˚C in My Cycler (Bio Rad, Hercules, CA, USA). DNA of pBKhg ASAL and pBK16.2 plasmids was taken as positive control for ASAL and cre gene respectively. DNA of control rice plant was taken as negative control. The segregation patterns of ASAL and cre genes in the T3 hybrid lines were observed by analysing the presence or absence of respective gene-specific amplicons resolved in 1.2% Agarose gel.
2.4. Western Blotting
Protein from leaves of 1 month old control and T3 progeny plants were extracted in Tris-cl buffer pH-7. After quantification using Bradford reagent [20], western blotting was carried out with total protein extracted from each of the T3 progeny plants probed with anti-ASAL antibody according to the protocol described by Dutta et al. 2005 [11]. About 50 µg of total plant protein re
Table 1. List of PCR primers.

solved in 15% SDS-PAGE was electrophoretically transferred to Hybond-C membrane (GE Healthcare, UK) using Hoeffer (Hoefer Inc., Holliston, MA, USA) electroblot apparatus. The membrane was transiently stained with Ponceau S (Sigma-Aldrich, USA) for confirming the transfer of proteins onto it. Blocking of membrane was done with 5% non-fat milk (Merck, Germany) dissolved in 1x PBST at 37˚C for 3 hours. After washing with 1x PBST, the membrane was then incubated with anti-ASAL polyclonal antiboby (1:4000) for 2 hours at 37˚C. Further probing of the primary antibody was done for 2 hours with anti-rabbit IgG-HRP conjugate (1:20,000) (Sigma-Aldrich, USA) and developed using Hybond ECL kit on Hybond ECL film (GE Healthcare, Germany). Purified native ASAL protein (0.5 µg) was used as positive control and total protein (50 µg) from untransformed plants was used as negative control.
2.5. ELISA
100 µg of crude protein extracted from T3 transgenic leaves of each plant were coated in the wells of microtitre plate with coating buffer (15 mM Sodium carbonate, 35 mM sodium bicarbonate, 3 mM sodium azide). After charging the protein extract with primary and secondary antibody, substrate [O-phenylen ediamine diamine hydrochloride (OPD, BRL)] dissolved in citrate buffer (pH –9.6) was added and coloured reaction was developed. Optical density was measured at 415 nm in a Bio-Rad microtitre plate reader model no. 680 [11].
2.6. Hemagglutinaton Assay
The heamagglutination assay was done with the total protein extracts (100 µg/well) of five T3 ASAL expressing lines. Blood sample was collected from the rabbit by a syringe and immediately transferred to a glass tube pre filled with 0.9% saline as described by Banerjee et al. [21]. The erythrocytes were incubated with plant extracts in microtiter well and after 1 hour of incubation at 37˚C, the hemagglutination activity was checked.
2.7. In planta Insect Bioassay
T3 plants expressing ASAL and control plants were subjected to insect bioassay with BPH as described by Sengupta et al. [17]. Five marker gene free plants (line no. SD2, SD4, SD5, SD7 and SD8) were tested with BPH nymph and compared the mortality of the insects incubated on control plants. Each set was infested with 20 BPH nymphs of first instar into an individual insectproof cage and survivability of insects was monitored at every 2 days from the release of insects up to 16 days. Effect of ASAL on fecundity of BPH was assessed by measuring total nymph production by adult insects on the transgenic rice plants (line no. SD2, SD4, SD5, SD7 and SD8). Each experimental set was repeated five times and statistical analysis was done in order to compare the significance of differential mortality among all transgenic and controls plants.
2.8. Ligand Blot Assay
Total brush border membrane vesicle (BBMV) from Nilaparvata lugens was isolated according to Bandopadhyay et al. [22] using MgCl2 sequential precipitation method suited for luminal membranes. About 20 µg of total BBMV resolved in 12% SDS-PAGE was electrophoretically transferred to Hybond-C membrane (GE Healthcare, UK) using Hoeffer (Hoefer Inc., Holliston, MA, USA) electroblot apparatus. The membrane was temporarily stained with Ponceau S (Sigma-Aldrich, USA) to ensure the protein transfer from gel to Hybond-C membrane. Blocking of membrane was done with 5% non-fat milk (Merck, Germany) dissolved in 1x PBST at 37˚C for 2 hours. The membrane was further subjected to incubation with 100 µg of total protein extracted from transgenic ASAL positive SD7 line (ASAL expression level ~5 ng∙µg−1) for 2 hours following extensive washing with 1x PBST. The membrane was then incubated with anti-ASAL antibody (1:4000) for 2 hours at 37˚C. Further probing of the secondary antibody was done with anti-rabbit IgG-HRP conjugate (1:20,000) (Sigma-Aldrich, USA) and film was developed using Hybond ECL kit (GE Healthcare, Germany) as described by Mondal et al. [23]. Ligand blot assay with total protein (100 µg) from non-transgenic rice plant served as negative control.
2.9. LC MS/MS Analysis of Insect Receptor
Receptor protein band detected via ligand blot assay when challenged with total protein from ASAL positive transgenic rice line were excised, trypsinised and processed for LC MS/MS analysis as described in details by Sarkar et al. [24]. The extracted peptides were analyzed by LC MS/MS on Orbitrap Velos. Individual MS/MS spectra were searched through Mascot search using UNIPROT_2011_08 database with preset parameters such as standard global modifications, minimum of 5 peptide match and 99% protein thresholds. The results were observed using Scaffold (version Scaffold_3.6.4, Proteome software Inc., Portland, OR).
3. Results
3.1. PCR Analysis
Seven T3 plants were taken from T2 parental line no. L03C04 (6) (randomly chosen) for multiplex PCR analysis. Five plants were found to be positive for ASAL and two plants were found to be negative suggesting about the segregation of ASAL in T3 generation (Figure 1). Absence of cre gene was also observed in the ASAL positive lines.
3.2. Expression of ASAL in T3 Progenies
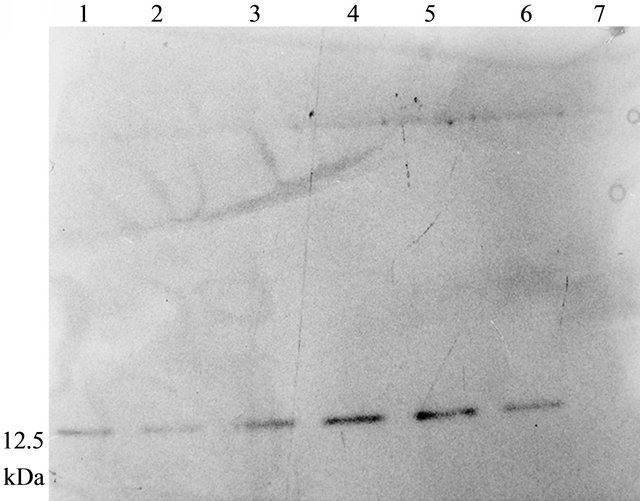
Western blot of total protein extract from 1 month old leaves of the five T3 lines were analysed which showed ASAL positive protein band at 12.5 kDa region (Figure 2(a)). ELISA result of five plants showed ASAL expression level in the range of 0.4% to 0.5% of the total soluble protein fraction (Figure 2(b)). Among these five lines, SD7 rice line showed highest expression level of ASAL (0.5%).
3.3. Agglutination Assay
In the heamagglutination assay, ASAL positive T3 lines showed agglutination of rabbit erythrocytes by forming a carpet of erythrocytes cells in the wells (Figure 3: Panel A, well 1 - 5) whereas the control line showed a tight button of rabbit erythrocytes cells at the centre of the well (Figure 3: Panel B, well 1 - 5) indicating the negative agglutination reaction. This result confirmed the retention of lectin activity of the expressed ASAL from transgenic rice lines.
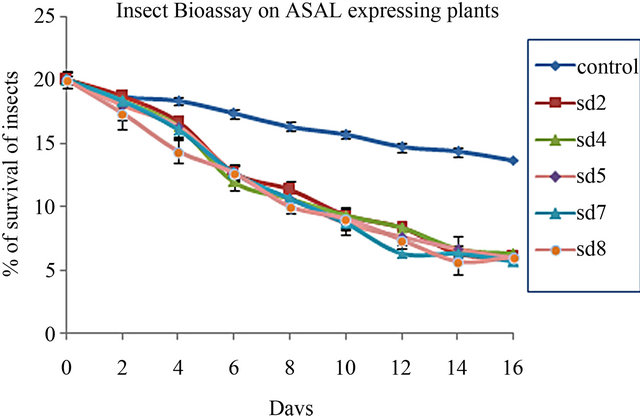
3.4. Insect Bioassay
In planta insect bioassay was carried out on 1-month-old five ASAL-expressing T3 progeny rice plants. All five transgenic plants inoculated with BPH, showed significant reduction in survivability and fecundity of the pest compared to that of the insects incubated on untrans-
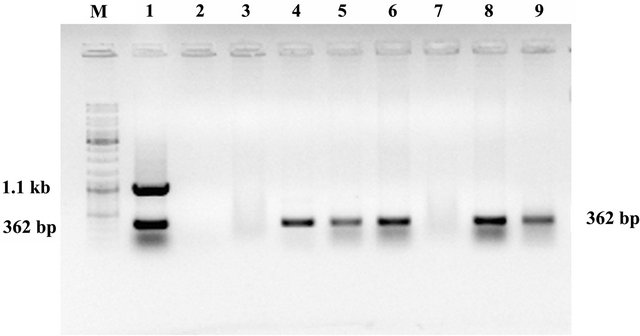
Figure 1. Molecular analysis of gene. Multiplex PCR analysis of T3 hybrid lines. Lane 1: Showing amplification of cre and ASAL gene at 1.1 kb and 362 bp respectively as positive controls. Lane 2: Represents negative control. Lane 3 - 9: Represents T3 progeny plants of line no. L03C04 (6) [SD1, SD2, SD4, SD5, SD6, SD7 and SD8] showing amplification of ASAL gene at 362 bp. Lane M. DNA molecular weight marker.
formed control plants. The number of living insect declined to 5.3 ± 0.33 (27%) to 6 ± 1.0 (30%) whereas in control plant the number of living insect was 13.66 ± 0.3 (68.3%) (Figure 4(a)). The mean number of survival of BPH on transgenic plants [t(0.01) 18.37] was significantly (p < 0.01) different than the control plants (unpaired t test). After 16 days of the experiment SD7 line represents maximum mortality of BPH. Fecundity of BPH on ASAL expressing plants was also examined by counting the total nymphs produced by the adult insects on transformed and untransformed control rice plants. After 30 days of assay period, the mean number of BPH nymphs on ASAL positive T3 plants was found to be 50 ± 2.8 (42.4%) to 40.67 ± 1.2 (35%) where as in control plants the number increased up to 118 ± 4.1 (100%) (Figure 4(b)) suggesting the lower reproductive rate of BPH fed on transgenic rice plants compared to the control rice plants.
3.5. Ligand Blots Assay and LC MS/MS Analysis
Approximately 56 kDa receptor protein from the brush border membrane vesicle (BBMV) of N. lugens was lightened up after ligand blot assay carried out with total protein from transgenic ASAL positive SD7 rice lines (Figure 5). Identified receptor protein band was subjected to LC MS/MS analysis. Sequence information of peptide fragments obtained through MS/MS were individually searched against Uniport database and receptor protein identified as NADH-quinone oxidoreductase from N. lugens [Accession no: Q5W1K6_NILLU] with a sequence coverage of 45% (Table 2, Supply Figure 1).
4. Discussion
Lectin has been defined as carbohydrate-binding protein or glycoprotein of non-immune origin which agglutinates or precipitates glycoconjugates linked to the erythrocytes [25]. Lectins are generally multivalent in nature and the specificity of which is largely determined by monosaccharide termini. Several plants possess lectin particularly in the seeds and tubers as storage protein. Some of which also serve as defence proteins when infested by enemies. A broad spectrum of plant lectins has been tested on several insect species more specifically on hemipterans. Usually the first experiments to check the effects of a particular lectin on an insect are conducted with an artificial diet. A major drawback of this approach is the composition of the artificial diet, which often differs considerably from the natural insect diet. Furthermore, for rearing of many insects no artificial diet had been worked out yet. However for controlling hemipteran pests, efforts have been made to target lectin ex-
 (a)
(a)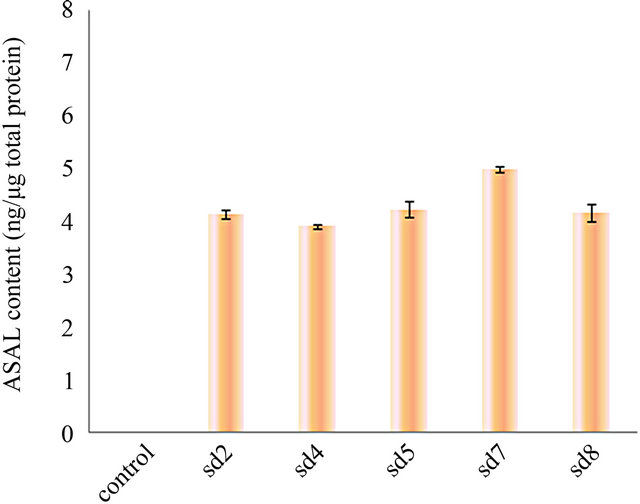 (b)
(b)
Figure 2. Expression analysis of ASAL in T3 hybrid plants. (a) Western blot analysis of protein (50 µg) extracted from leaves of T3 hybrid rice plants and control plant. Lane 1 to 5: Transgenic ASAL cassette bearing rice lines like SD2, SD4, SD5, SD7 and SD8. Lane 6: ~0.5 μg purified native ASAL used as positive control. Lane 7: Represents crude protein from untransformed plant used as negative control; (b) ELISA analysis for expression of ASAL in total soluble protein of T3 lines.
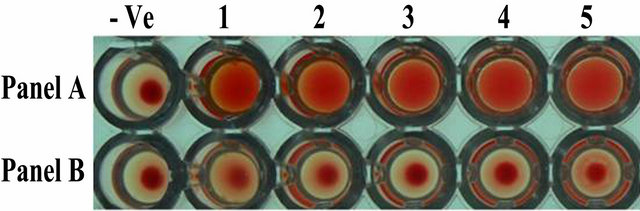
Figure 3. Agglutination assay of ASAL protein from expressed plants. Panel A, Well 1 to 5: Represents rabbit erythrocytes incubated with total protein (100 µg) extracted from transformed ASAL expressing plant lines. Panel B, Well 1 to 5: Total protein (100 µg) from control plants. Well (−Ve): no protein control.
pression specifically to the phloem sap of transgenic plants [26].
Rice is one of the few plants which were subjected to foreign gene insertion (ASAL, GNA etc.) and the transgenic plants showed significant efficacy against various insects including planthoppers. Recent studies by Sengupta et al. [17] reported that marker free ASAL positive rice lines are resistant to a significant extent to leafhoppers like GLH, BPH. But the basis of this resistance is not yet documented. In the present report, we focus to address this aspect.
4.1. Molecular Analysis of Transgenic T3 Lines
Analysing T3 marker free transgenic rice lines, it was found that 71% of the transgenic rice plants showed presence of ASAL gene and absence of cre gene in this generation suggesting about segregation of ASAL among T3 progenies which is close to attain homozygosity. Significant level of ASAL expression was observed in T3 rice lines revealed by ELISA and western blot assay.
4.2. Insecticidal Activity of T3 ASAL Positive Rice Lines
In planta insect bioassay experiments demonstrated significant insecticidal efficacy of different transgenic ASAL positive rice lines against BPH. Furthermore, fecundity rate was also observed to be reduced among the insects (BPH) fed on transgenic rice plants which seem to be comparable with the earlier reports from the present investigating group [13,17].
4.3. Basis behind the Observed Toxicity of Expressed ASAL against N. lugens
The insecticidal efficacy of ASAL in SD2, SD4, SD5, SD7 and SD8 T3 rice lines had already been documented by in planta insect bioassay experiment. To understand the basis of this toxic effect, ligand blot assay had been done and ~56 kDa putative receptor protein band from N. lugens BBMV fraction was detected. LC MS/MS analysis has shown the receptor protein band (56 kDa) was of NADH quinone oxidoreductase enzyme of N. lugens [27].
NADH quinone oxidoreductase (NQO) is the first enzyme of electron transport respiratory chain. It contributes to ATP synthesis. Studies revealed that some secondary metabolites like rotenone and piericidin A from microbial and plant sources are important components that act on the respiratory chain by inhibiting NQO [28]. It was also documented by earlier investigators that NQO gene expression was enhanced in BPH, feeding on B5 (BPH resistance) rice lines, indicating that there might be some compounds having a similar structure and function as rotenone or piericidin A generated in B5 plants, which might inhibit the respiratory chain enzymes in BPH to
 (a)
(a)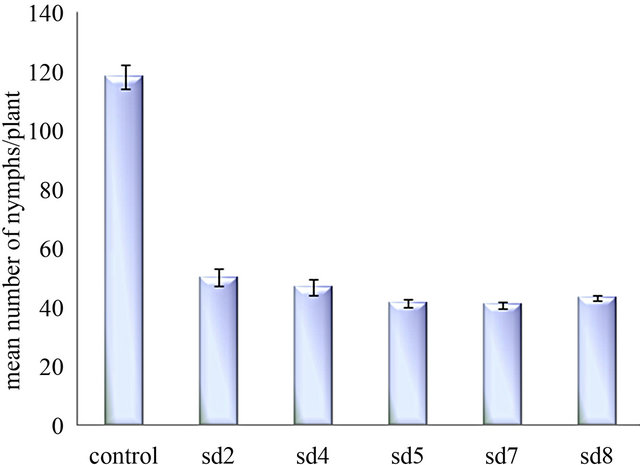 (b)
(b)
Figure 4. In planta insect bioassay. (a) The graph shows the percentage of survival of BPH on untransformed control plant and five T3 progeny plants SD2, SD4, SD5, SD7, SD8; (b) Bar diagram showing fecundity pattern of BPH fed on untransformed (control) and ASAL expressing rice plants (SD2, SD4, SD5, SD7, SD8).
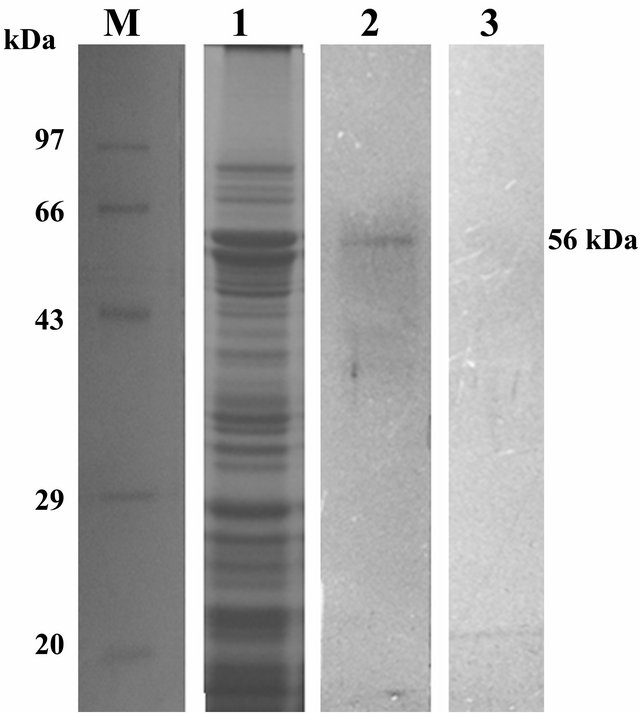
Figure 5. SDS-PAGE (12%) profile and subsequent ligand blot assay of total BBMV protein from N. lugens. Lane M: Standard protein molecular weight marker. Lane 1: SDS PAGE profile of total BBMV protein fraction of N. lugens stained by Coomasie brilliant blue; Lane 2: Ligand blot analysis of total BBMV protein of N. lugens challenged with total protein from SD7 transgenic rice lines showing putative ~56 KDa receptor protein band of ASAL. Lane 3: Ligand blot analysis of total BBMV protein of N. lugens challenged with Control non-transgenic rice line (negative control).
certain degree. This result emphasises that NQO may play important role in the defence response of BPH against resistant rice lines [27]. So it is quite obvious that binding of ASAL to NQO may upset insect defence mechanism and costs its survival.
It has also been reported in Drosophila melanogaster that the mitochondrial acyl carrier protein (mtACP), a subunit of NADH ubiquinone oxidoreductase is involved in de novo fatty acid synthesis in the mitochondrion. APelement-induced loss-of-function mutation in the mtacp1 gene resulted in lethality, indicating that the gene is essential for viability of the insect [29]. Additional studies showed that mtacp1 is expressed at higher levels during late embryogenesis, in the pupa and adult mosquitoes, indicating its requirement for both male and female gametogenesis and possibly for tracheal development in Drosophila. Correlating the earlier observations with our present findings, it is assumed that ASAL binding to NQO of BPH can affect normal development and gametogenesis, which results into loss of fecundity of insects feeding on presently, mentioned ASAL rice lines.
5. Conclusion
Antibiotic resistant marker gene free ASAL expressing rice lines are found to be highly resistant to BPH. ASAL, having binding affinity to NQO (a respiratory chain enzyme) of BPH, exerts its toxicity resulting in high mortality and loss of fecundity. Above findings provide adequate information about the potentiality of ASAL expressing advanced progenies of rice plants to minimize the crop loss due to sucking insect attack.
6. Acknowledgements
Authors are thankful to Bose Institute for infrastructural facility. AB & NB are thankful to Department of Biotechnology, Government of India and Swiss Agency for Development and Cooperation, Government of Switzerland under the Indo-Swiss Collaboration in Biotechnology for financial support. AR is thankful to Council of Scientific and Industrial Research for providing fellowship. Technical support of Mr. Arup Kumar Dey, Mr.
Table 2. Receptor identification through LC MS/MS.

Swarnava Das and Mr. Sudipta Basu are also duly acknowledged.
REFERENCES
- G. S. Khush, “Green Revolution: The Way Forward,” Nature Reviews Genetics, Vol. 2, No. 10, 2001, pp. 815- 822. doi:10.1038/35093585
- P. Brookes and G. B. Barfoot, “GM Rice, Will This Be the Way for Global Acceptance of GM Crop Technology,” ISAAA Briefs No. 28, International Service for the Aquisition of Agri-Biotech Application, Ithaca, 2003.
- “Threats of Insecticide Misuse in Rice Ecosystems— Exploring Options for Mitigation,” International Conference Announcement, Hanoi, 16 December 2011. www.spipm.cgiar.org/c/document_library
- X. Foissac, N. T. Loc, P. Choursistou, A. M. R. Gatehouse and J. A. Gatehouse, “Resistance to Green Leafhopper (Nephotettix virescens) and Brown Planthopper (Nilaparvata lugens) in Transgenic Rice Expressing Snowdrop Lectin (Galanthus nivalis agglutinin; GNA),” Journal of Insect Physiology, Vol. 46, No. 4, 2000, pp. 573- 583. doi:10.1016/S0022-1910(99)00143-2
- M. B. Cohen, S. N. Alam, E. B. Medina and C. C. Bernal, “Brown planthopper, Nilaparvata lugens, Resistance in Rice Cultivar IR64 Mechanism and Role in Successful N. Lugens Management in Central Luzon, the Philippines, ” Entomologia Experimentalis et Applicata, Vol. 85, No. 3, 1997, pp. 221-229. doi:10.1046/j.1570-7458.1997.00252.x
- K. D. Gallagher, P. E. Kenmore and K. Sogawa, “Judicial Use Insecticides Deter Planthopper Outbreaks and Extend the Life of Resistant Varieties in Southeast Asia Rice,” In: R. K. Denno and T. J. Perfect, Eds., Planthopper: Their Ecology and Management, Chapman and Hall, London, 1994, pp. 599-614.
- K. V. Rao, K. S. Rathore, T. K. Hodges, X. Fu, E. Stoger, S. Sudhakar, P. Williams, P. Choursistou, M. Bharathi, D. P. Bown, K. S. Powell, J. Spence, A. M. R. Gatehouse and J. A. Gatehouse, “Expression of Snowdrop Lectin (GNA) in Transgenic Plants Confers Resistance to Rice Brown Planthopper,” Plant Journal, Vol. 15, No. 4, 1998, pp. 469-477. doi:10.1046/j.1365-313X.1998.00226.x
- S. Ramesh, D. Nagadhara, V. D. Reddy and K. V. Rao, “Production of Transgenic Indica Rice Resistant to Yellow Stem Borer and Sap Sucking Insects, Using Superbinary Vectors of Agrobacterium tumafaciens,” Plant Science, Vol. 166, No. 4, 2004, pp. 1077-1085. doi:10.1016/j.plantsci.2003.12.028
- K. S. Powell, A. M. R. Gatehouse, V. A. Hilder and A. J. Gatehouse, “Antifeedant Effects of Plant Lectins and an Enzyme on the Adult Stage of the Rice Brown Planthopper, Nilaparvata lugens,” Entomologia Experimentalis et Applicata, Vol. 75, No. 1, 1995, pp. 51-59. doi:10.1111/j.1570-7458.1995.tb01909.x
- P. Majumder, S. Banerjee and S. Das, “Identification of Receptors Responsible for Binding of the Mannose Specific Lectin to the Gut Epithelial Membrane of the Target Insects,” Glycoconjugate Journal, Vol. 20, No. 9, 2004, pp. 525-530.
- I. Dutta, P. Saha, P. Majumder, A. Sarkar, D. Chakraborti, S. Banerjee and S. Das, “The Efficacy of a Novel Insecticidal Protein, Allium sativum leaf lectin (ASAL) against Homopteran Insect Monitored in Transgenic Tobacco,” Plant Biotechnology Journal, Vol. 3, No. 6, 2005, pp. 601- 611. doi:10.1111/j.1467-7652.2005.00151.x
- I. Dutta, P. Majumder, P. Saha, K. Ray and S. Das, “Constitutive and Phloem Specific Expression of Allium sativum leaf agglutinin (ASAL) to Engineer Aphid (Lipaphis erysimi) Resistance in Transgenic Indian Mustard (Brassica juncea),” Plant Science, Vol. 169, No. 6, 2005, pp. 996-1007. doi:10.1016/j.plantsci.2005.05.016
- P. Saha, P. Majumder, I. Dutta, T. Ray, S. C. Roy and S. Das, “Transgenic Rice Expressing Allium sativum Leaf Lectin with Enhanced Resistance against Sap-Sucking Insect Pests,” Planta, Vol. 223, No. 6, 2006, pp. 1329- 1343. doi:10.1007/s00425-005-0182-z
- E. C. Dale and D. W. Ow, “Gene Transfer with Subsequent Removal of the Selection Gene from the Host Genome,” Proceedings of National Academy of Science USA, Vol. 88, No. 23, 1991, pp. 10558-10562. doi:10.1073/pnas.88.23.10558
- Y. Wang, B. Chen, Y. Hu, J. Li and Z. Lin, “Inducible Excision of Selectable Marker Gene from Transgenic Plants by the Cre/Lox Site-Specific Recombination System,” Transgenic Research, Vol. 14, No. 5, 2005, pp. 605- 614. doi:10.1007/s11248-005-0884-9
- J. Zuo, Q. W. Niu, S. G. Moller and N. H. Chua, “ChemicalRegulated, Site-Specific DNA Excision in Transgenic Plants,” Nature Biotechnology, Vol. 19, No. 2, 2001, pp. 157-161. doi:10.1038/84428
- S. Sengupta, D. Chakraborti, H. A. Mondal and S. Das, “Selectable Antibiotic Resistance Marker Gene Free Transgenic Rice Harbouring the Garlic Leaf Lectin Gene Ex Hibits Resistance to Sap Sucking Planthoppers,” Plant Cell Report, Vol. 29, No. 3, 2010, pp. 261-271. doi:10.1007/s00299-010-0819-7
- T. Murashige and F. Skoog, “A Revised Method for Rapid Growth and Bioassays with Tobacco Tissue Cultures,” Plant Physiology, Vol. 15, No. 43, 1962, pp. 473- 497. doi:10.1111/j.1399-3054.1962.tb08052.x
- M. A. Saghai-Maroof, K. M. Soliman, R. A. Jorgensen and R. W. Allard, “Ribosomal DNA Spacer-Length Polymorphisms in Barley: Mendelian Inheritance, Choursomosomal Location and Population Dynamics,” Proceedings of the National Academy of Sciences USA, Vol. 81, No. 24, 1984, pp. 8014-8018. doi:10.1073/pnas.81.24.8014
- M. M. Bradford, “Rapid and Sensitive Method for Quan Titation of Microgram Quantities of Protein Utilizing Principle of Protein Dye Binding,” Annual Review of Biochemistry, Vol. 72, No. 1-2, 1976, pp. 248-254. doi:10.1016/0003-2697(76)90527-3
- N. Banerjee, S. Sengupta, A. Roy, P. Ghosh, K. Das and S. Das, “Functional Alteration of a Dimeric Insecticidal Lectin to a Monomeric Antifungal Protein Correlated to Its Oligomeric Status,” PLoS One, Vol. 6, No. 4, 2011, Article ID: e18593. doi:10.1371/journal.pone.0018593
- S. Bandyopadhyay, A. Roy and S. Das, “Binding of Garlic (Allium sativum) Leaf Lectin to the Gut Receptors of Hemipteran Pests Is Correlated to Its Insecticidal Activity,” Plant Science, Vol. 161, No. 5, 2001, pp. 1025-1033. doi:10.1016/S0168-9452(01)00507-6
- A. Sarkar, D. Hess, H. A. Mondal, S. Banerjee, H. C. Sharma and S. Das, “Homodimeric Alkaline Phosphatase Located at Helicoverpa armigera Midgut, a Putative Receptor of Cry1Ac Contains #-GalNAc in Terminutesal Glycan Structure as Interactive Epitope,” Journal Proteome Research, Vol. 8, No. 4, 2009, pp. 1838-1848. doi:10.1021/pr8006528
- H. A. Mondal, D. Chakraborti, P. Majumder, P. Roy, A. Roy and S. Das, “Allergenicity Assessment of Allium sativum leaf agglutinin, a Potential Candidate Protein for Developing Sap Sucking Insect Resistant Food Crops,” PLoS One, Vol. 6, No. 11, 2011, Article ID: e27716. doi:10.1371/journal.pone.0027716
- I. J. Goldstein, R. C. Hughes, M. Monsigny, T. Osawa and N. Sharon, “What Should Be Called a Lectin?” Nature, Vol. 285, No. 5760, 1980, p. 66. doi:10.1038/285066b0
- Y. Shi, M. B. Wang, K. S. Powell, E. Van Damme, V. A. Hilder, A. M. R. Gatehouse, D. Boulter and J. A. Gatehouse, “Use of the Rice Sucrose Synthase-1 Promoter to Direct Phloem-Specific Expression of β-Glucuronidase and Snowdrop Lectin Genes in Transgenic Tobacco Plants,” Journal of Experimental Botany, Vol. 45, No. 5, 1994, pp. 623-631. doi:10.1093/jxb/45.5.623
- Z. Yang, F. Zhang, Q. He and G. He, “Ribosomal DNA Spacer-Length Polymorphisms in Barley: Mendelian Inheritance, Choursomosomal Location and Population Dynamics,” Archives of Insect Biochemistry and Physiology, Vol. 59, No. 2, 2005, pp. 59-66. doi:10.1002/arch.20055
- P. Lümmen, “Complex I Inhibitors as Insecticides and Acaricides,” Biochimica et Biophysica Acta, Vol. 1364, No. 2, 1998, pp. 287-296. doi:10.1016/S0005-2728(98)00034-6
- G. Ragoneá, R. Caizziá, R. Moschettiá, P. Barsanti, V. De Pintoá and C. Caggese, “The Drosophila Melanogaster Gene for the NADH: Ubiquinone Oxidoreductase Acyl Carrier Protein: Developmental Expression Analysis and Evidence for Alternatively Spliced Forms,” Molecular and General Genetics, Vol. 261, No. 4-5, 1999, pp. 690- 697. doi:10.1007/s004380050012
Supply Figure 1

NOTES
*Equal Contribution.
#Corresponding author.

